Prototype Gluten-Free Breads from Processed Durum Wheat: Use of Monovarietal Flours and Implications for Gluten Detoxification Strategies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Proteolytic Mixture
2.2. Prototype-Breads Preparation
2.3. Gluten Quantification by Immune-Enzymatic Assay (R5 Competitive ELISA)
2.4. Proteomic Characterization by High Resolution Tandem Mass Spectrometry (HR-MS/MS) Analysis
2.4.1. Sample Preparation Protocol
2.4.2. Discovery HR-MS/MS Analysis and Protein/Peptides Identification
2.5. In Vitro-Simulated Human Gastroduodenal Digestion Experiments
3. Results and Discussion
3.1. Prototype Bread Sample Preparation
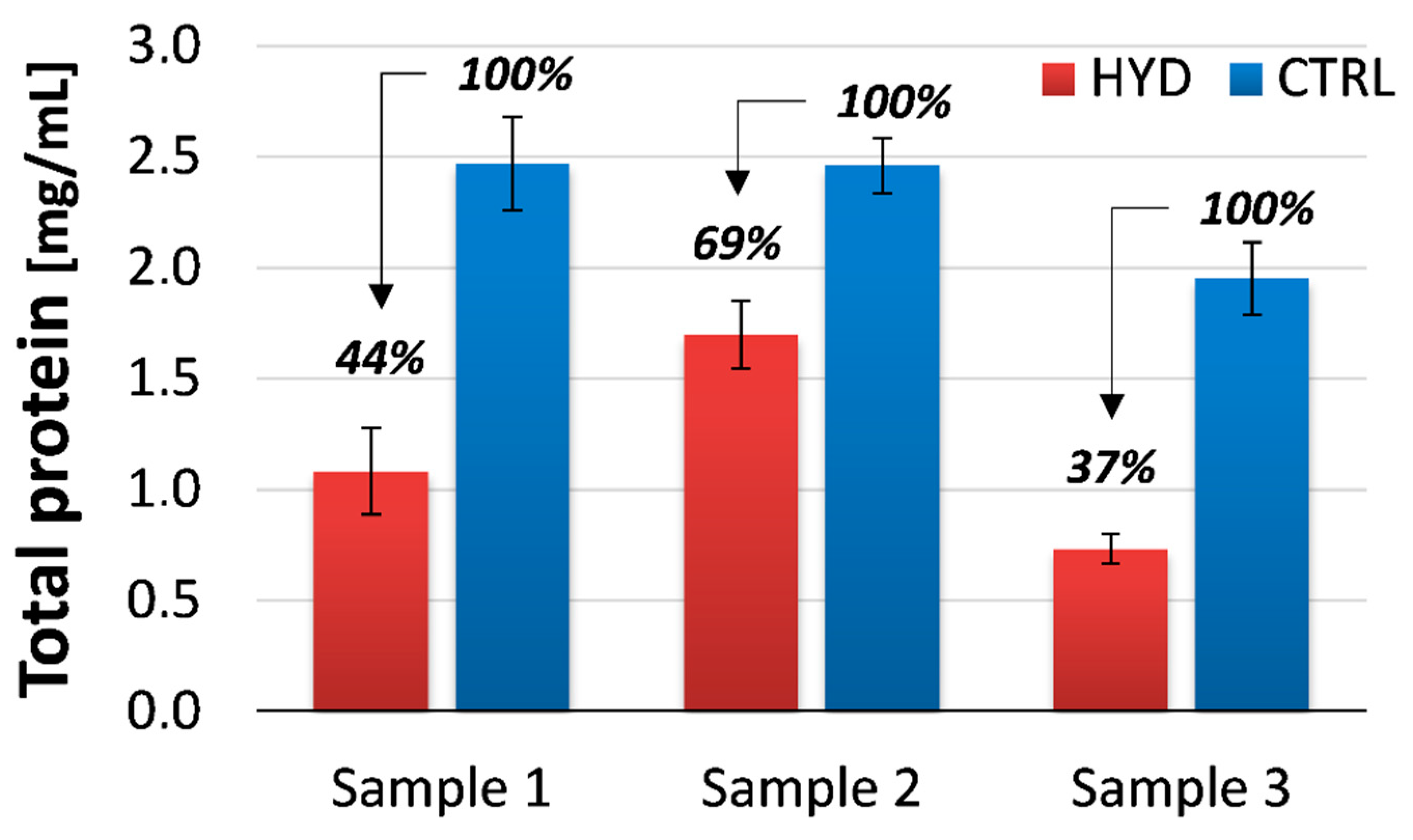
3.2. Gluten Quantitation and Detoxification Efficiency
3.3. Proteome Profiling and Resistant Epitope Matching
3.4. In Vitro-Simulated Human Gastroduodenal Digestion Experiments and In-Silico Evaluation of the Toxicity Risk for Celiac Disease Patients
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ribeiro, M.; Nunes, F.M. We might have got it wrong: Modern wheat is not more toxic for celiac patients. Food Chem. 2019, 278, 820–822. [Google Scholar] [CrossRef]
- Ribeiro, M.; Rodríguez-Quijano, M.; Nunes, F.M.; Carrillo, J.M.; Branlard, G.; Igrejas, G. New insights into wheat toxicity: Breeding did not seem to contribute to a prevalence of potential celiac disease’s immunostimulatory epitopes. Food Chem. 2016, 213, 8–18. [Google Scholar] [CrossRef]
- Pilolli, R.; Gadaleta, A.; Mamone, G.; Nigro, D.; De Angelis, E.; Montemurro, N.; Monaci, L. Scouting for naturally low-toxicity wheat genotypes by a multidisciplinary approach. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Escarnot, E.; Gofflot, S.; Sinnaeve, G.; Dubois, B.; Bertin, P.; Mingeot, D. Reactivity of gluten proteins from spelt and bread wheat accessions towards A1 and G12 antibodies in the framework of celiac disease. Food Chem. 2018, 268, 522–532. [Google Scholar] [CrossRef]
- Malalgoda, M.; Meinhardt, S.W.; Simsek, S. Detection and quantitation of immunogenic epitopes related to celiac disease in historical and modern hard red spring wheat cultivars. Food Chem. 2018, 264, 101–107. [Google Scholar] [CrossRef]
- Prandi, B.; Tedeschi, T.; Folloni, S.; Galaverna, G.; Sforza, S. Peptides from gluten digestion: A comparison between old and modern wheat varieties. Food Res. Int. 2017, 91, 92–102. [Google Scholar] [CrossRef]
- Schalk, K.; Lang, C.; Wieser, H.; Koehler, P.; Scherf, K.A. Quantitation of the immunodominant 33-mer peptide from α-gliadin in wheat flours by liquid chromatography tandem mass spectrometry. Sci. Rep. 2017, 7, 45092. [Google Scholar] [CrossRef]
- De Santis, M.A.; Giuliani, M.M.; Giuzio, L.; De Vita, P.; Lovegrove, A.; Shewry, P.R.; Flagella, Z. Differences in gluten protein composition between old and modern durum wheat genotypes in relation to 20th century breeding in Italy. Eur. J. Agron. 2017, 87, 19–29. [Google Scholar] [CrossRef]
- Picascia, S.; Camarca, A.; Malamisura, M.; Mandile, R.; Galatola, M.; Cielo, D.; Gazza, L.; Mamone, G.; Auricchio, S.; Troncone, R.; et al. In celiac disease patients the in vivo challenge with the diploid Triticum monococcum elicits a reduced immune response compared to hexaploid wheat. Mol. Nutr. Food Res. 2020, 64, e1901032. [Google Scholar] [CrossRef]
