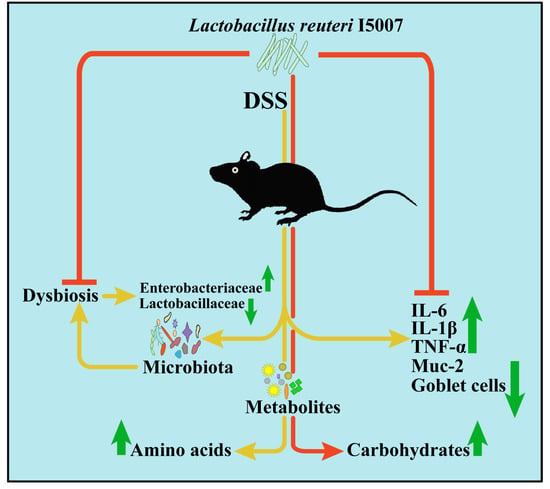
Lactobacillus reuteri Ameliorates Intestinal Inflammation and Modulates Gut Microbiota and Metabolic Disorders in Dextran Sulfate Sodium-Induced Colitis in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Preparation of Bacteria
2.3. Cell Culture and Treatment
2.4. Animals and Treatments
2.5. Histological and Goblet Cell Evaluation
2.6. Quantitative Real-Time Polymerase Chain Reaction (PCR) Analysis
2.7. Assay of Inflammatory Cytokines
2.8. Bioinformatics Analysis of 16S rRNA Gene Profiling
2.9. Untargeted Metabolomic Study
2.10. Statistical Analysis
3. Results
3.1. Effects of L. reuteri I5007 on Inflammatory Cytokines in HT-29 Cells Challenged with LPS
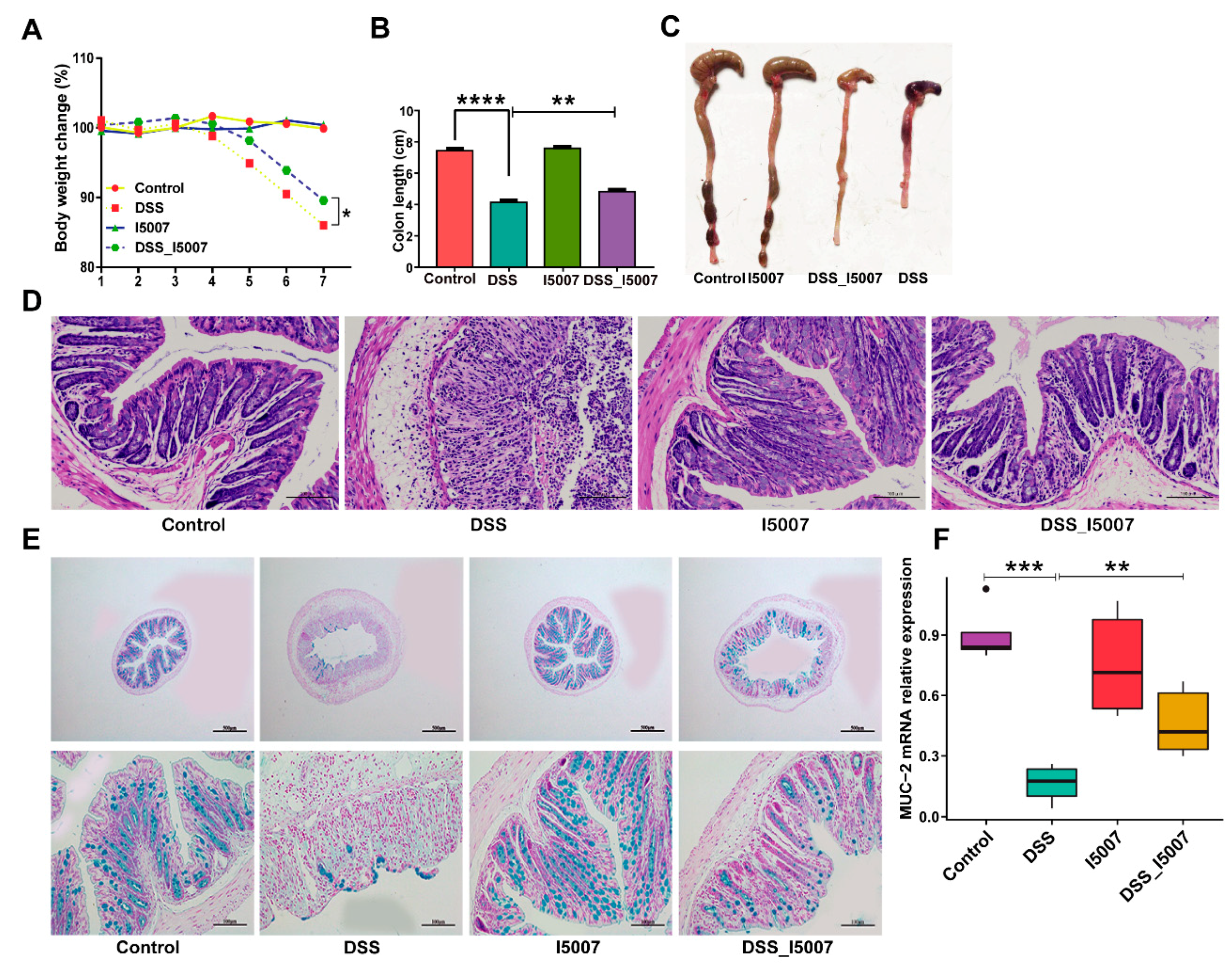
3.2. Effects of L. reuteri I5007 on Colitis Symptoms
3.3. Effects of L. reuteri I5007 on Cytokines Expression in Colon Tissue and Serum
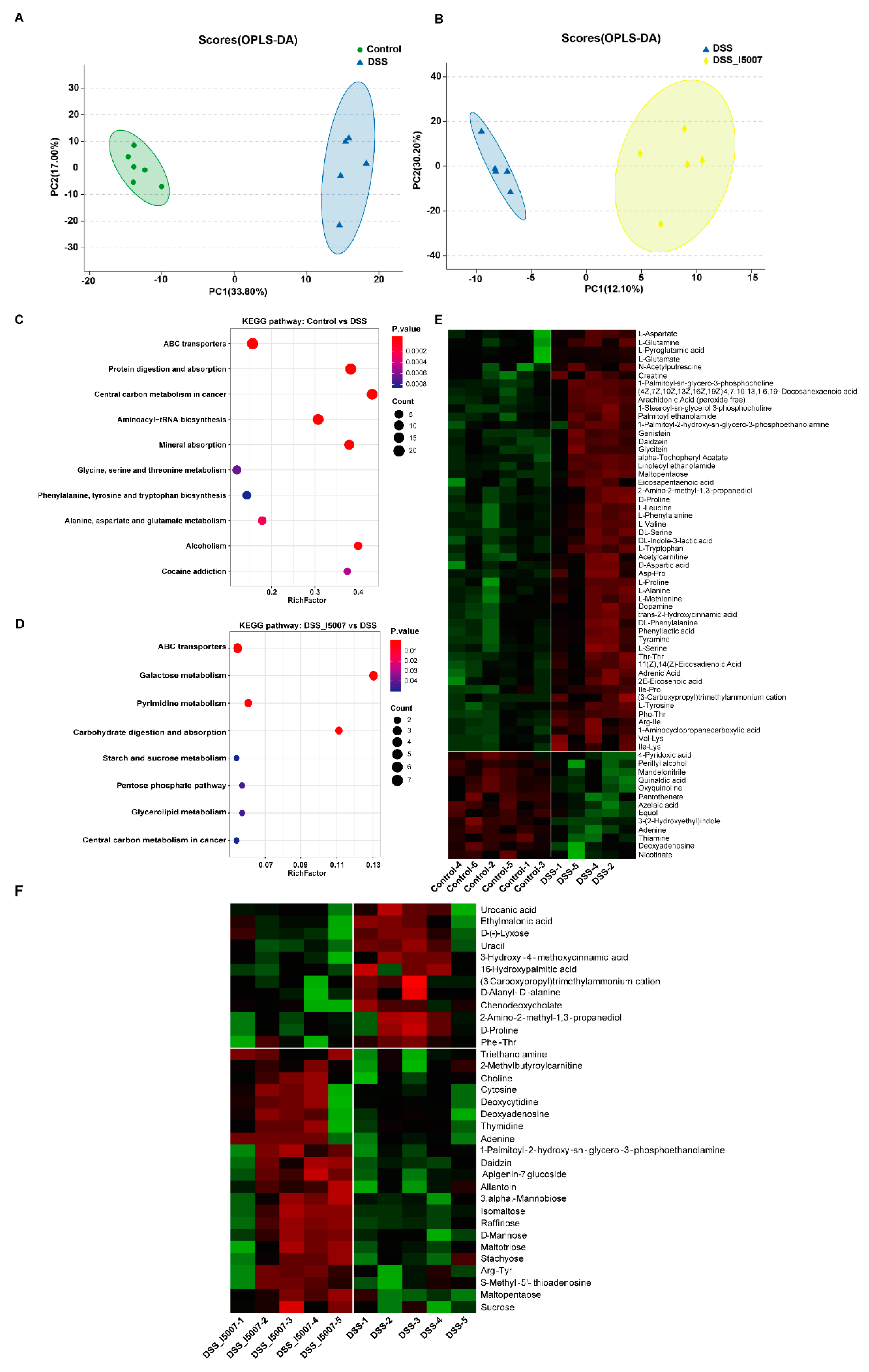
3.4. Effects of L. reuteri I5007 on Microbiota Composition in Colon Contents
3.5. Effects of L. reuteri I5007 on Microbiota Functional Composition in Colonic Contents
3.6. Effects of L. reuteri I5007 Metabolite Composition in Colonic Contents
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| IFN-γ | Interferon-gamma |
| TNF-α | Tumour necrosis factor-alpha |
| IL-6 | Interleukin-6 |
| IL-1 | Interleukin-10 |
| IL-17A | Interleukin-17A |
| IL-17F | Interleukin-17F |
| IL-22 | Interleukin-22 |
| IL-21 | Interleukin-21 |
| LPS | Lipopolysaccharides |
| PcoA | Principal coordinate analysis |
| LefSe | Linear discriminant analysis effect size |
References
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.Y.; Chan, F.K.L.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet 2018, 390, 2769–2778. [Google Scholar] [CrossRef]
