Discovery of a Novel Multi-Strains Probiotic Formulation with Improved Efficacy toward Intestinal Inflammation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Probiotics
2.2. Animals and Colitis Protocols
2.3. Histology
2.4. Quantification of Fecal Lcn-2 by ELISA
2.5. Isolation of Lamina Propria Cells
2.6. Flow Cytometry
2.7. Reverse Transcription of mRNA and Real-Time PCR
2.8. Metagenomic Analysis
2.9. Statistical Analysis
3. Results
3.1. Effects of the Probiotics Formulations in Mouse Model of Acute Colitis
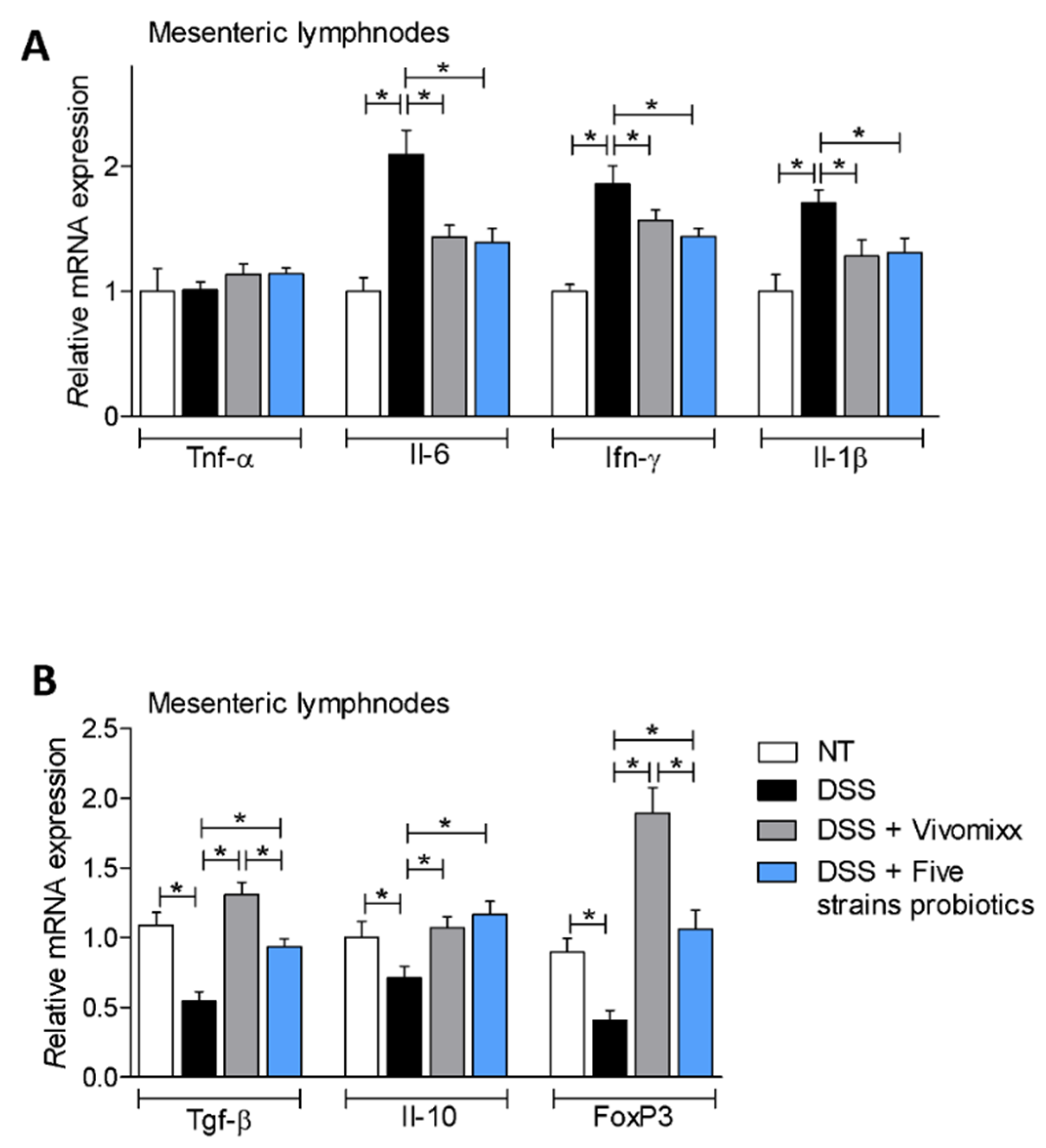
3.2. Effects of the Probiotic Formulations in a Model of Chronic Colitis
3.3. Impact of the two Probiotic Formulations on the Composition of the Intestinal Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviation
| TNBS | 2,4,6-Trinitrobenzenesulfonic acid |
| DSS | dextran sodium sulfate |
| H&E | Hematoxilin and Eosin |
| CDAI | colitis disease activity index |
| Treg | Regulatory T lymphocytes |
| mLN | mesenteric lymph nodes |
References
- De Souza, H.S.; Fiocchi, C. Immunopathogenesis of IBD: Current state of the art. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; St Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, A.W.; Sanderson, J.D.; Churcher, C.; Parkes, G.C.; Hudspith, B.N.; Rayment, N.; Brostoff, J.; Parkhill, J.; Dougan, G.; Petrovska, L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011, 11, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sartor, R.B.; Wu, G.D. Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology 2017, 152, 327.e4–339.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danese, S.; Panés, J. Development of drugs to target interactions between leukocytes and endothelial cells and treatment algorithms for inflammatory bowel diseases. Gastroenterology 2014, 147, 981–989. [Google Scholar] [CrossRef]
- Mosli, M.H.; Rivera-Nieves, J.; Feagan, B.G. T-cell trafficking and anti-adhesion strategies in inflammatory bowel disease: Current and future prospects. Drugs 2014, 74, 297–311. [Google Scholar] [CrossRef]
- Fiorucci, S.; Biagioli, M.; Distrutti, E. Immunephenotype Predicts Response to Vedolizumab: Integrating Clinical and Biochemical Biomarkers in the Treatment of Inflammatory Bowel Diseases. Dig. Dis. Sci. 2018, 63, 2168–2171. [Google Scholar] [CrossRef] [Green Version]
- Oka, A.; Sartor, R.B. Microbial-Based and Microbial-Targeted Therapies for Inflammatory Bowel Diseases. Dig. Dis. Sci. 2020, 65, 757–788. [Google Scholar] [CrossRef] [Green Version]
- Korada, S.K.; Yarla, N.S.; Mishra, V.; Daim, M.A.; Sharma, B.; Gm, A.; R, R.; M, P.; Peluso, I.; Kamal, M.A. Single Probiotic versus Multiple Probiotics—A Debate On Current Scenario for Alleviating Health Benefits. Curr. Pharm. Des. 2018, 24, 4150–4153. [Google Scholar] [CrossRef]
- Rondanelli, M.; Faliva, M.A.; Perna, S.; Giacosa, A.; Peroni, G.; Castellazzi, A.M. Using probiotics in clinical practice: Where are we now? A review of existing meta-analyses. Gut Microbes 2017, 8, 521–543. [Google Scholar] [CrossRef] [Green Version]