- Jouanin, A.; Gilissen, L.W.J.; Boyd, L.A.; Cockram, J.; Leigh, F.J.; Wallington, E.J.; van den Broeck, H.C.; van der Meer, I.M.; Schaart, J.G.; Visser, R.G.F.; et al. Food processing and breeding strategies for coeliac-safe and healthy wheat products. Food Res. Int. 2018, 110, 11–21. [Google Scholar] [CrossRef] [Green Version]
- Gianfrani, C.; Camarca, A.; Mazzarella, G.; Di Stasio, L.; Giardullo, N.; Ferranti, P.; Picariello, G.; Aufiero, V.R.; Picascia, S.; Troncone, R.; et al. Extensive in vitro gastrointestinal digestion markedly reduces the immune-toxicity of Triticum monococcum wheat: Implication for celiac disease. Mol. Nutr. Food Res. 2015, 59, 1844–1854. [Google Scholar] [CrossRef] [PubMed]
- Boukid, F.; Mejri, M.; Pellegrini, N.; Sforza, S.; Prandi, B. How looking for celiac-safe wheat can influence its technological properties. Compr. Rev. Food Sci. Food Saf. 2017, 16, 797–807. [Google Scholar] [CrossRef] [Green Version]
- Goesaert, H.; Brijs, K.; Veraverbeke, W.; Courtin, C.; Gebruers, K.; Delcour, J.A. Wheat flour constituents: How they impact bread quality, and how to impact their functionality. Trends Food Sci. Technol. 2005, 16, 12–30. [Google Scholar] [CrossRef]
- Ribeiro, M.; Nunes, F.M.; Rodríguez-Quijano, M.; Carrillo, J.M.; Branlard, G.; Igrejas, G. Next-generation therapies for celiac disease: The gluten-targeted approaches. Trends Food Sci. Technol. 2018, 75, 56–71. [Google Scholar] [CrossRef]
- Luongo, D.; Maurano, F.; Bergamo, P.; Rossi, M. Microbial transglutaminase: A biotechnological tool to manage gluten intolerance. Anal. Biochem. 2020, 592, 113584. [Google Scholar] [CrossRef] [PubMed]
- Gobbetti, M.; Pontonio, E.; Filannino, P.; Rizzello, C.G.; De Angelis, M.; Di Cagno, R. How to improve the gluten-free diet: The state of the art from a food science perspective. Food Res. Int. 2018, 110, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Rai, S.; Kaur, A.; Chopra, C.S. Gluten-free products for celiac susceptible people. Front. Nutr. 2018, 5, 116. [Google Scholar] [CrossRef] [PubMed]
- Gianfrani, C.; Siciliano, R.A.; Facchiano, A.M.; Camarca, A.; Mazzeo, M.F.; Costantini, S.; Salvati, V.M.; Maurano, F.; Mazzarella, G.; Iaquinto, G.; et al. Transamidation of wheat flour inhibits the response to gliadin of intestinal T cells in celiac disease. Gastroenterology 2007, 133, 780–789. [Google Scholar] [CrossRef]
- Sharma, N.; Bhatia, S.; Chunduri, V.; Kaur, S.; Sharma, S.; Kapoor, P.; Kumari, A.; Garg, M. Pathogenesis of celiac disease and other gluten related disorders in wheat and strategies for mitigating them. Front. Nutr. 2020, 7, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, G.; Helmerhorst, E.J.; Darwish, G.; Blumenkranz, G.; Schuppan, D. Gluten degrading enzymes for treatment of celiac disease. Nutrients 2020, 12, 2095. [Google Scholar] [CrossRef]
- De Angelis, M.; Rizzello, C.G.; Fasano, A.; Clemente, M.G.; De Simone, C.; Silano, M.; De Vincenzi, M.; Losito, I.; Gobbetti, M. VSL#3 probiotic preparation has the capacity to hydrolyze gliadin polypeptides responsible for Celiac Sprue probiotics and gluten intolerance. Biochim. Biophys. Acta Mol. Basis Dis. 2006, 1762, 80–93. [Google Scholar] [CrossRef] [Green Version]
- Gerez, C.L.; Dallagnol, A.; Rollán, G.; Font de Valdez, G. A combination of two lactic acid bacteria improves the hydrolysis of gliadin during wheat dough fermentation. Food Microbiol. 2012, 32, 427–430. [Google Scholar] [CrossRef]