- Moradkhani, A.; Beckman, L.J.; Tabibian, J.H. Health-related quality of life in inflammatory bowel disease: Psychosocial, clinical, socioeconomic, and demographic predictors. J. Crohn’s Colitis 2013, 7, 467–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, L.E.; Sturino, J.M.; Carroll, R.J.; Rooney, L.W.; Azcarate-Peril, M.A.; Turner, N.D. Polyphenol-rich sorghum brans alter colon microbiota and impact species diversity and species richness after multiple bouts of dextran sodium sulfate-induced colitis. FEMS Microbiol. Ecol. 2015, 91, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tremaroli, V.; Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Manichanh, C.; Borruel, N.; Casellas, F.; Guarner, F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 599–608. [Google Scholar] [CrossRef]
- Nagalingam, N.A.; Lynch, S.V. Role of the microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 2012, 18, 968–984. [Google Scholar] [CrossRef]
- Goyal, N.; Shukla, G. Probiotic Lactobacillus rhamnosus GG modulates the mucosal immune response in Giardia intestinalis-infected BALB/c mice. Dig. Dis. Sci. 2013, 58, 1218–1225. [Google Scholar] [CrossRef]
- Llopis, M.; Antolin, M.; Carol, M.; Borruel, N.; Casellas, F.; Martinez, C.; Espín-Basany, E.; Guarner, F.; Malagelada, J.-R. Lactobacillus casei downregulates commensals’ inflammatory signals in Crohn’s disease mucosa. Inflamm. Bowel Dis. 2009, 15, 275–283. [Google Scholar] [CrossRef]
- Gionchetti, P.; Rizzello, F.; Venturi, A.; Brigidi, P.; Matteuzzi, D.; Bazzocchi, G.; Poggioli, G.; Miglioli, M.; Campieri, M. Oral bacteriotherapy as maintenance treatment in patients with chronic pouchitis: A double-blind, placebo-controlled trial. Gastroenterology 2000, 119, 305–309. [Google Scholar] [CrossRef]
