The Combination of Fasting, Acute Resistance Exercise, and Protein Ingestion Led to Different Responses of Autophagy Markers in Gastrocnemius and Liver Samples
Abstract
1. Introduction
2. Material and Methods
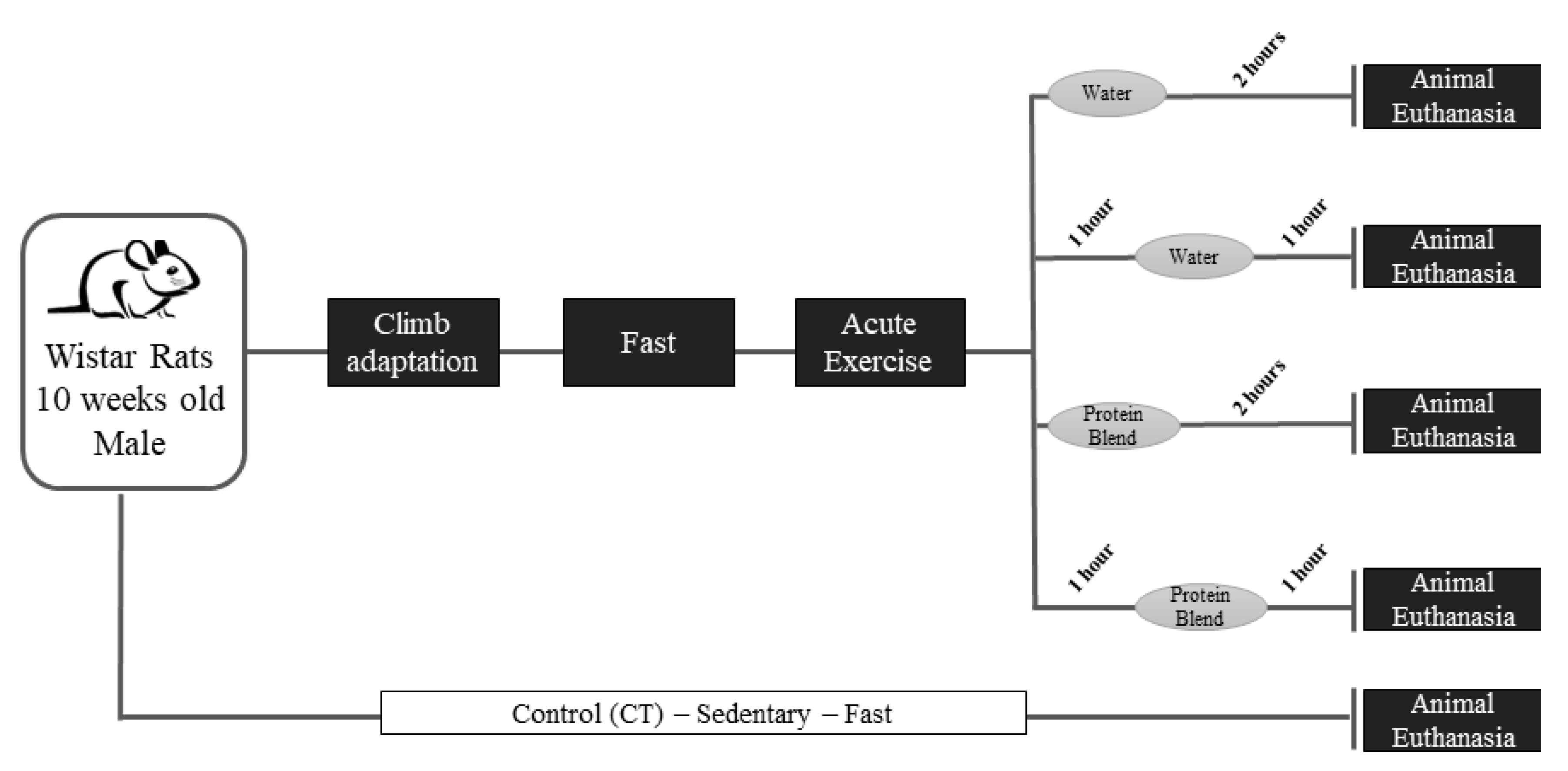
2.1. Experimental Animals
2.2. Acute Resistance Exercise Protocol and Supplementation
2.3. Skeletal Muscle and Liver Extraction
2.4. Biochemical Analysis
2.5. Leucine Dosage
Reverse Transcription-Quantitative Polymerase Chain Reaction (RTq-PCR)
2.6. Immunoblotting Technique
2.7. Statistical Analysis
3. Results
3.1. Body Weight, Gastrocnemius Weight, and Supplementation
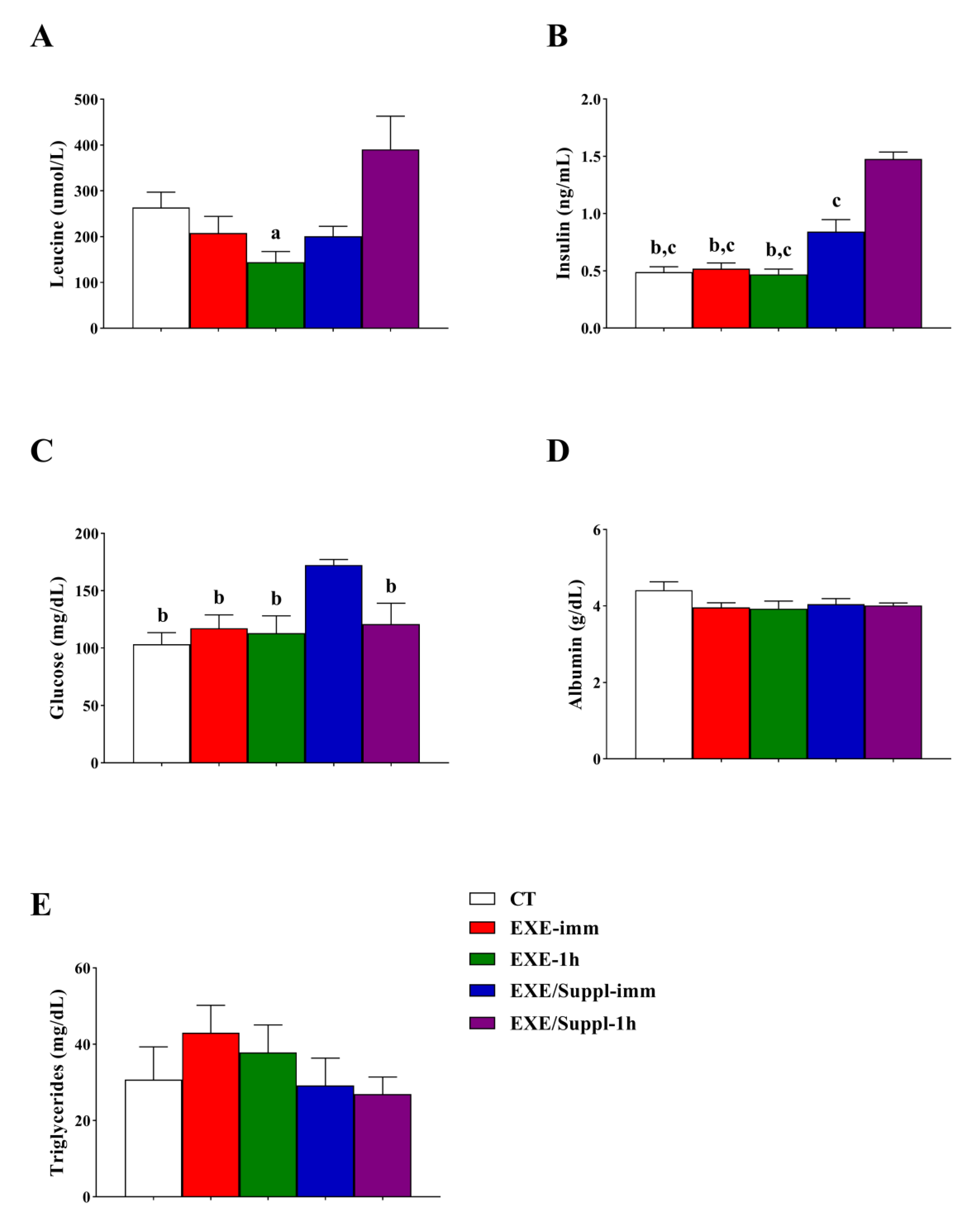
3.2. Insulin, Leucine, and Biochemical Analysis
3.3. Gene and Proteins Related to the Autophagy Pathway in Gastrocnemius
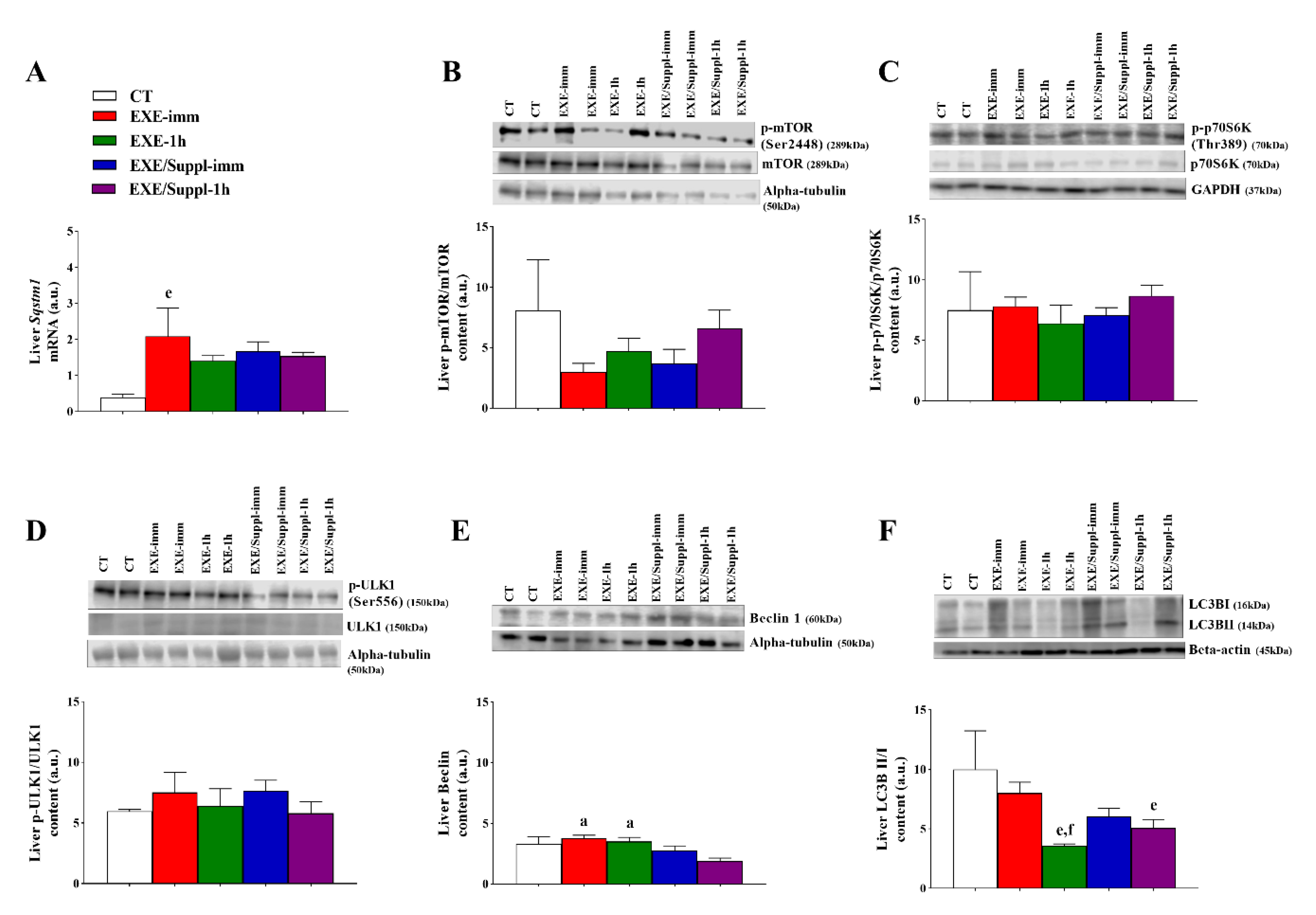
3.4. Gene and Proteins Related to the Autophagy Pathway in the Liver
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Campbell, W.W.; Haub, M.D.; Wolfe, R.R.; Ferrando, A.A.; Sullivan, D.H.; Apolzan, J.W.; Iglay, H.B. Resistance training preserves fat-free mass without impacting changes in protein metabolism after weight loss in older women. Obesity 2009, 17, 1332–1339. [Google Scholar] [CrossRef]
- Carroll, B.; Korolchuk, V.I.; Sarkar, S. Amino acids and autophagy: Cross-talk and co-operation to control cellular homeostasis. Amino Acids 2015, 47, 2065–2088. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; Marino, G.; Kroemer, G. Autophagy and Aging. Cell 2011, 146, 682–695. [Google Scholar] [CrossRef]
- Kuma, A.; Mizushima, N. Physiological role of autophagy as an intracellular recycling system: With an emphasis on nutrient metabolism. Semin. Cell Dev. Biol. 2010, 21, 683–690. [Google Scholar] [CrossRef]
- Cui, J.; Gong, Z.; Shen, H.M. The role of autophagy in liver cancer: Molecular mechanisms and potential therapeutic targets. Biochim. Biophys. Acta 2013, 1836, 15–26. [Google Scholar] [CrossRef]
- Yang, Z.; Klionsky, D.J. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010, 22, 124–131. [Google Scholar] [CrossRef]
- Halling, J.F.; Ringholm, S.; Olesen, J.; Prats, C.; Pilegaard, H. Exercise training protects against aging-induced mitochondrial fragmentation in mouse skeletal muscle in a PGC-1α dependent manner. Exp. Gerontol. 2017, 96, 1–6. [Google Scholar] [CrossRef]