- Di Cagno, R.; De Angelis, M.; Lavermicocca, P.; De Vincenzi, M.; Giovannini, C.; Faccia, M.; Gobbetti, M. Proteolysis by sourdough lactic acid bacteria: Effects on wheat flour protein fractions and gliadin peptides involved in human cereal intolerance. Appl. Environ. Microbiol. 2002, 68, 623–633. [Google Scholar] [CrossRef] [Green Version]
- Di Cagno, R.; De Angelis, M.; Auricchio, S.; Greco, L.; Clarke, C.; De Vincenzi, M.; Giovannini, C.; D’Archivio, M.; Landolfo, F.; Parrilli, G.; et al. Sourdough bread made from wheat and nontoxic flours and started with selected lactobacilli is tolerated in celiac sprue patients. Appl. Environ. Microbiol. 2004, 70, 1088–1096. [Google Scholar] [CrossRef] [Green Version]
- Pilolli, R.; Gadaleta, A.; Di Stasio, L.; Lamonaca, A.; De Angelis, E.; Nigro, D.; De Angelis, M.; Mamone, G.; Monaci, L. A Comprehensive peptidomic approach to characterize the protein profile of selected durum wheat genotypes: Implication for coeliac disease and wheat allergy. Nutrients 2019, 11, 2321. [Google Scholar] [CrossRef] [Green Version]
- Colgrave, M.L.; Goswami, H.; Byrne, K.; Blundell, M.; Howitt, C.A.; Tanner, G.J. Proteomic profiling of 16 cereal grains and the application of targeted proteomics to detect wheat contamination. J. Proteome Res. 2015, 14, 2659–2668. [Google Scholar] [CrossRef]
- Naegeli, H.; Birch, A.N.; Casacuberta, J.; De Schrijver, A.; Gralak, M.A.; Guerche, P.; Jones, H.; Manachini, B.; Messéan, A.; Nielsen, E.E.; et al. Guidance on allergenicity assessment of genetically modified plants. EFSA J. 2017, 15, 04862. [Google Scholar] [CrossRef] [Green Version]
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 2019, 51, 885–895. [Google Scholar] [CrossRef] [Green Version]
- Pilolli, R.; De Angelis, E.; Monaci, L. In house validation of a high resolution mass spectrometry Orbitrap-based method for multiple allergen detection in a processed model food. Anal. Bioanal. Chem. 2018, 410, 5653–5662. [Google Scholar] [CrossRef]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised staticin vitrodigestion method suitable for food—An international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef] [Green Version]
- Fiorda, F.A.; Soares, M.S.; Da Silva, F.A.; Grosmann, M.V.; Souto, L.R. Microestructure, texture and colour of gluten-free pasta made with amaranth flour, cassava starch and cassava bagasse. LWT 2013, 54, 132–138. [Google Scholar] [CrossRef]
- Sandhu, K.S.; Kaur, M. Mukesh Studies on noodle quality of potato and rice starches and their blends in relation to their physicochemical, pasting and gel textural properties. LWT 2010, 43, 1289–1293. [Google Scholar] [CrossRef]
- Hager, A.-S.; Wolter, A.; Czerny, M.; Bez, J.; Zannini, E.; Arendt, E.K.; Czerny, M. Investigation of product quality, sensory profile and ultrastructure of breads made from a range of commercial gluten-free flours compared to their wheat counterparts. Eur. Food Res. Technol. 2012, 235, 333–344. [Google Scholar] [CrossRef]
- Kadan, R.; Robinson, M.G.; Thibodeaux, D.P.; Pepperman, A.B. Texture and other physicochemical properties of whole rice bread. J. Food Sci. 2006, 66, 940–944. [Google Scholar] [CrossRef]
- See, J.A.; Kaukinen, K.; Makharia, G.K.; Gibson, P.R.; Murray, J.A. Practical insights into gluten-free diets. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 580–591. [Google Scholar] [CrossRef]
- Rizzello, C.G.; Montemurro, M.; Gobbetti, M. Characterization of the bread made with durum wheat semolina rendered gluten free by sourdough biotechnology in comparison with commercial gluten-free products. J. Food Sci. 2016, 81, H2263–H2272. [Google Scholar] [CrossRef]
- Calvo-Lerma, J.; Crespo-Escobar, P.; Martínez-Barona, S.; Fornés-Ferrer, V.; Donat, E.; Ribes-Koninckx, C. Differences in the macronutrient and dietary fibre profile of gluten-free products as compared to their gluten-containing counterparts. Eur. J. Clin. Nutr. 2019, 73, 930–936. [Google Scholar] [CrossRef]
- Papillo, V.A.; Agostoni, C. Nutritional aspects of gluten-free products. J. Sci. Food Agric. 2015, 95, 2380–2385. [Google Scholar] [CrossRef]
- Rizzello, C.G.; De Angelis, M.; Di Cagno, R.; Camarca, A.; Silano, M.; Losito, I.; De Vincenzi, M.; De Bari, M.D.; Palmisano, F.; Maurano, F.; et al. Highly efficient gluten degradation by lactobacilli and fungal proteases during food processing: New perspectives for celiac disease. Appl. Environ. Microbiol. 2007, 73, 4499–4507. [Google Scholar] [CrossRef] [Green Version]
- De Angelis, M.; Cassone, A.; Rizzello, C.G.; Gagliardi, F.; Minervini, F.; Calasso, M.; Di Cagno, R.; Francavilla, R.; Gobbetti, M. Mechanism of degradation of immunogenic gluten epitopes from Triticum turgidum L. var. durum by sourdough lactobacilli and fungal proteases. Appl. Environ. Microbiol. 2009, 76, 508–518. [Google Scholar] [CrossRef] [Green Version]
- Di Cagno, R.; Barbato, M.; Di Camillo, C.; Rizzello, C.G.; De Angelis, M.; Giuliani, G.; De Vincenzi, M.; Gobbetti, M.; Cucchiara, S. Gluten-free sourdough wheat baked goods appear safe for young celiac patients: A pilot study. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 777–783. [Google Scholar] [CrossRef]
- Greco, L.; Gobbetti, M.; Auricchio, R.; Di Mase, R.; Landolfo, F.; Paparo, F.; Di Cagno, R.; De Angelis, M.; Rizzello, C.G.; Cassone, A.; et al. Safety for patients with celiac disease of baked goods made of wheat flour hydrolyzed during food processing. Clin. Gastroenterol. Hepatol. 2011, 9, 24–29. [Google Scholar] [CrossRef]
- Curiel, J.A.; Coda, R.; Limitone, A.; Katina, K.; Raulio, M.; Giuliani, G.; Rizzello, C.G.; Gobbetti, M. Manufacture and characterization of pasta made with wheat flour rendered gluten-free using fungal proteases and selected sourdough lactic acid bacteria. J. Cereal Sci. 2014, 59, 79–87. [Google Scholar] [CrossRef]
- Giuliani, G.; Benedusi, A.; Di Cagno, R.; Rizzello, C.G.; De Angelis, M.; Gobbetti, M.; Cassone, A. Process of Microbic Biotechnology for Completely Degrading Gluten in Flours. W.O. Patent 2010073283-A2, 1 July 2010. [Google Scholar]
- Kunji, E.R.; Mierau, I.; Hagting, A.; Poolman, B.; Konings, W.N. The proteotytic systems of lactic acid bacteria. Antonie Leeuwenhoek 1996, 70, 187–221. [Google Scholar] [CrossRef]
- Collin, P.; Thorell, L.; Kaukinen, K.; Mäki, M. The safe threshold for gluten contamination in gluten-free products. Can trace amounts be accepted in the treatment of coeliac disease? Aliment. Pharmacol. Ther. 2004, 19, 1277–1283. [Google Scholar] [CrossRef]
- European Commission. Commission Regulation (EC) No. 41/2009 concerning the composition and labelling of foodstuffs suitable for people intolerant to gluten. Off. J. Eur. Union 2009, 16, 3–5. [Google Scholar]