- Grimm, V.; Riedel, C.U. Manipulation of the Microbiota Using Probiotics. Adv. Exp. Med. Biol. 2016, 902, 109–117. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mennigen, R.; Nolte, K.; Rijcken, E.; Utech, M.; Bruewer, M. Probiotic mixture VSL#3 protects the epithelial barrier by maintaining tight junction protein expression and preventing apoptosis in a murine model of colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2009, 296, G1140–G1149. [Google Scholar] [CrossRef] [PubMed]
- Miele, E.; Pascarella, F.; Giannetti, E.; Quaglietta, L.; Baldassano, R.N.; Staiano, A. Effect of a probiotic preparation (VSL#3) on induction and maintenance of remission in children with ulcerative colitis. Am. J. Gastroenterol. 2009, 104, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Geier, M.S.; Butler, R.N.; Giffard, P.M.; Howarth, G.S. Lactobacillus fermentum BR11, a potential new probiotic, alleviates symptoms of colitis induced by dextran sulfate sodium (DSS) in rats. Int. J. Food Microbiol. 2007, 114, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Bron, P.A.; Kleerebezem, M.; Brummer, R.J.; Cani, P.D.; Mercenier, A.; Macdonald, T.T.; Garcia-Ródenas, C.L.; Wells, J.M. Can probiotics modulate human disease by impacting intestinal barrier function? Br. J. Nutr. 2017, 117, 93–107. [Google Scholar] [CrossRef]
- Valeur, N.; Engel, P.; Carbajal, N.; Connolly, E.; Ladefoged, K. Colonization and immunomodulation by Lactobacillus reuteri ATCC 55730 in the human gastrointestinal tract. Appl. Environ. Microbiol. 2004, 70, 1176–1181. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Fatheree, N.Y.; Mangalat, N.; Rhoads, J.M. Human-derived probiotic Lactobacillus reuteri strains differentially reduce intestinal inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G1087–G1096. [Google Scholar] [CrossRef] [Green Version]