- Naseer, M.; Poola, S.; Ali, S.; Samiullah, S.; Tahan, V. Prebiotics and Probiotics in Inflammatory Bowel Disease (IBD): Where Are We Now And Where Are We Going? Funders. Curr. Clin. Pharmacol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.L.; Li, W.S.; Xu, D.N.; Zheng, W.W.; Liu, Y.; Chen, J.; Qiu, Z.B.; Dorfman, R.G.; Zhang, J.; Liu, J. Mucosa-reparing and microbiota-balancing therapeutic effect of. Exp. Ther. Med. 2016, 12, 2554–2562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, G.; Wang, K.; Li, Z.; Tao, F.; Xu, Y.; Lan, J.; Chen, G.; Yang, C. Ameliorates Dextran Sulfate Sodium-Induced Colitis by Improving Gut Microbial Dysbiosis in Mice Model. Front. Microbiol. 2018, 9, 3260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Ouyang, M.; Guo, Q.; Jia, J.; Liu, R.; Jiang, Y.; Wu, M.; Shen, S. Changes in the intestinal microecology induced by bacillus subtilis inhibit the occurrence of ulcerative colitis and associated cancers: A study on the mechanisms. Am. J. Cancer Res. 2019, 9, 872–886. [Google Scholar] [PubMed]
- Jing, Y.; Liu, H.; Xu, W.; Yang, Q. Amelioration of the DSS-induced colitis in mice by pretreatment with 4,4′-diaponeurosporene-producing. Exp. Ther. Med. 2017, 14, 6069–6073. [Google Scholar] [CrossRef] [Green Version]
- Gong, Y.; Li, H.; Li, Y. Effects of Bacillus subtilis on Epithelial Tight Junctions of Mice with Inflammatory Bowel Disease. J. Interf. Cytokine Res. 2016, 36, 75–85. [Google Scholar] [CrossRef]
- Bailey, J.R.; Vince, V.; Williams, N.A.; Cogan, T.A. Streptococcus thermophilus NCIMB 41856 ameliorates signs of colitis in an animal model of inflammatory bowel disease. Benef. Microbes 2017, 8, 605–614. [Google Scholar] [CrossRef] [Green Version]
- Wasilewska, E.; Zlotkowska, D.; Wroblewska, B. Yogurt starter cultures of Streptococcus thermophilus and Lactobacillus bulgaricus ameliorate symptoms and modulate the immune response in a mouse model of dextran sulfate sodium-induced colitis. J. Dairy Sci. 2019, 102, 37–53. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zhang, M.; Ren, F. A Role of Exopolysaccharide Produced by. Molecules 2019, 24, 513. [Google Scholar] [CrossRef] [Green Version]
- Ito, M.; Ohishi, K.; Yoshida, Y.; Okumura, T.; Sato, T.; Yokoi, W.; Sawada, H. Preventive effect of Streptococcus thermophilus YIT 2001 on dextran sulfate sodium-induced colitis in mice. Biosci. Biotechnol. Biochem. 2008, 72, 2543–2547. [Google Scholar] [CrossRef] [Green Version]
- Thakur, B.K.; Saha, P.; Banik, G.; Saha, D.R.; Grover, S.; Batish, V.K.; Das, S. Live and heat-killed probiotic Lactobacillus casei Lbs2 protects from experimental colitis through Toll-like receptor 2-dependent induction of T-regulatory response. Int. Immunopharmacol. 2016, 36, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Bellavia, M.; Rappa, F.; Lo Bello, M.; Brecchia, G.; Tomasello, G.; Leone, A.; Spatola, G.; Uzzo, M.L.; Bonaventura, G.; David, S.; et al. Lactobacillus casei and bifidobacterium lactis supplementation reduces tissue damage of intestinal mucosa and liver after 2,4,6-trinitrobenzenesulfonic acid treatment in mice. J. Biol. Regul. Homeost. Agents 2014, 28, 251–261. [Google Scholar] [PubMed]
- Zhang, Y.; Hou, Q.; Ma, C.; Zhao, J.; Xu, H.; Li, W.; Wang, Y.; Ma, H.; Zhang, H.; Sun, Z. Lactobacillus casei protects dextran sodium sulfate- or rapamycin-induced colonic inflammation in the mouse. Eur. J. Nutr. 2019. [Google Scholar] [CrossRef] [PubMed]
- Jacouton, E.; Chain, F.; Sokol, H.; Langella, P.; Bermúdez-Humarán, L.G. Probiotic Strain. Front. Immunol. 2017, 8, 1553. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Perez, N.G.; Lozano-Ojalvo, D.; Maiga, M.A.; Hazebrouck, S.; Adel-Patient, K. Intragastric administration of Lactobacillus casei BL23 induces regulatory FoxP3+RORγt+ T cells subset in mice. Benef. Microbes 2017, 8, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Van Bergenhenegouwen, J.; Overbeek, S.; Van de Kant, H.J.; Garssen, J.; Folkerts, G.; Vos, P.; Morgan, M.E.; Kraneveld, A.D. Bifidobacterium breve attenuates murine dextran sodium sulfate-induced colitis and increases regulatory T cell responses. PLoS ONE 2014, 9, e95441. [Google Scholar] [CrossRef] [Green Version]
- Jeon, S.G.; Kayama, H.; Ueda, Y.; Takahashi, T.; Asahara, T.; Tsuji, H.; Tsuji, N.M.; Kiyono, H.; Ma, J.S.; Kusu, T.; et al. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 2012, 8, e1002714. [Google Scholar] [CrossRef] [Green Version]
- Heuvelin, E.; Lebreton, C.; Grangette, C.; Pot, B.; Cerf-Bensussan, N.; Heyman, M. Mechanisms involved in alleviation of intestinal inflammation by bifidobacterium breve soluble factors. PLoS ONE 2009, 4, e5184. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.; Chung, W.H.; Lim, T.J.; Lim, S.; Nam, Y.D. Complete genome sequence of the. New Microbes New Infect. 2017, 19, 34–37. [Google Scholar] [CrossRef]
- Chae, J.M.; Heo, W.; Cho, H.T.; Lee, D.H.; Kim, J.H.; Rhee, M.S.; Park, T.S.; Kim, Y.K.; Lee, J.H.; Kim, Y.J. Erratum to: Orally-Administered. J. Microbiol. Biotechnol. 2019, 29, 665. [Google Scholar] [CrossRef] [Green Version]
- Paveljšek, D.; Juvan, P.; Košir, R.; Rozman, D.; Hacin, B.; Ivičak-Kocjan, K.; Rogelj, I. Lactobacillus fermentum L930BB and Bifidobacterium animalis subsp. animalis IM386 initiate signalling pathways involved in intestinal epithelial barrier protection. Benef. Microbes 2018, 9, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Khatri, I.; Sharma, G.; Subramanian, S. Composite genome sequence of Bacillus clausii, a probiotic commercially available as Enterogermina. BMC Microbiol. 