- Panda, R.; Garber, E.A.E. Detection and quantitation of gluten in fermented-hydrolyzed foods by antibody-based methods: Challenges, progress, and a potential path forward. Front. Nutr. 2019, 6, 97. [Google Scholar] [CrossRef] [PubMed]
- Sollid, L.M.; Tye-Din, J.A.; Qiao, S.-W.; Anderson, R.P.; Gianfrani, C.; Koning, F. Update 2020: Nomenclature and listing of celiac disease–relevant gluten epitopes recognized by CD4+ T cells. Immunogenetics 2019, 72, 85–88. [Google Scholar] [CrossRef]
- Wang, D.; Li, D.; Wang, J.; Zhao, Y.; Wang, Z.; Yue, G.; Liu, X.; Qin, H.; Zhang, K.; Dong, L.; et al. Genome-wide analysis of complex wheat gliadins, the dominant carriers of celiac disease epitopes. Sci. Rep. 2017, 7, 44609. [Google Scholar] [CrossRef] [Green Version]
- Karell, K.; Louka, A.S.; Moodie, S.J.; Ascher, H.; Clot, F.; Greco, L.; Ciclitira, P.J.; Sollid, L.M.; Partanen, J. Hla types in celiac disease patients not carrying the DQA1*05-DQB1*02 (DQ2) heterodimer: Results from the European genetics cluster on celiac disease. Hum. Immunol. 2003, 64, 469–477. [Google Scholar] [CrossRef]

| Sample Code | Gluten [mg/kg] | Relative Standard Deviation % | Degradation Efficiency |
|---|---|---|---|
| HYD-1 * | 11.3 ± 1.3 | 11% | 99.5% |
| CTRL-1 * | 2490 ± 80 | 3% | |
| HYD-2 * | 7600 ± 700 | 9% | 22.0% |
| CTRL-2 * | 9700 ± 1100 | 11% | |
| HYD-3 * | 36 ± 7 | 19% | 99.6% |
| CTRL-3 * | 9800 ±1100 | 11% |
| Sample Type | Peptides Count | HYD-1 | CTRL-1 | HYD-2 | CTRL-2 | HYD-3 | CTRL-3 | |
|---|---|---|---|---|---|---|---|---|
| Low molecular weight (LMW) fraction, < 3 kDa | total identified | 312 | 4 | 242 | 4 | 384 | 7 | |
| hazard peptides with intact celiac disease (CD) epitope | - | - | 7 | - | 6 | - | ||
| Protein distribution of hazard peptides * | γ-gliadin | - | - | 7 | - | 6 | - | |
| ω-gliadin | - | - | 2 | - | 1 | - | ||
| High molecular weight (HMW) fraction, chymotrypsin digest | total identified | 614 | 1394 | 1097 | 1671 | 663 | 1599 | |
| hazard peptides with intact CD epitope | - | 69 | 46 | 83 | 1 | 92 | ||
| Protein distribution of hazard peptides * | α-gliadin | - | 14 | 11 | 19 | - | 23 | |
| γ-gliadin | - | 22 | 20 | 34 | - | 32 | ||
| ω-gliadin | - | 14 | 7 | 14 | 1 | 24 | ||
| LMW-glutenin | - | 25 | 11 | 21 | - | 12 | ||
| HMW-glutenin | - | 1 | 1 | 1 | - | 1 | ||
| AAI domain containing | - | 8 | 9 | 11 | - | 21 | ||
| Epitopes Search * | N° of Hazard Peptides/Sample ** | |||||||
|---|---|---|---|---|---|---|---|---|
| ID | Type | Toxicity *** | HLA-DQ | Sequence | Core T-Cell Epitope | CTRL-1 | CTRL-2 | CTRL-3 |
| 55 | α-gliadin | I | DQ2 | PQPQLPYPQPQLPY | DQ2.5-glia-α1b, DQ2.5-glia-α2 | 0 | 1 | 1 |
| 64 | α-gliadin | I | DQ2 | PQPQLPYPQPQL | DQ2.5-glia-α2 | 0 | 1 | 1 |
| 66 | α-gliadin | I | DQ2 | PQPQLPYPQPQ | DQ2.5-glia-α2 | 0 | 1 | 1 |
| 68 | α-2 gliadin | I | DQ2.5 | PQPQLPYPQ | DQ2.5-glia-α2 | 0 | 1 | 1 |
| 72 | α-gliadin | I | DQ2 | PQLPYPQPQLPY | DQ2.5-glia-α1b | 0 | 1 | 1 |
| 84 | α-3 gliadin | I | DQ2.5 | PYPQPQLPY | DQ2.5-glia-α1b | 0 | 1 | 1 |
| 93 | α-20 gliadin | I | DQ2.5 | FRPQQPYPQ | DQ2.5-glia-α3 | 1 | 1 | 1 |
| 119 | α-gliadin | I | DQ8 | GSFQPSQQNPQAQGS | 0 | 1 | 0 | |
| 140 | α-gliadin | I | DQ8 | QLIPCMDVVL | 1 | 0 | 1 | |
| 182 | α-gliadin | I | DQ2 | LQPFPQPQPFLPQLPYPQPQ | 1 | 1 | 1 | |
| 188 | α-gliadin | I | DQ2 | FPGQQQQFPPQQPYPQPQPF | 1 | 0 | 1 | |
| 221 | ω-II gliadin | I | DQ2 | PQPQQPFPW | DQ2.5-glia-ω2 | 0 | 1 | 0 |
| 222 | ω-gliadin | I | DQ2 | PFPWQPQQPFPQ | 1 | 1 | 0 | |
| 226 | ω-gliadin | I | DQ2 | QQPQQPFPQPQLPFPQQSEQ | DQ2.5-glia-γ4c/DQ8-glia-γ1a | 1 | 1 | 0 |
| 231 | ω-gliadin | I | DQ2 | PFPQPQQPIPV | 1 | 1 | 1 | |
| 236 | ω-gliadin | I | DQ2 | PFPLQPQQPFPQ | DQ2.5-glia- γ4e | 0 | 0 | 1 |
| 463 | γ-gliadin | I | DQ8 (DQ2/8) | QQPYPQQPQQPFPQ | DQ2.5-glia-γ4c/DQ8-glia-γ1a | 0 | 1 | 1 |
| 501 | γ1-gliadin | I | DQ2 | PQQPFPQPQQTFPQQPQLPF | 0 | 0 | 1 | |
| 502 | γ1-gliadin | I | DQ2, DQ8 | PFPQPQQTFPQQPQLPFPQQ | 0 | 0 | 1 | |
| 504 | γ1-gliadin | I | DQ2 | PQQTFPQQPQLP | 0 | 0 | 1 | |
| 523 | γ1-gliadin | I | DQ2 | QQPQQSFPQQQ | DQ2.5-glia-γ1/DQ8.5-glia-γ1/DQ8-glia-γ2 | 1 | 3 | 1 |
| 524 | γ1-gliadin | I | DQ2 | QPQQSFPQQQ | DQ2.5-glia-γ1/DQ8.5-glia-γ1/DQ8-glia-γ2 | 2 | 3 | 1 |
| 530 | γ-gliadin | I | DQ8 (DQ2/8) | QFPQTQQPQQPFPQ | DQ2.5-glia-γ4c/DQ8-glia-γ1a | 0 | 0 | 1 |
| 536 | γ-gliadin | I | DQ2, DQ8 | QQPQLPFPQQPQQPFPQPQQ | DQ2.5-glia-γ4c/DQ8-glia-γ1a | 1 | 0 | 0 |
| 537 | γ-gliadin | I | DQ8 (DQ2/8) | QLPFPQQPQQPFPQ | DQ2.5-glia-γ4c/DQ8-glia-γ1a | 1 | 2 | 0 |
| 573 | γ-gliadin | I | DQ2 | FPQPQQQFPQPQ | DQ2.5-glia-γ4b | 0 | 0 | 1 |
| 577 | γ-gliadin | I | DQ2.5 | PQPQQQFPQ | DQ2.5-glia-γ4b | 0 | 0 | 1 |
| 583 | γ-1 gliadin | I | DQ2.5/DQ8 | PQQSFPQQQ | DQ2.5-glia-γ1/DQ8.5-glia-γ1/DQ8-glia-γ2 | 2 | 4 | 2 |
| 611 | γ-gliadin | I | DQ2 (DQ2.5) | PHQPQQQVPQPQQPQQPF | 0 | 1 | 0 | |
| 617 | γ-gliadin | I | DQ8 (DQ2/8) | PFPQLQQPQQPFPQ | DQ2.5-glia-γ4c/DQ8-glia-γ1a | 1 | 1 | 0 |
| 640 | γ-gliadin | I | DQ2 | QPQQSFPQQQRP | DQ2.5-glia-γ1/DQ8.5-glia-γ1/DQ8-glia-γ2 | 1 | 0 | 0 |
| 721 | LMW glutenin | I | DQ2 | QQQQPPFSQQQQSPFSQQQQ | DQ2.5-glut-L2 | 1 | 1 | 1 |
| 729 | LMW glutenin | I | DQ2 | QQPPFSQQQQSPFSQ | DQ2.5-glut-L2 | 2 | 1 | 1 |
| 731 | LMW glutenin | I | DQ2 | QQPPFSQQQQSP | 5 | 1 | 2 | |
| 733 | LMW glutenin | I | DQ2 | QPPFSQQQQSPFSQ | DQ2.5-glut-L2 | 3 | 2 | 1 |
| 734 | LMW glutenin | I | DQ2 | PPFSQQQQSPFSQQQ | DQ2.5-glut-L2 | 2 | 1 | 1 |
| 736 | LMW glutenin | I | DQ2 | PFSQQQQSPFSQQQQ | DQ2.5-glut-L2 | 2 | 1 | 1 |
| 738 | LMW glutenin | I | DQ2 | PFSQQQQSPF | DQ2.5-glut-L2 | 6 | 2 | 2 |
| 747 | glut-L2 | I | DQ2.5 | FSQQQQSPF | DQ2.5-glut-L2 | 6 | 2 | 2 |
| 835 | Hordein | I | DQ2 | QPFPQPQQPFPL | DQ2.5-glia-ω1 | 1 | 1 | 1 |
| 867 | hor-1 | I | DQ2.5 | PFPQPQQPF | DQ2.5-glia-ω1 | 1 | 1 | 4 |
| 878 | Hordein | I | DQ2 | QPFPQPQQPFSW | DQ2.5-glia-ω1 | 0 | 0 | 1 |
| 886 | γ-hordein | I | DQ2 | QQFPQPQQPFPQQP | DQ2.5-hor-2 | 0 | 0 | 1 |
| 890 | γ-hordein | I | DQ2 | QQFPQPQQPFPQ | DQ2.5-hor-2 | 0 | 0 | 1 |
| 891 | hor-2 | I | DQ2.5 | PQPQQPFPQ | DQ2.5-hor-2 | 0 | 1 | 3 |
| 930 | γ-secalin | I | DQ2 | QSIPQPQQPFPQ | DQ2.5-hor-2 | 0 | 0 | 1 |
| 950 | ω-Secalin | I | DQ2 | QPFPQPQQPIPQ | 1 | 1 | 0 | |
| 973 | ω-Secalin | I | DQ2 | IIPQQPQQPFPL | 0 | 1 | 1 | |
| 1040 | glia-ω 3 | I | DQ2.5 | PFPQPQQPI | 2 | 2 | 1 | |
| 1042 | glia-ω 4 | I | DQ2.5 | PQPQQPIPV | 1 | 1 | 1 | |
| 1044 | glia-ω 5 | I | DQ2.5 | LQPQQPFPQ | DQ2.5-glia-γ4e | 4 | 1 | 4 |
| Epitopes Search * | N° of Hazard Peptides/Sample ** | |||||||
|---|---|---|---|---|---|---|---|---|
| ID | Type | Toxicity *** | HLA-DQ | Epitope Sequence | Core T-Cell Epitope | HYD-1 | HYD-2 | HYD-3 |
| 1 | α-gliadin | T | Unknown | VPVPQLQPQNPSQQQPQEQVPL | - | 0 | 1 | 0 |
| 3 | α-gliadin | I | DQ2 | VRVPVPQLQPQNPSQQQPQ | - | 0 | 1 | 0 |
| 5 | α-gliadin | I | DQ2 | FPGQQQPFPPQQPYPQPQPF | - | 0 | 1 | 0 |
| 7 | α-gliadin | I, T | HLA-DR | PQPQPFPSQQPY | - | 0 | 3 | 0 |