- Escobar, K.A.; Cole, N.H.; Mermier, C.M.; VanDusseldorp, T.A. Autophagy and aging: Maintaining the proteome through exercise and caloric restriction. Aging Cell 2019, 18, e12876. [Google Scholar] [CrossRef]
- Mooren, F.C.; Krüger, K. Exercise, Autophagy, and Apoptosis. Prog. Mol. Biol. Transl. Sci. 2015, 135, 407–422. [Google Scholar] [CrossRef]
- Grumati, P.; Coletto, L.; Schiavinato, A.; Castagnaro, S.; Bertaggia, E.; Sandri, M.; Bonaldo, P. Physical exercise stimulates autophagy in normal skeletal muscles but is detrimental for collagen VI-deficient muscles. Autophagy 2011, 7, 1415–1423. [Google Scholar] [CrossRef]
- Jamart, C.; Francaux, M.; Millet, G.Y.; Deldicque, L.; Frère, D.; Féasson, L. Modulation of autophagy and ubiquitin-proteasome pathways during ultra-endurance running. J. Appl. Physiol. 2012, 112, 1529–1537. [Google Scholar] [CrossRef] [PubMed]
- Jamart, C.; Benoit, N.; Raymackers, J.M.; Kim, H.J.; Kim, C.K.; Francaux, M. Autophagy-related and autophagy-regulatory genes are induced in human muscle after ultraendurance exercise. Eur. J. Appl. Physiol. 2012, 112, 3173–3177. [Google Scholar] [CrossRef] [PubMed]
- Schwalm, C.; Jamart, C.; Benoit, N.; Naslain, D.; Premont, C.; Prevet, J.; Van Thienen, R.; Deldicque, L.; Francaux, M. Activation of autophagy in human skeletal muscle is dependent on exercise intensity and AMPK activation. FASEB J. 2015, 29, 3515–3526. [Google Scholar] [CrossRef]
- Smiles, W.J.; Areta, J.L.; Coffey, V.G.; Phillips, S.M.; Moore, D.R.; Stellingwerff, T.; Burke, L.M.; Hawley, J.A.; Camera, D.M. Modulation of autophagy signaling with resistance exercise and protein ingestion following short-term energy deficit. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015, 309, R603–R612. [Google Scholar] [CrossRef]
- Sieghart, W.; Fuereder, T.; Schmid, K.; Cejka, D.; Werzowa, J.; Wrba, F.; Wang, X.; Gruber, D.; Rasoul-Rockenschaub, S.; Peck-Radosavljevic, M.; et al. Mammalian target of rapamycin pathway activity in hepatocellular carcinomas of patients undergoing liver transplantation. Transplantation 2007, 83, 425–432. [Google Scholar] [CrossRef]