- Thomas, C.M.; Hong, T.; Pijkeren, J.P.V.; Hemarajata, P.; Dan, V.T.; Hu, W.; Britton, R.A.; Kalkum, M.; Versalovic, J. Histamine Derived from Probiotic Lactobacillus reuteri Suppresses TNF via Modulation of PKA and ERK Signaling. PLoS ONE 2012, 7, e31951. [Google Scholar] [CrossRef] [Green Version]
- Preidis, G.A.; Saulnier, D.M.; Blutt, S.E.; Mistretta, T.-A.; Riehle, K.P.; Major, A.M.; Venable, S.F.; Barrish, J.P.; Finegold, M.J.; Petrosino, J.F.; et al. Host response to probiotics determined by nutritional status of rotavirus-infected neonatal mice. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 299–307. [Google Scholar] [CrossRef] [Green Version]
- Hou, C.; Wang, Q.; Zeng, X.; Yang, F.; Zhang, J.; Liu, H.; Ma, X.; Qiao, S. Complete genome sequence of Lactobacillus reuteri I5007, a probiotic strain isolated from healthy piglet. J. Biotechnol. 2014, 179, 63–64. [Google Scholar] [CrossRef]
- Wang, X.; Yang, F.; Liu, C.; Zhou, H.; Wu, G.; Qiao, S.; Li, D.; Wang, J. Dietary supplementation with the probiotic Lactobacillus fermentum I5007 and the antibiotic aureomycin differentially affects the small intestinal proteomes of weanling piglets. J. Nutr. 2012, 142, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Li, X.J.; Yue, L.Y.; Guan, X.F.; Qiao, S.Y. The adhesion of putative probiotic lactobacilli to cultured epithelial cells and porcine intestinal mucus. J. Appl. Microbiol. 2008, 104, 1082–1091. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.; Liu, H.; Zhang, J.; Zhang, S.; Yang, F.; Zeng, X.; Thacker, P.A.; Zhang, G.; