Metabolite Profiling of Rambutan (Nephelium lappaceum L.) Seeds Using UPLC-qTOF-MS/MS and Senomorphic Effects in Aged Human Dermal Fibroblasts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Instruments
2.2. Plant Material
2.3. UHPLC-qTOF-MS2 Experiments
2.4. Sample Preparation of Nephelium lappaceum for MS/MS Analysis
2.5. Extraction and Isolation of Flavonoid Glycosides
2.6. Sugar Analysis
2.7. Cloning of the Human p16INK4A and GLB1 Promoter in a Reporter Plasmid
2.8. Cell Culture
2.9. Cell Viability Assay
2.10. Luciferase Reporter Assay
2.11. Quantitative Reverse Transcription-Polymerase Chain Reaction
2.12. Senescence-Associated β-Galactosidase Activity
2.13. Statistical Analysis
3. Results
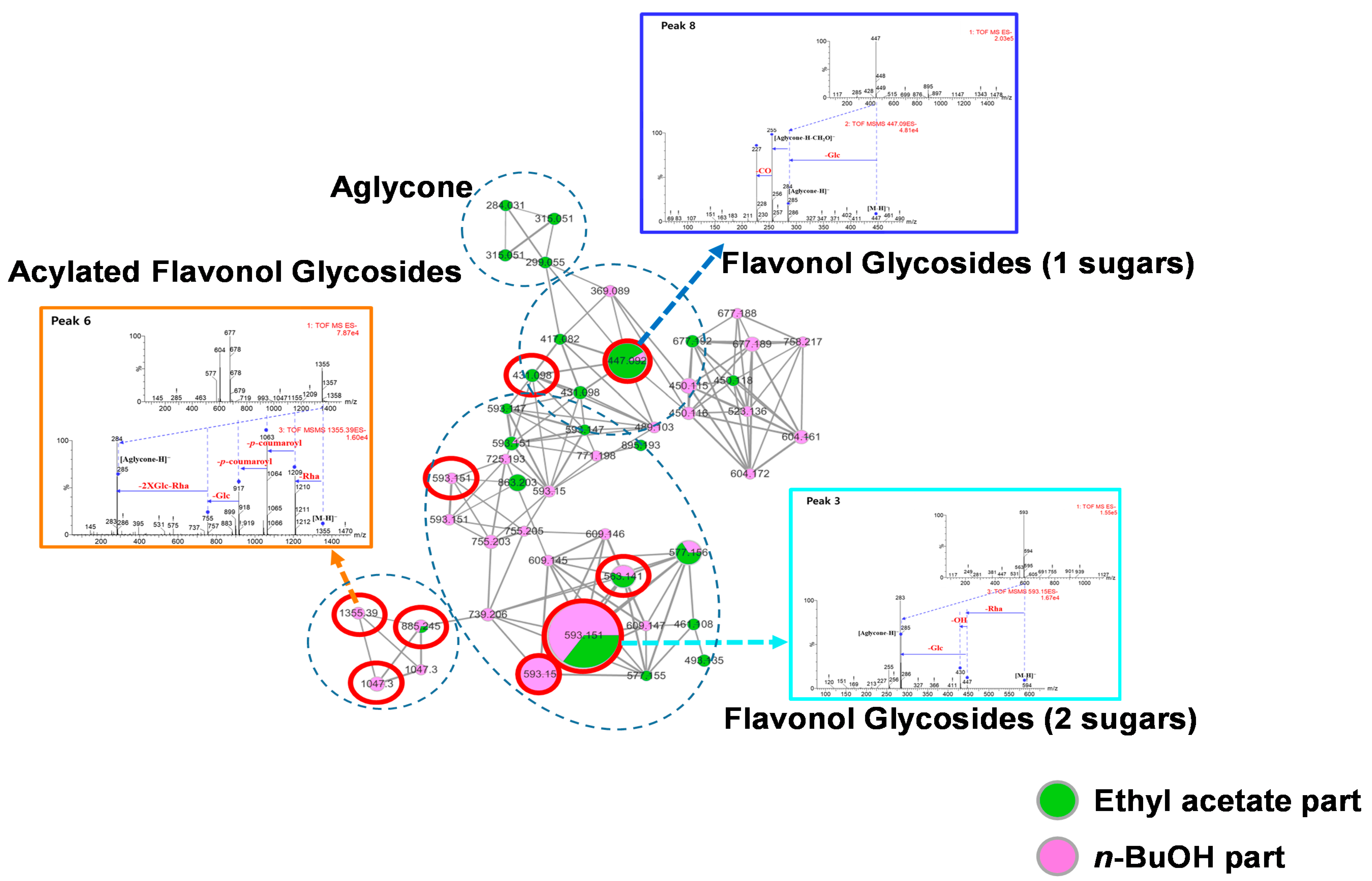
3.1. Molecular Networking of the Seed, Pulp and Peel Extracts of N. lappaceum Fruits
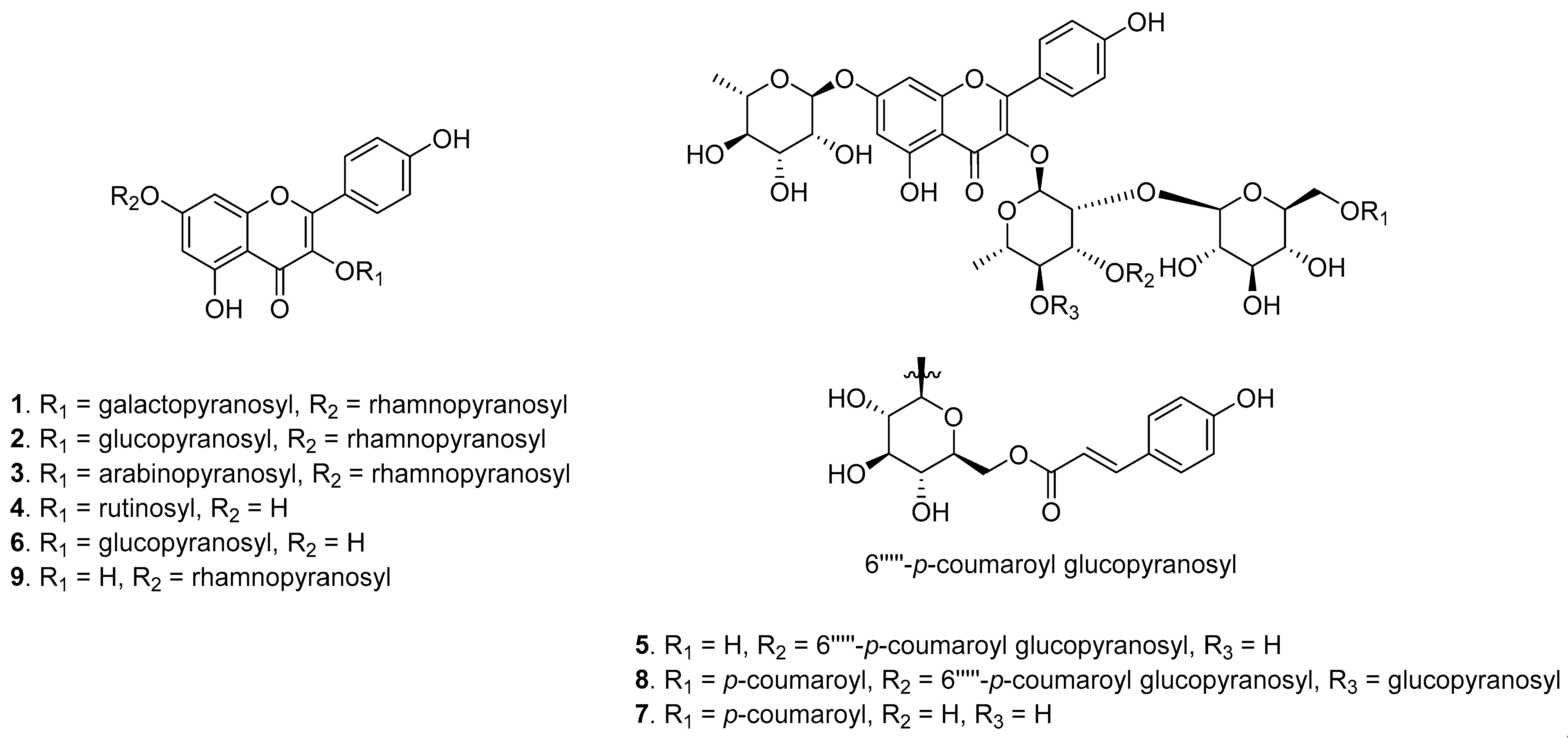
3.2. Bioactivity-Guided Isolation and Dereplication of the Active Compounds
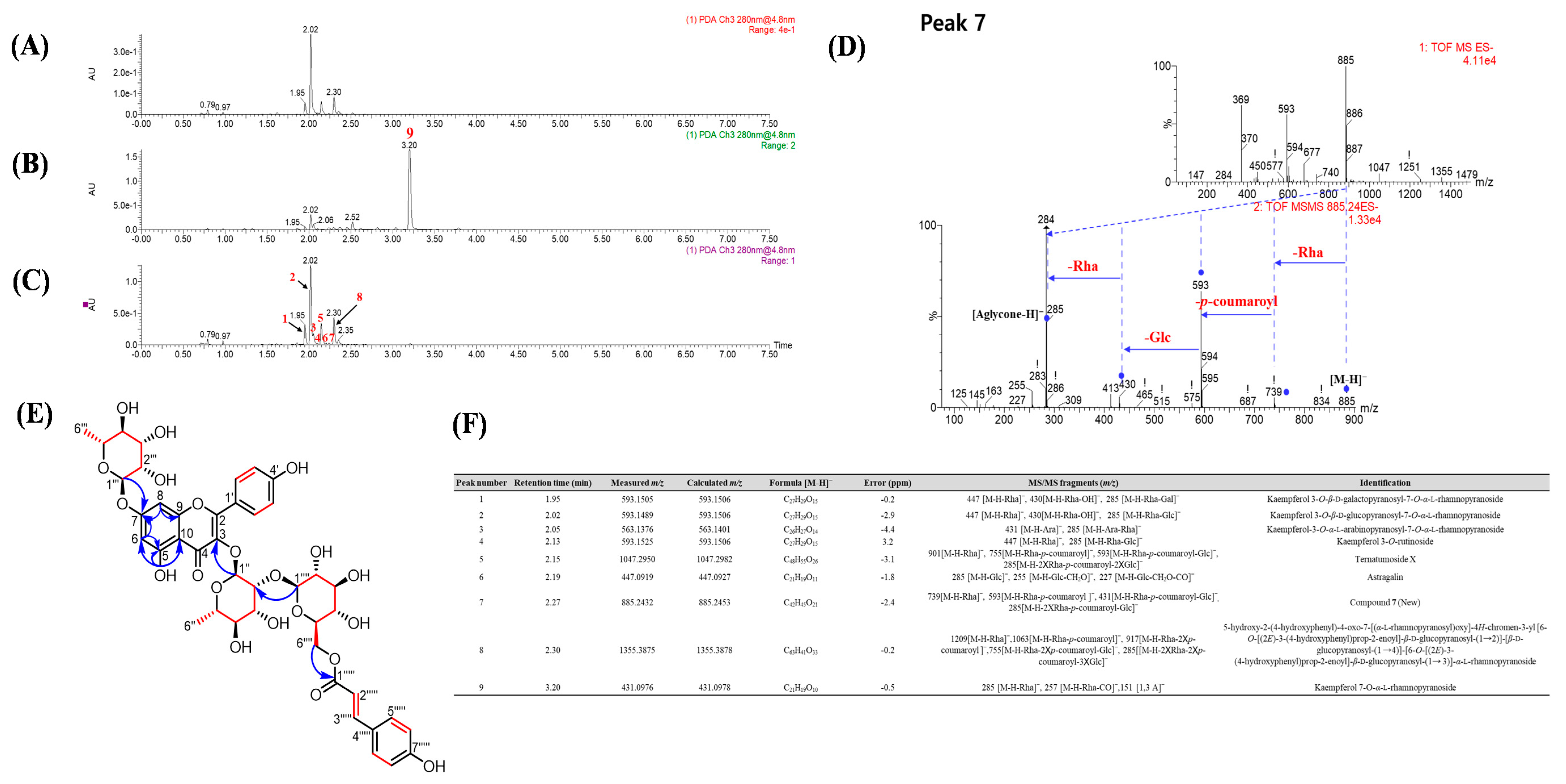
3.3. Isolation and Identification of New Compound 7
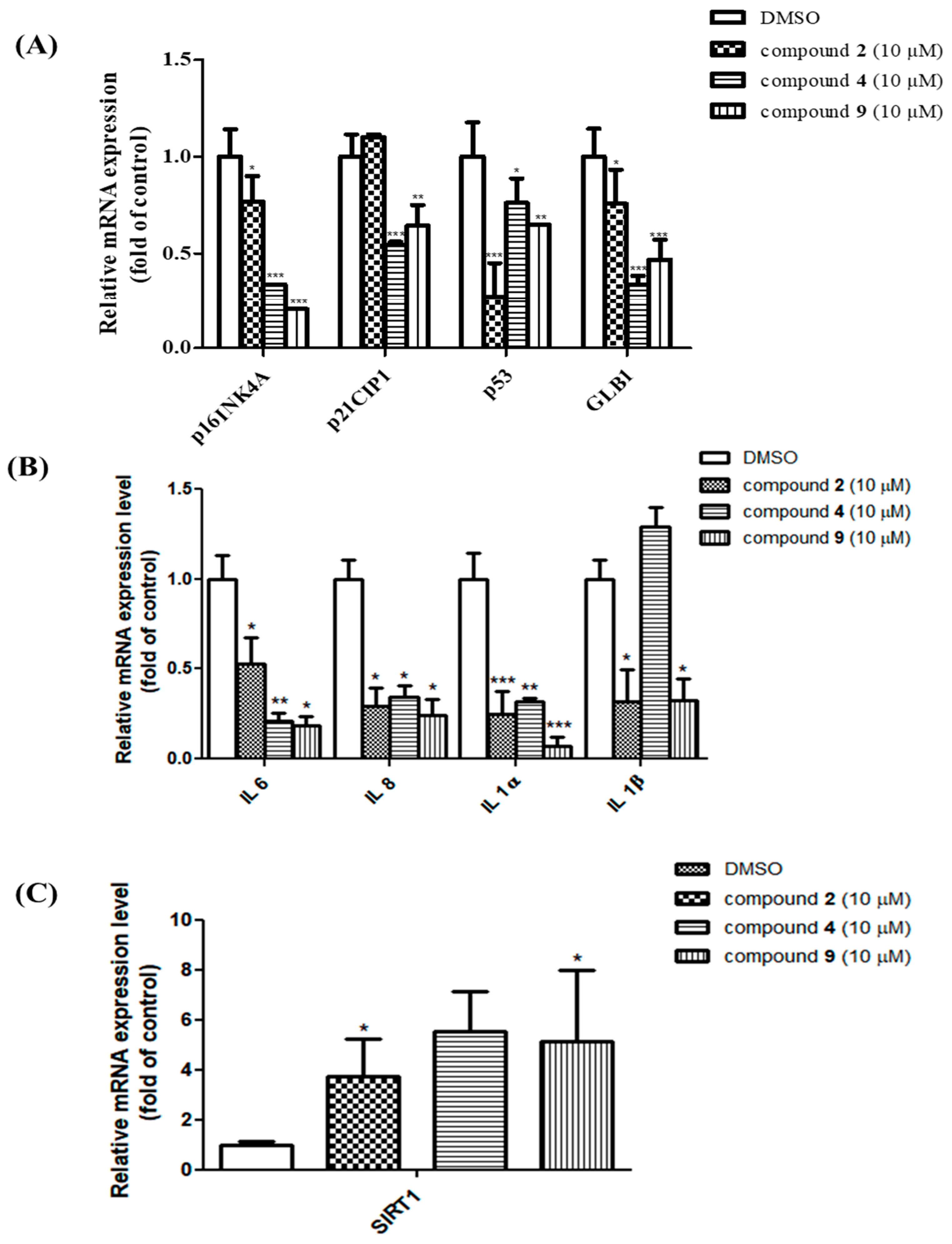
3.4. Effect of the Fractions and Compounds 1–9 at the Transcriptional Level of p16INK4A and Senescence-Associated-β-Galactosidase