- Fry, C.S.; Drummond, M.J.; Glynn, E.L.; Dickinson, J.M.; Gundermann, D.M.; Timmerman, K.L.; Walker, D.K.; Volpi, E.; Rasmussen, B.B. Skeletal muscle autophagy and protein breakdown following resistance exercise are similar in younger and older adults. J. Gerontol. A Biol. Sci. Med. Sci. 2013, 68, 599–607. [Google Scholar] [CrossRef]
- Kwon, I.; Jang, Y.; Cho, J.Y.; Jang, Y.C.; Lee, Y. Long-term resistance exercise-induced muscular hypertrophy is associated with autophagy modulation in rats. J. Physiol. Sci. 2018, 68, 269–280. [Google Scholar] [CrossRef]
- Jamart, C.; Naslain, D.; Gilson, H.; Francaux, M. Higher activation of autophagy in skeletal muscle of mice during endurance exercise in the fasted state. Am. J. Physiol. Endocrinol. Metab. 2013, 305, E964–E974. [Google Scholar] [CrossRef]
- Impey, S.G.; Hammond, K.M.; Naughton, R.; Langan-Evans, C.; Shepherd, S.O.; Sharples, A.P.; Cegielski, J.; Smith, K.; Jeromson, S.; Hamilton, D.L.; et al. Whey Protein Augments Leucinemia and Postexercise p70S6K1 Activity Compared With a Hydrolyzed Collagen Blend When in Recovery From Training With Low Carbohydrate Availability. Int. J. Sport Nutr. Exerc. Metab. 2018, 28, 651–659. [Google Scholar] [CrossRef]
- Coffey, V.G.; Moore, D.R.; Burd, N.A.; Rerecich, T.; Stellingwerff, T.; Garnham, A.P.; Phillips, S.M.; Hawley, J.A. Nutrient provision increases signalling and protein synthesis in human skeletal muscle after repeated sprints. Eur. J. Appl. Physiol. 2011, 111, 1473–1483. [Google Scholar] [CrossRef]
- Hammond, K.M.; Impey, S.G.; Currell, K.; Mitchell, N.; Shepherd, S.O.; Jeromson, S.; Hawley, J.A.; Close, G.L.; Hamilton, L.D.; Sharples, A.P.; et al. Postexercise High-Fat Feeding Suppresses p70S6K1 Activity in Human Skeletal Muscle. Med. Sci. Sports Exerc. 2016, 48, 2108–2117. [Google Scholar] [CrossRef]
- Rowlands, D.S.; Thomson, J.S.; Timmons, B.W.; Raymond, F.; Fuerholz, A.; Mansourian, R.; Zwahlen, M.C.; Métairon, S.; Glover, E.; Stellingwerff, T.; et al. Transcriptome and translational signaling following endurance exercise in trained skeletal muscle: Impact of dietary protein. Physiol. Genom. 2011, 43, 1004–1020. [Google Scholar] [CrossRef]
- Sato, T.; Ito, Y.; Nagasawa, T. Dietary L-Lysine Suppresses Autophagic Proteolysis and Stimulates Akt/mTOR Signaling in the Skeletal Muscle of Rats Fed a Low-Protein Diet. J. Agric. Food Chem. 2015, 63, 8192–8198. [Google Scholar] [CrossRef]
- Madrigal-Matute, J.; Cuervo, A.M. Regulation of Liver Metabolism by Autophagy. Gastroenterology 2016, 150, 328–339. [Google Scholar] [CrossRef]
- Schneider, J.L.; Cuervo, A.M. Liver autophagy: Much more than just taking out the trash. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 187–200. [Google Scholar] [CrossRef]
- Komatsu, M. Liver autophagy: Physiology and pathology. J. Biochem. 2012, 152, 5–15. [Google Scholar] [CrossRef]
- Kanda, A.; Nakayama, K.; Sanbongi, C.; Nagata, M.; Ikegami, S.; Itoh, H. Effects of Whey, Caseinate, or Milk Protein Ingestion on Muscle Protein Synthesis after Exercise. Nutrients 2016, 8, 339. [Google Scholar] [CrossRef]
- Boirie, Y.; Dangin, M.; Gachon, P.; Vasson, M.P.; Maubois, J.L.; Beaufrère, B. Slow and fast dietary proteins differently modulate postprandial protein accretion. Proc. Natl. Acad. Sci. USA 1997, 94, 14930–14935. [Google Scholar] [CrossRef]
- Aragon, A.A.; Schoenfeld, B.J. Nutrient timing revisited: Is there a post-exercise anabolic window? J. Int. Soc. Sports Nutr. 2013, 10, 5. [Google Scholar] [CrossRef]
- Doering, T.M.; Reaburn, P.R.; Phillips, S.M.; Jenkins, D.G. Postexercise Dietary Protein Strategies to Maximize Skeletal Muscle Repair and Remodeling in Masters Endurance Athletes: A Review. Int. J. Sport Nutr. Exerc. Metab. 2016, 26, 168–178. [Google Scholar] [CrossRef]
- Dijk, F.J.; van Dijk, M.; Walrand, S.; van Loon, L.J.C.; van Norren, K.; Luiking, Y.C. Differential effects of leucine and leucine-enriched whey protein on skeletal muscle protein synthesis in aged mice. Clin. Nutr. ESPEN 2018, 24, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Deyl, Z.; Hyanek, J.; Horakova, M. Profiling of amino acids in body fluids and tissues by means of liquid chromatography. J. Chromatogr. 1986, 379, 177–250. [Google Scholar] [CrossRef]
- Pereira, B.C.; da Rocha, A.L.; Pinto, A.P.; Pauli, J.R.; de Souza, C.T.; Cintra, D.E.; Ropelle, E.R.; de Freitas, E.C.; Zagatto, A.M.; da Silva, A.S.R. Excessive eccentric exercise-induced overtraining model leads to endoplasmic reticulum stress in mice skeletal muscles. Life Sci. 2016, 145, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Da Rocha, A.L.; Pereira, B.C.; Pauli, J.R.; De Souza, C.T.; Teixeira, G.R.; Lira, F.S.; Cintra, D.E.; Ropelle, E.R.; Carlos, R.B., Jr.; Da Silva, A.S.R. Downhill Running Excessive Training Inhibits Hypertrophy in Mice Skeletal Muscles with Different Fiber Type Composition. J. Cell Physiol. 2016, 231, 1045–1056. [Google Scholar] [CrossRef]
- Reidy, P.T.; Walker, D.K.; Dickinson, J.M.; Gundermann, D.M.; Drummond, M.J.; Timmerman, K.L.; Fry, C.S.; Borack, M.S.; Cope, M.B.; Mukherjea, R.; et al. Protein blend ingestion following resistance exercise promotes human muscle protein synthesis. J. Nutr. 2013, 143, 410–416. [Google Scholar] [CrossRef]
- Tang, J.E.; Phillips, S.M. Maximizing muscle protein anabolism: The role of protein quality. Curr. Opin. Clin. Nutr. Metab. Care 2009, 12, 66–71. [Google Scholar] [CrossRef]
- Kato, H.; Suzuki, H.; Inoue, Y.; Takimoto, T.; Suzuki, K.; Kobayashi, H. Co-ingestion of carbohydrate with leucine-enriched essential amino acids does not augment acute postexercise muscle protein synthesis in a strenuous exercise-induced hypoinsulinemic state. Springerplus 2016, 5, 1299. [Google Scholar] [CrossRef]
- Kramer, I.F.; Verdijk, L.B.; Hamer, H.M.; Verlaan, S.; Luiking, Y.; Kouw, I.W.; Senden, J.M.; van Kranenburg, J.; Gijsen, A.P.; Poeze, M.; et al. Impact of the Macronutrient Composition of a Nutritional Supplement on Muscle Protein Synthesis Rates in Older Men: A Randomized, Double Blind, Controlled Trial. J. Clin. Endocrinol. Metab. 2015, 100, 4124–4132. [Google Scholar] [CrossRef]
- Esmarck, B.; Andersen, J.L.; Olsen, S.; Richter, E.A.; Mizuno, M.; Kjaer, M. Timing of postexercise protein intake is important for muscle hypertrophy with resistance training in elderly humans. J. Physiol. 2001, 535, 301–311. [Google Scholar] [CrossRef]
- Cohen, A.; Hall, M.N. An amino acid shuffle activates mTORC1. Cell 2009, 136, 399–400. [Google Scholar] [CrossRef]
- Naito, T.; Kuma, A.; Mizushima, N. Differential contribution of insulin and amino acids to the mTORC1-autophagy pathway in the liver and muscle. J. Biol. Chem. 2013, 288, 21074–21081. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.H.; Ro, S.H.; Cao, J.; Otto, N.M.; Kim, D.H. mTOR regulation of autophagy. FEBS Lett. 2010, 584, 1287–1295. [Google Scholar] [CrossRef]
- Ogasawara, R.; Suginohara, T. Rapamycin-insensitive mechanistic target of rapamycin regulates basal and resistance exercise-induced muscle protein synthesis. FASEB J. 2018, fj201701422R. [Google Scholar] [CrossRef] [PubMed]