2019, 19, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biagioli, M.; Capobianco, D.; Carino, A.; Marchianò, S.; Fiorucci, C.; Ricci, P.; Distrutti, E.; Fiorucci, S. Divergent Effectiveness of Multispecies Probiotic Preparations on Intestinal Microbiota Structure Depends on Metabolic Properties. Nutrients 2019, 11, 325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masco, L.; Ventura, M.; Zink, R.; Huys, G.; Swings, J. Polyphasic taxonomic analysis of Bifidobacterium animalis and Bifidobacterium lactis reveals relatedness at the subspecies level: Reclassification of Bifidobacterium animalis as Bifidobacterium animalis subsp. animalis subsp. nov. and Bifidobacterium lactis as Bifidobacterium animalis subsp. lactis subsp. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 1137–1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turroni, F.; Foroni, E.; Pizzetti, P.; Giubellini, V.; Ribbera, A.; Merusi, P.; Cagnasso, P.; Bizzarri, B.; De’Angelis, G.L.; Shanahan, F.; et al. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol. 2009, 75, 1534–1545. [Google Scholar] [CrossRef] [Green Version]
- Loquasto, J.R.; Barrangou, R.; Dudley, E.G.; Stahl, B.; Chen, C.; Roberts, R.F. Bifidobacterium animalis subsp. lactis ATCC 27673 is a genomically unique strain within its conserved subspecies. Appl. Environ. Microbiol. 2013, 79, 6903–6910. [Google Scholar] [CrossRef] [Green Version]
- Biagioli, M.; Laghi, L.; Carino, A.; Cipriani, S.; Distrutti, E.; Marchianò, S.; Parolin, C.; Scarpelli, P.; Vitali, B.; Fiorucci, S. Metabolic Variability of a Multispecies Probiotic Preparation Impacts on the Anti-inflammatory Activity. Front. Pharmacol. 2017, 8, 505. [Google Scholar] [CrossRef] [Green Version]
- Talero, E.; Bolivar, S.; Ávila-Román, J.; Alcaide, A.; Fiorucci, S.; Motilva, V. Inhibition of chronic ulcerative colitis-associated adenocarcinoma development in mice by VSL#3. Inflamm. Bowel Dis. 2015, 21, 1027–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mencarelli, A.; Distrutti, E.; Renga, B.; D’Amore, C.; Cipriani, S.; Palladino, G.; Donini, A.; Ricci, P.; Fiorucci, S. Probiotics modulate intestinal expression of nuclear receptor and provide counter-regulatory signals to inflammation-driven adipose tissue activation. PLoS ONE 2011, 6, e22978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erben, U.; Loddenkemper, C.; Doerfel, K.; Spieckermann, S.; Haller, D.; Heimesaat, M.M.; Zeitz, M.; Siegmund, B.; Kühl, A.A. A guide to histomorphological evaluation of intestinal inflammation in mouse models. Int. J. Clin. Exp. Pathol. 2014, 7, 4557–4576. [Google Scholar] [PubMed]
- Chassaing, B.; Srinivasan, G.; Delgado, M.A.; Young, A.N.; Gewirtz, A.T.; Vijay-Kumar, M. Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS ONE 2012, 7, e44328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bain, C.C.; Mowat, A.M. The monocyte-macrophage axis in the intestine. Cell. Immunol. 2014, 291, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Cerovic, V.; Bain, C.C.; Mowat, A.M.; Milling, S.W. Intestinal macrophages and dendritic cells: What’s the difference? Trends Immunol. 2014, 35, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef] [Green Version]