3.5. Senomorphic Effects of Compounds 2, 4 and 9 in Rescuing Replicative Senescence and Inhibiting SASP
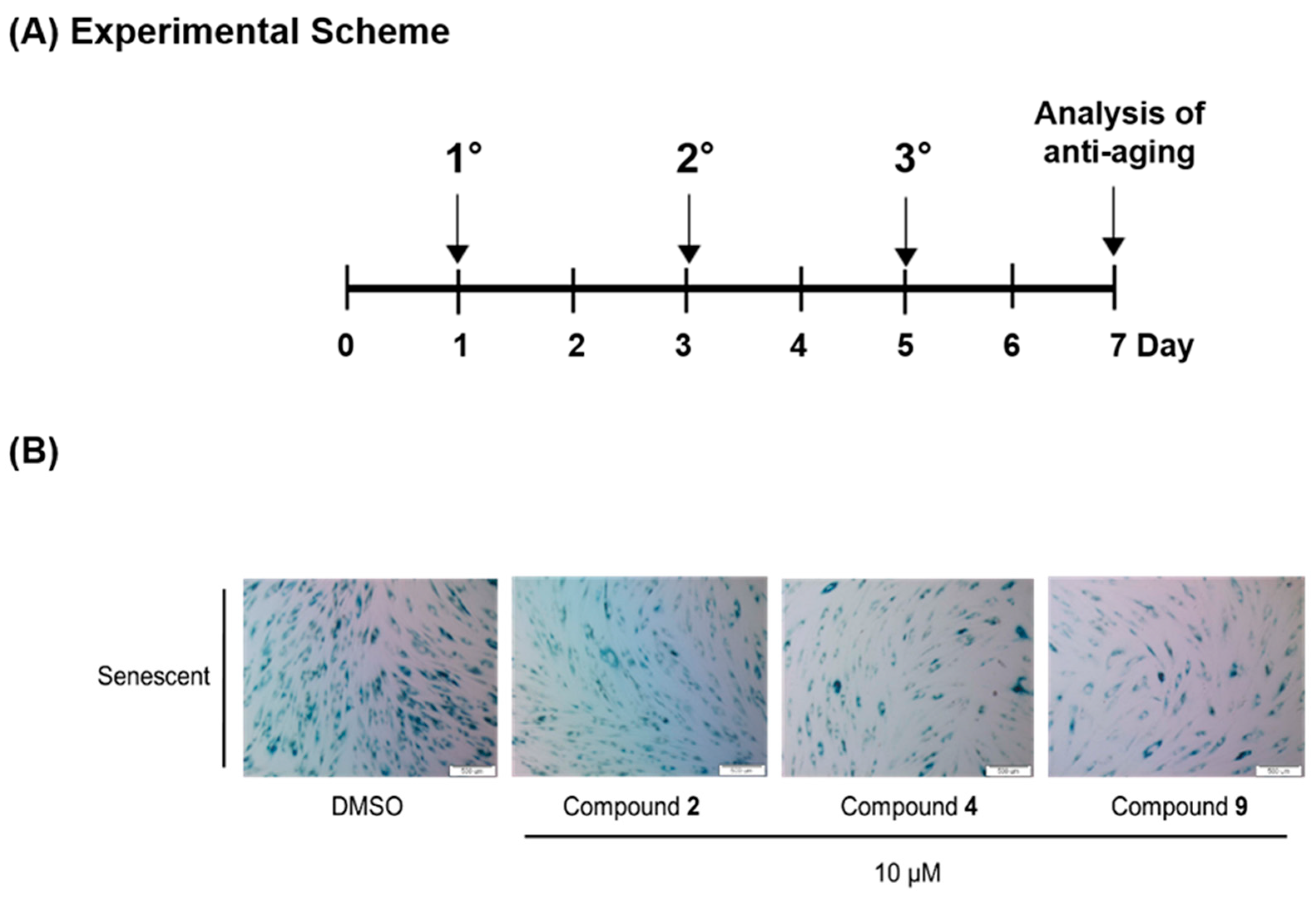
3.6. Compounds 2, 4 and 9 Attenuate the Senescence Phenotype
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Xu, M.; Pirtskhalava, T.; Farr, J.N.; Weigand, B.M.; Palmer, A.K.; Weivoda, M.M.; Inman1, C.L.; Ogrodnik1, M.B.; Hachfeld, C.M.; Fraser, D.G.; et al. Senolytics improve physical function and increase lifespan in old age. Nat. Med. 2018, 24, 1246–1256. [Google Scholar] [CrossRef] [PubMed]
- Myrianthopoulos, V. The emerging field of senotherapeutic drugs. Biochem. Pharmacol. 2018, 10, 2369–2372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Tchkonia, T.; Pirtskhalava, T.; Gower, A.C.; Ding, H.; Giorgadze, N.; Palmer, A.K.; Ikeno, Y.; Hubbard, G.B.; Lenburg, M.; et al. The Achilles’ heel of senescent cells: From transcriptome to senolytic drugs. Aging Cell 2015, 14, 644–658. [Google Scholar] [CrossRef]
- Zhu, Y.; Doornebal, E.J.; Pirtskhalava, T.; Giorgadze, N.; Wentworth, M.; Fuhrmann-Stroissnigg, H.; Niedernhofer, L.J.; Robbins, P.D.; Tchkonia, T.; Kirkland, J.L. New agents that target senescent