- Verdijk, L.B.; Jonkers, R.A.; Gleeson, B.G.; Beelen, M.; Meijer, K.; Savelberg, H.H.; Wodzig, W.K.; Dendale, P.; van Loon, L.J. Protein supplementation before and after exercise does not further augment skeletal muscle hypertrophy after resistance training in elderly men. Am. J. Clin. Nutr. 2009, 89, 608–616. [Google Scholar] [CrossRef] [PubMed]
- Kitaoka, Y.; Nakazato, K.; Ogasawara, R. Combined effects of resistance training and calorie restriction on mitochondrial fusion and fission proteins in rat skeletal muscle. J. Appl. Physiol. 2016, 121, 806–810. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Bassik, M.C.; Moresi, V.; Sun, K.; Wei, Y.; Zou, Z.; An, Z.; Loh, J.; Fisher, J.; Sun, Q.; et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 2012, 481, 511–515. [Google Scholar] [CrossRef]
- Ogata, T.; Oishi, Y.; Higuchi, M.; Muraoka, I. Fasting-related autophagic response in slow- and fast-twitch skeletal muscle. Biochem. Biophys. Res. Commun. 2010, 394, 136–140. [Google Scholar] [CrossRef]
- Kholodenko, B.N. Cell-signalling dynamics in time and space. Nat. Rev. Mol. Cell. Biol. 2006, 7, 165–176. [Google Scholar] [CrossRef]
- Møller, A.B.; Vendelbo, M.H.; Christensen, B.; Clasen, B.F.; Bak, A.M.; Jørgensen, J.O.; Møller, N.; Jessen, N. Physical exercise increases autophagic signaling through ULK1 in human skeletal muscle. J. Appl. Physiol. 2015, 118, 971–979. [Google Scholar] [CrossRef]
- Kruse, R.; Pedersen, A.J.; Kristensen, J.M.; Petersson, S.J.; Wojtaszewski, J.F.; Højlund, K. Intact initiation of autophagy and mitochondrial fission by acute exercise in skeletal muscle of patients with Type 2 diabetes. Clin. Sci. 2017, 131, 37–47. [Google Scholar] [CrossRef]
- Mônico-Neto, M.; Antunes, H.K.; Lee, K.S.; Phillips, S.M.; Giampá, S.Q.; Souza, H.e.S.; Dáttilo, M.; Medeiros, A.; de Moraes, W.M.; Tufik, S.; et al. Resistance training minimizes catabolic effects induced by sleep deprivation in rats. Appl. Physiol. Nutr. Metab. 2015, 40, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, C.M.; Olsen, M.A.; Jessen, H.; Brandt, N.; Meldgaard, J.N.; Pilegaard, H. PGC-1 in exercise and fasting-induced regulation of hepatic UPR in mice. Pflug. Arch. Eur. J. Phys. 2018, 470, 1431–1447. [Google Scholar] [CrossRef] [PubMed]