| 14 | α-gliadin | I | DQ2 | LQLQPFPQPQLPY | DQ2.5-glia-α1a | 0 | 1 | 0 |
| 24 | α-gliadin | I | DQ2 | QLQPFPQPQLPY | DQ2.5-glia-α1a | 0 | 1 | 0 |
| 32 | γ-gliadin | I | DQ2 | PQQPFPQQPQQ | DQ2.5-glia-γ5 | 0 | 0 | 0 (1) |
| 36 | α-gliadin | I | DQ2 | LQPFPQPQLPY | DQ2.5-glia-α1a | 0 | 1 | 0 |
| 42 | α-gliadin | I | DQ2 | QPFPQPQLPY | DQ2.5-glia-α1a | 0 | 1 | 0 |
| 53 | α-9 gliadin | I | DQ2.5 | PFPQPQLPY | DQ2.5-glia-α1a | 0 | 1 | 0 |
| 95 | α-gliadin | I | DQ8 (DQ2/8, DQ1/8) | QQPQQQYPSGQGSFQPSQQNPQAQG | DQ8-glia-α1 | 0 | 1 | 0 |
| 96 | α-gliadin | I | DQ8 | QQPQQQYPSGQGSFQPSQQNPQAQ | DQ8-glia-α1 | 0 | 1 | 0 |
| 100 | α-gliadin | I | DQ8 | QPQQQYPSGQGSFQPSQQNP | DQ8-glia-α1 | 0 | 1 | 0 |
| 101 | α-gliadin | I | DQ8 (DQ2/8, DQ1/8) | QQYPSGQGSFQPSQQNPQ | DQ8-glia-α1 | 0 | 1 | 0 |
| 102 | α-gliadin | I | DQ8 | QYPSGQGSFQPSQQNPQA | DQ8-glia-α1 | 0 | 1 | 0 |
| 104 | α-gliadin | I | DQ8 | YPSGQGSFQPSQQNP | DQ8-glia-α1 | 0 | 1 | 0 |
| 105 | α-gliadin | I | DQ8 (DQ2/8) | PSGQGSFQPSQQNPQAQG | DQ8-glia-α1 | 0 | 1 | 0 |
| 106 | α-gliadin | I | DQ8 (DQ2/8) | PSGQGSFQPSQQ | DQ8-glia-α1 | 0 | 1 | 0 |
| 107 | α-gliadin | I | DQ8 (DQ2/8) | PSGQGSFQPSQ | - | 0 | 1 | 0 |
| 108 | α-gliadin | I | DQ8 (DQ2/8) | SGQGSFQPSQQN | DQ8-glia-α1 | 0 | 1 | 0 |
| 113 | α-gliadin | I | DQ8 (DQ2/8) | GQGSFQPSQ | - | 0 | 1 | 0 |
| 115 | α2 gliadin | I | DQ8 (DQ2/8) | QGSFQPSQQ | DQ8-glia-α1 | 0 | 1 | 0 |
| 138 | α-gliadin | I | DQ2 | PQQPYPQPQPQ | - | 0 | 1 | 0 |
| 146 | α-gliadin | I | DQ2 | QVPLVQQQQFLGQQQPFPPQ | - | 0 | 1 | 0 |
| 149 | α-gliadin | I, T | Unknown | LGQQQPFPPQQPYPQPQPFPSQQPY | - | 0 | 1 | 0 |
| 150 | α-gliadin | I, T | DQ2 (α1*0501, α1*0201) | LGQQQPFPPQQPYPQPQPF | - | 0 | 1 | 0 |
| 151 | α-gliadin | I | DQ2 (α1*0501, α1*0201) | LGQQQPFPPQQPYPQPQ | - | 0 | 1 | 0 |
| 152 | α-gliadin | I, T | HLA-DR | LGQQQPFPPQQPY | - | 0 | 2 | 0 |
| 185 | α-gliadin | I | DQ2 | QPQPFLPQLPYPQP | - | 0 | 1 | 0 |
| 187 | α-gliadin | I | DQ2 | PQPFLPQLPYPQ | - | 0 | 1 | 0 |
| 195 | ω-gliadin | I | DQ2 | PQQPFPQQPQQP | DQ2.5-glia-γ5 | 0 | 2 (2) | 0 |
| 227 | ω-gliadin | I | DQ2 | QPFPQPQLPFPQ | 0 | 1 | 0 | |
| 229 | ω-gliadin | I | DQ2 | PFPQQPQQPFPQ | DQ2.5-glia-γ4c/DQ8-glia-γ1a | 0 | 3 (1) | 0 (1) |
| 246 | ω5-gliadin/LMW glutenin | I | DQ2 | QQQQIPQQPQQF | - | 0 | 1 | 0 |
| 252 | ω5-gliadin/LMW glutenin | I | DQ2 | QIPQQPQQF | - | 0 | 2 | 0 |
| 426 | γ-gliadin | I | DQ2 | PQQPFPQQPQQPYPQQP | DQ2.5-glia-γ3/DQ8-glia-γ1b, DQ2.5-glia-γ5 | 0 | 1 | 0 |
| 427 | γ-gliadin | I | DQ2 | PQQPFPQQPQQPY | DQ2.5-glia-γ5 | 0 | 1 | 0 |
| 432 | γ-gliadin | I | DQ2 | PQQPFPQQPQQ | DQ2.5-glia-γ5 | 0 | 2 (2) | 0 |
| 437 | γ-gliadin | I | DQ2 | QQPFPQQPQQPYPQ | DQ2.5-glia-γ3/DQ8-glia-γ1b, DQ2.5-glia-γ5 | 0 | 1 | 0 |
| 438 | γ5 gliadin | I | DQ2.5 | QQPFPQQPQ | DQ2.5-glia-γ5 | 0 | 4 (2) | 0 (1) |
| 441 | γ-gliadin | I | DQ2 | PFPQQPQQPYPQQPQ | DQ2.5-glia-γ3/DQ8-glia-γ1b | 0 | 1 | 0 |
| 445 | γ-gliadin | I | DQ2 | PFPQQPQQPYPQ | DQ2.5-glia-γ3/DQ8-glia-γ1b | 0 | 1 | 0 |
| 446 | γ-gliadin | I | DQ8, DQ2 | FPQQPQQPYPQQPQQ | DQ2.5-glia-γ3/DQ8-glia-γ1b | 0 | 1 | 0 |
| 451 | γ-gliadin | I | DQ2 | FPQQPQQPYPQQP | DQ2.5-glia-γ3/DQ8-glia-γ1b | 0 | 1 | 0 |
| 454 | γ-gliadin | I | DQ2 | FPQQPQQPYPQQ | DQ2.5-glia-γ3/DQ8-glia-γ1b | 0 | 1 | 0 |
| 458 | γ1 and γ5 gliadin | I | DQ2.5/DQ8 | QQPQQPYPQ | DQ2.5-glia-γ3/DQ8-glia-γ1b | 0 | 2 (1) | 0 (1) |
| 464 | γ-gliadin | I | DQ2 | QQPYPQQPQ | - | 0 | 1 (1) | 0 (1) |