cells: The flavone, fisetin, and the BCL-XL inhibitors, A1331852 and A1155463. Aging (Albany NY) 2017, 9, 955–963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayakawa, T.; Iwai, M.; Aoki, S.; Takimoto, K.; Maruyama, M.; Maruyama, W.; Motoyama, N. SIRT1 suppresses the senescence-associated secretory phenotype through epigenetic gene regulation. PLoS ONE 2015, 10, e0116480. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.Y.; Han, J.A.; Im, J.S.; Morrone, A.; Johung, K.; Goodwin, E.C.; Kleijet, W.J.; DiMaio, D.; Hwang, E.S. Senescence-associated β-galactosidase is lysosomal β-galactosidase. Aging Cell 2006, 5, 187–195. [Google Scholar] [CrossRef]
- Wang, A.S.; Dreesen, O. Biomarkers of Cellular Senescence and Skin Aging. Front. Genet. 2018, 9, 247–260. [Google Scholar] [CrossRef]
- Lim, H.; Park, H.; Kim, H.P. Effects of flavonoids on senescence-associated secretory phenotype formation from bleomycin-induced senescence in BJ fibroblasts. Biochem. Pharmacol. 2015, 96, 337–348. [Google Scholar] [CrossRef]
- Jang, H.J.; Yang, K.E.; Oh, W.K.; Lee, S.I.; Hwang, I.H.; Ban, K.T.; Yoo, H.S.; Choi, J.S.; Yeo, E.J.; Jang, I.S. Nectandrin B-mediated activation of the AMPK pathway prevents cellular senescence in human diploid fibroblasts by reducing intracellular ROS levels. Aging (Albany NY) 2019, 11, 3731–3749. [Google Scholar] [CrossRef]
- Thitilertdecha, N.; Teerawutgulrag, A.; Kilburn, J.D.; Rakariyatham, N. Identification of major phenolic compounds from Nephelium lappaceum L. and their antioxidant activities. Molecules 2010, 15, 1453–1465. [Google Scholar] [CrossRef] [Green Version]
- Palanisamy, U.; Ming, C.H.; Masilamani, T.; Subramaniam, T.; Teng, L.L. Radhakrishnan. Rind of the rambutan, Nephelium lappaceum, a potential source of natural antioxidants. Food Chem. 2008, 1090, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.M.; Ha, T.K.Q.; Dang, L.H.; Pham, H.T.T.; Tran, V.O.; Huh, J.; An, J.P.; Oh, W.K. Prenylated phenolic compounds from the leaves of Sabia limoniacea and their antiviral activities against porcine epidemic diarrhea virus. J. Nat. Prod. 2019, 82, 702–713. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, J.; Takamori, Y.; Tanaka, Y. Studies on the constituents of Acanthopanax sciadophylloides FR. Et SAV. Leaves. Yakugaku Zasshi J. Pharm. Soc. Jpn. 1989, 109, 188–191. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Kim, J.; Whang, W. Chemical constituents of Smilax china L. stems and their inhibitory activities against glycation, aldose reductase, α-glucosidase, and lipase. Molecules 2017, 22, 451. [Google Scholar] [CrossRef] [Green Version]
- Dong, L.; Yin, L.; Quan, H.; Chu, Y.; Lu, J. Hepatoprotective effects of kaempferol-3-O-α-l-arabinopyranosyl-7-O-α-l-rhamnopyranoside on d-galactosamine and lipopolysaccharide caused hepatic failure in mice. Molecules 2017, 22, 1755. [Google Scholar] [CrossRef] [Green Version]
- Kazuma, K.; Noda, N.; Suzuki, M. Malonylated flavonol glycosides from the petals of Clitoria ternatea. Phytochemistry 2003, 62, 229–237. [Google Scholar] [CrossRef]
- Warashina, T.; Umehara, K.; Miyase, T. Flavonoid glycosides from Botrychium ternatum. Chem. Pharm Bull. 2012, 60, 1561–1573. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Xie, Q.; Fisher, D.; Sutherland, L.A. Separation of patuletin-3-O-glucoside, astragalin, quercetin, kaempferol and isorhamnetin from Flaveria bidentis (L.) Kuntze by elution-pump-out high-performance counter-current chromatography. J. Chromatogr. A 2011, 36, 6206–6211. [Google Scholar] [CrossRef]
- Khan, A.; Ahmad, V.U.; Farooq, U. Two new acylated flavonol glycosides from the roots of Otostegia limbata. Hel. Chim. Acta. 2009, 92, 731–739. [Google Scholar] [CrossRef]
- Seo, K.H.; Jung, J.W.; Thi, N.N.; Lee, Y.H.; Baek, N.I. Flavonoid glycosides from the flowers of Pulsatilla koreana Nakai. Nat Prod Sci. 2016, 22, 41–45. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Ikeda, T.; Matsuoka, N.; Nohara, T.; Zhang, H.; Sakamoto, T.; Nonaka, G.I. Cucurbitane glycosides from unripe fruits of Lo Han Kuo (Siraitia grosvenori). Chem. Pharm. Bull. 2006, 54, 1425–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latorre, E.; Birar, V.C.; Sheerin, A.N.; Jeynes, J.C.C.; Hooper, A.; Dawe, H.R.; Melze, D.; Cox, L.S.; Faragher, R.G.; Ostler, E.; et al. Small molecule modulation of splicing factor expression is associated with rescue from cellular senescence. BMC Cell Biol. 2017, 18, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.Q.; Guan, J.T.; Xu, X.H.; Fu, Y.C. Senescence-associated beta-galactosidase activity expression in aging hippocampal neurons. Biochem. Biophys. Res. Commun. 2010, 396, 866–869. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.R.; Cho, H.M.; Park, E.J.; Zhang, M.; Doan, T.P.; Lee, B.W.; Cho, K.A.; Oh, W.K. Metabolite Profiling of Rambutan (Nephelium lappaceum L.) Seeds Using UPLC-qTOF-MS/MS and Senomorphic Effects in Aged Human Dermal Fibroblasts. Nutrients 2020, 12, 1430. https://doi.org/10.3390/nu12051430
Lee YR, Cho HM, Park EJ, Zhang M, Doan TP, Lee BW, Cho KA, Oh WK. Metabolite Profiling of Rambutan (Nephelium lappaceum L.) Seeds Using UPLC-qTOF-MS/MS and Senomorphic Effects in Aged Human Dermal Fibroblasts. Nutrients. 2020; 12(5):1430. https://doi.org/10.3390/nu12051430
Chicago/Turabian StyleLee, Yae Rin, Hyo Moon Cho, Eun Jin Park, Mi Zhang, Thi Phuong Doan, Ba Wool Lee, Kyung A Cho, and Won Keun Oh. 2020. "Metabolite Profiling of Rambutan (Nephelium lappaceum L.) Seeds Using UPLC-qTOF-MS/MS and Senomorphic Effects in Aged Human Dermal Fibroblasts" Nutrients 12, no. 5: 1430. https://doi.org/10.3390/nu12051430
APA StyleLee, Y. R., Cho, H. M., Park, E. J., Zhang, M., Doan, T. P., Lee, B. W., Cho, K. A., & Oh, W. K. (2020). Metabolite Profiling of Rambutan (Nephelium lappaceum L.) Seeds Using UPLC-qTOF-MS/MS and Senomorphic Effects in Aged Human Dermal Fibroblasts. Nutrients, 12(5), 1430. https://doi.org/10.3390/nu12051430