- Kwon, I.; Song, W.; Jang, Y.; Choi, M.D.; Vinci, D.M.; Lee, Y. Elevation of hepatic autophagy and antioxidative capacity by endurance exercise is associated with suppression of apoptosis in mice. Ann. Hepatol. 2019. [Google Scholar] [CrossRef]
- Nicklin, P.; Bergman, P.; Zhang, B.; Triantafellow, E.; Wang, H.; Nyfeler, B.; Yang, H.; Hild, M.; Kung, C.; Wilson, C.; et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 2009, 136, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, M.M.; Kristensen, C.M.; Tøndering, A.S.; Lassen, S.B.; Ringholm, S.; Pilegaard, H. Impact of liver PGC-1α on exercise and exercise training-induced regulation of hepatic autophagy and mitophagy in mice on HFF. Physiol. Rep. 2018, 6, e13731. [Google Scholar] [CrossRef] [PubMed]
- Santos-Alves, E.; Marques-Aleixo, I.; Rizo-Roca, D.; Torrella, J.R.; Oliveira, P.J.; Magalhães, J.; Ascensão, A. Exercise modulates liver cellular and mitochondrial proteins related to quality control signaling. Life Sci. 2015, 135, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Lira, V.A.; Okutsu, M.; Zhang, M.; Greene, N.P.; Laker, R.C.; Breen, D.S.; Hoehn, K.L.; Yan, Z. Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance. FASEB J. 2013, 27, 4184–4193. [Google Scholar] [CrossRef]
- Dong, G.; Zhang, Z.; Duan, K.; Shi, W.; Huang, R.; Wang, B.; Luo, L.; Zhang, Y.; Ruan, H.; Huang, H. Beclin 1 deficiency causes hepatic cell apoptosis via endoplasmic reticulum stress in zebrafish larvae. FEBS Lett. 2019. [Google Scholar] [CrossRef]
- Wang, N.; Tan, H.Y.; Li, S.; Feng, Y. Atg9b Deficiency Suppresses Autophagy and Potentiates Endoplasmic Reticulum Stress-Associated Hepatocyte Apoptosis in Hepatocarcinogenesis. Theranostics 2017, 7, 2325–2338. [Google Scholar] [CrossRef]


| Protein Blend | |
|---|---|
| Total (Casein + Whey Protein) | |
| Aspartic Acid | 6.92 (2.59 + 4.33) |
| Glutamic Acid | 11.96 (5.01 + 6.95) |
| Alanine | 5.85 (3.91 + 1.94) |
| Arginine | 5.12 (4.0 + 1.12) |
| Cystine | 0.73 (0.00 + 0.73) |
| Phenylalanine | 2.19 (0.88 + 1.31) |
| Glycine | 8.48 (7.70 + 0.78) |
| Histidine | 1.13 (0.35 + 0.78) |
| Isoleucine | 2.96 (0.57 + 2.39) |
| Leucine | 5.74 (1.36 + 4.38) |
| Lysine | 5.58 (1.84 + 3.74) |
| Methionine | 1.85 (0.39 +1.46) |
| Proline | 6.68 (5.90 + 0.78) |
| Serine | 2.81 (1.40 + 1.41) |