- Hoarau, G.; Mukherjee, P.K.; Gower-Rousseau, C.; Hager, C.; Chandra, J.; Retuerto, M.A.; Neut, C.; Vermeire, S.; Clemente, J.; Colombel, J.F.; et al. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Ohkusa, T.; Okayasu, I.; Ogihara, T.; Morita, K.; Ogawa, M.; Sato, N. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut 2003, 52, 79–83. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.C.; Tonkonogy, S.L.; Albright, C.A.; Tsang, J.; Balish, E.J.; Braun, J.; Huycke, M.M.; Sartor, R.B. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology 2005, 128, 891–906. [Google Scholar] [CrossRef]
- Britton, G.J.; Contijoch, E.J.; Mogno, I.; Vennaro, O.H.; Llewellyn, S.R.; Ng, R.; Li, Z.; Mortha, A.; Merad, M.; Das, A.; et al. Microbiotas from Humans with Inflammatory Bowel Disease Alter the Balance of Gut Th17 and RORγt. Immunity 2019, 50, 212–224.e214. [Google Scholar] [CrossRef] [Green Version]
- Laurell, A.; Sjöberg, K. Prebiotics and synbiotics in ulcerative colitis. Scand. J. Gastroenterol. 2017, 52, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Imdad, A.; Nicholson, M.R.; Tanner-Smith, E.E.; Zackular, J.P.; Gomez-Duarte, O.G.; Beaulieu, D.B.; Acra, S. Fecal transplantation for treatment of inflammatory bowel disease. Cochrane Database Syst. Rev. 2018, 11, CD012774. [Google Scholar] [CrossRef] [PubMed]
- Moayyedi, P.; Surette, M.G.; Kim, P.T.; Libertucci, J.; Wolfe, M.; Onischi, C.; Armstrong, D.; Marshall, J.K.; Kassam, Z.; Reinisch, W.; et al. Fecal Microbiota Transplantation Induces Remission in Patients With Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology 2015, 149, 102–109.e106. [Google Scholar] [CrossRef] [Green Version]
- Cohen, L.J.; Cho, J.H.; Gevers, D.; Chu, H. Genetic Factors and the Intestinal Microbiome Guide Development of Microbe-Based Therapies for Inflammatory Bowel Diseases. Gastroenterology 2019, 156, 2174–2189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganji-Arjenaki, M.; Rafieian-Kopaei, M. Probiotics are a good choice in remission of inflammatory bowel diseases: A meta analysis and systematic review. J. Cell. Physiol. 2018, 233, 2091–2103. [Google Scholar] [CrossRef]
- Peng, L.; Zhong, Y.; Wang, A.; Jiang, Z. Probiotics combined with aminosalicylic acid affiliates remission of ulcerative colitis: A meta-analysis of randomized controlled trial. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [Green Version]
- Distrutti, E.; Cipriani, S.; Mencarelli, A.; Renga, B.; Fiorucci, S. Probiotics VSL#3 protect against development of visceral pain in murine model of irritable bowel syndrome. PLoS ONE 2013, 8, e63893. [Google Scholar] [CrossRef]
- Wirtz, S.; Popp, V.; Kindermann, M.; Gerlach, K.; Weigmann, B.; Fichtner-Feigl, S.; Neurath, M.F. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat. Protoc. 2017, 12, 1295–1309. [Google Scholar] [CrossRef]
- Vlamakis, H.; Chai, Y.; Beauregard, P.; Losick, R.; Kolter, R. Sticking together: Building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 2013, 11, 157–168. [Google Scholar] [CrossRef] [Green Version]