| 468 | γ-gliadin | I | DQ2 (DQ2.2 and DQ2.5) | PYPQQPQQP | - | 0 | 1 | 0 |
| 472 | γ-gliadin | I | DQ2.5/DQ8 | QQPQQPFPQ | DQ2.5-glia-γ4c/DQ8-glia-γ1a | 0 | 8 (4) | 0 (4) |
| 479 | γ1-gliadin | I | DQ2 | QVDPSGQVQWPQ | - | 0 | 3 | 0 |
| 503 | γ1-gliadin | I | DQ2 | PFPQPQQTFPQ | - | 0 | 1 | 0 |
| 538 | γ-gliadin | I | DQ2 | PFPQQPQQPF | - | 0 | 3 (1) | 0 (1) |
| 542 | γ-gliadin | I | DQ2 | FPQQPQQPF | - | 0 | 4 (1) | 0 (1) |
| 553 | γ-gliadin | I | DQ8 (DQ2/8) | PFPQTQQPQQPFPQ | DQ2.5-glia-γ4c/DQ8-glia-γ1a | 0 | 1 | 0 |
| 555 | γ-gliadin | I | DQ8 (DQ2/8) | PFPQSQQPQQPFPQ | DQ2.5-glia-γ4c/DQ8-glia-γ1a | 0 | 1 | 0 |
| 587 | γ-gliadin | I | DQ2 | VQGQGIIQPQQPAQL | DQ2.5-glia-γ2 | 0 | 3 (1) | 0 |
| 593 | γ-gliadin | I | DQ2 | GIIQPQQPAQL | DQ2.5-glia-γ2 | 0 | 4 (1) | 0 |
| 595 | γ-gliadin | I | DQ2 | IIQPQQPAQL | DQ2.5-glia-γ2 | 0 | 4 (1) | 0 |
| 597 | γ-gliadin | I | DQ2 | IIQPQQPAQ | - | 0 | 6 (1) | 0 |
| 599 | γ5 gliadin | I | DQ2.5 | IQPQQPAQL | DQ2.5-glia-γ2 | 0 | 4 (1) | 0 |
| 612 | γ-gliadin | I | DQ8 (DQ2/8) | QQPFPQQPQQPFPQ | DQ2.5-glia-γ4c/DQ8-glia-γ1a, DQ2.5-glia-γ5 | 0 | 3 (1) | 0 |
| 650 | γ-gliadin | I | DQ2 | QPFPQLQQPQQP | - | 0 | 1 | 0 |
| 659 | LMW glutenin | I | DQ2 | QAFPQPQQTFPH | - | 0 | 1 | 0 |
| 701 | γ-gliadin or LMW glutenin | I | DQ2 | QQPPFSQQQQPVLPQ | DQ2.5-glut-L1/DQ2.2-glut-L1 | 0 | 3 | 0 |
| 706 | Glut-L1 | I | DQ2.2 | PFSQQQQPV | DQ2.5-glut-L1/DQ2.2-glut-L1 | 0 | 7 | 0 |
| 720 | LMW glutenin | I | DQ2 | QQPPFSQQQQPPFSQ | - | 0 | 2 | 0 |
| 762 | LMW glutenin | I | DQ2 | QQPPFSQQQQQPILL | - | 0 | 1 | 0 |
| 763 | LMW glutenin | I | DQ2 | QPPFSQQQQQPILL | - | 0 | 1 | 0 |
| 781 | HMW-Glutenin | I | DQ8 (DQ2/8) | GQPGYYPTSPQQPGQ | - | 0 | 1 | 0 |
| 903 | Secalin | I | DQ2 | PQQSFPQQP | - | 0 | 0 | 1 |
| 926 | γ-secalin | I | DQ2 | PQTQQPQQPFPQ | DQ2.5-glia-γ4c/DQ8-glia-γ1a | 0 | 1 | 0 |
| 928 | γ-secalin | I | DQ2 | PQSQQPQQPFPQ | DQ2.5-glia-γ4c/DQ8-glia-γ1a | 0 | 1 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pilolli, R.; De Angelis, M.; Lamonaca, A.; De Angelis, E.; Rizzello, C.G.; Siragusa, S.; Gadaleta, A.; Mamone, G.; Monaci, L. Prototype Gluten-Free Breads from Processed Durum Wheat: Use of Monovarietal Flours and Implications for Gluten Detoxification Strategies. Nutrients 2020, 12, 3824. https://doi.org/10.3390/nu12123824
Pilolli R, De Angelis M, Lamonaca A, De Angelis E, Rizzello CG, Siragusa S, Gadaleta A, Mamone G, Monaci L. Prototype Gluten-Free Breads from Processed Durum Wheat: Use of Monovarietal Flours and Implications for Gluten Detoxification Strategies. Nutrients. 2020; 12(12):3824. https://doi.org/10.3390/nu12123824
Chicago/Turabian StylePilolli, Rosa, Maria De Angelis, Antonella Lamonaca, Elisabetta De Angelis, Carlo Giuseppe Rizzello, Sonya Siragusa, Agata Gadaleta, Gianfranco Mamone, and Linda Monaci. 2020. "Prototype Gluten-Free Breads from Processed Durum Wheat: Use of Monovarietal Flours and Implications for Gluten Detoxification Strategies" Nutrients 12, no. 12: 3824. https://doi.org/10.3390/nu12123824
APA StylePilolli, R., De Angelis, M., Lamonaca, A., De Angelis, E., Rizzello, C. G., Siragusa, S., Gadaleta, A., Mamone, G., & Monaci, L. (2020). Prototype Gluten-Free Breads from Processed Durum Wheat: Use of Monovarietal Flours and Implications for Gluten Detoxification Strategies. Nutrients, 12(12), 3824. https://doi.org/10.3390/nu12123824