| Tyrosine | 1.26 (0.09 + 1.17) |
| Threonine | 3.52 (0.70 + 2.82) |
| Tryptophan | 0.63 (0.00 + 0.63) |
| Valine | 3.38 (1.05 + 2.33) |
| Group | Body Weight (g) | Gastrocnemius Weight (g) | Supplementation (g) | |
|---|---|---|---|---|
| Casein | Whey Protein | |||
| CT (n = 4) | 417 ± 0.02 | 2.42 ± 0.16 | --- | ---- |
| EXE-imm (n = 4) | 436 ± 0.03 | 2.88 ± 0.32 | --- | ---- |
| EXE-1h (n = 4) | 427 ± 0.11 | 2.65 ± 0.23 | ---- | ---- |
| EXE/Suppl-imm (n = 5) | 449 ± 0.11 | 2.47 ± 0.08 | 0.85 ± 0.25 | 0.93 ± 0.05 |
| EXE/Suppl-1h (n = 5) | 472 ± 0.20 | 2.76 ± 0.14 | 0.94 ± 0.02 | 0.98 ± 0.05 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinto, A.P.; Vieira, T.S.; Marafon, B.B.; Batitucci, G.; Cabrera, E.M.B.; da Rocha, A.L.; Kohama, E.B.; Rodrigues, K.C.C.; de Moura, L.P.; Pauli, J.R.; et al. The Combination of Fasting, Acute Resistance Exercise, and Protein Ingestion Led to Different Responses of Autophagy Markers in Gastrocnemius and Liver Samples. Nutrients 2020, 12, 641. https://doi.org/10.3390/nu12030641
Pinto AP, Vieira TS, Marafon BB, Batitucci G, Cabrera EMB, da Rocha AL, Kohama EB, Rodrigues KCC, de Moura LP, Pauli JR, et al. The Combination of Fasting, Acute Resistance Exercise, and Protein Ingestion Led to Different Responses of Autophagy Markers in Gastrocnemius and Liver Samples. Nutrients. 2020; 12(3):641. https://doi.org/10.3390/nu12030641
Chicago/Turabian StylePinto, Ana P., Tales S. Vieira, Bruno B. Marafon, Gabriela Batitucci, Elisa M. B. Cabrera, Alisson L. da Rocha, Eike B. Kohama, Kellen C. C. Rodrigues, Leandro P. de Moura, José R. Pauli, and et al. 2020. "The Combination of Fasting, Acute Resistance Exercise, and Protein Ingestion Led to Different Responses of Autophagy Markers in Gastrocnemius and Liver Samples" Nutrients 12, no. 3: 641. https://doi.org/10.3390/nu12030641
APA StylePinto, A. P., Vieira, T. S., Marafon, B. B., Batitucci, G., Cabrera, E. M. B., da Rocha, A. L., Kohama, E. B., Rodrigues, K. C. C., de Moura, L. P., Pauli, J. R., Cintra, D. E., Ropelle, E. R., de Freitas, E. C., & da Silva, A. S. R. (2020). The Combination of Fasting, Acute Resistance Exercise, and Protein Ingestion Led to Different Responses of Autophagy Markers in Gastrocnemius and Liver Samples. Nutrients, 12(3), 641. https://doi.org/10.3390/nu12030641