- Hong, H.A.; Khaneja, R.; Tam, N.M.; Cazzato, A.; Tan, S.; Urdaci, M.; Brisson, A.; Gasbarrini, A.; Barnes, I.; Cutting, S.M. Bacillus subtilis isolated from the human gastrointestinal tract. Res. Microbiol. 2009, 160, 134–143. [Google Scholar] [CrossRef]
- Fritze, D. Taxonomy of the genus bacillus and related genera: The aerobic endospore-forming bacteria. Phytopathology 2004, 94, 1245–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.T.; Lee, F.L.; Tai, C.J.; Kuo, H.P. Bacillus velezensis is a later heterotypic synonym of Bacillus amyloliquefaciens. Int. J. Syst. Evol. Microbiol. 2008, 58, 671–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woese, C.R. Bacterial evolution. Microbiol. Rev. 1987, 51, 221–271. [Google Scholar] [CrossRef] [PubMed]
- Tam, N.K.; Uyen, N.Q.; Hong, H.A.; Duc, l.H.; Hoa, T.T.; Serra, C.R.; Henriques, A.O.; Cutting, S.M. The intestinal life cycle of Bacillus subtilis and close relatives. J. Bacteriol. 2006, 188, 2692–2700. [Google Scholar] [CrossRef] [Green Version]
- Shan, M.; Gentile, M.; Yeiser, J.R.; Walland, A.C.; Bornstein, V.U.; Chen, K.; He, B.; Cassis, L.; Bigas, A.; Cols, M.; et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science 2013, 342, 447–453. [Google Scholar] [CrossRef] [Green Version]
- Permpoonpattana, P.; Hong, H.A.; Phetcharaburanin, J.; Huang, J.M.; Cook, J.; Fairweather, N.F.; Cutting, S.M. Immunization with Bacillus spores expressing toxin A peptide repeats protects against infection with Clostridium difficile strains producing toxins A and B. Infect. Immun. 2011, 79, 2295–2302. [Google Scholar] [CrossRef] [Green Version]
- Stasiłojć, M.; Hinc, K.; Peszyńska-Sularz, G.; Obuchowski, M.; Iwanicki, A. Recombinant Bacillus subtilis Spores Elicit Th1/Th17-Polarized Immune Response in a Murine Model of Helicobacter pylori Vaccination. Mol. Biotechnol. 2015, 57, 685–691. [Google Scholar] [CrossRef] [Green Version]








© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biagioli, M.; Carino, A.; Di Giorgio, C.; Marchianò, S.; Bordoni, M.; Roselli, R.; Distrutti, E.; Fiorucci, S. Discovery of a Novel Multi-Strains Probiotic Formulation with Improved Efficacy toward Intestinal Inflammation. Nutrients 2020, 12, 1945. https://doi.org/10.3390/nu12071945
Biagioli M, Carino A, Di Giorgio C, Marchianò S, Bordoni M, Roselli R, Distrutti E, Fiorucci S. Discovery of a Novel Multi-Strains Probiotic Formulation with Improved Efficacy toward Intestinal Inflammation. Nutrients. 2020; 12(7):1945. https://doi.org/10.3390/nu12071945
Chicago/Turabian StyleBiagioli, Michele, Adriana Carino, Cristina Di Giorgio, Silvia Marchianò, Martina Bordoni, Rosalinda Roselli, Eleonora Distrutti, and Stefano Fiorucci. 2020. "Discovery of a Novel Multi-Strains Probiotic Formulation with Improved Efficacy toward Intestinal Inflammation" Nutrients 12, no. 7: 1945. https://doi.org/10.3390/nu12071945




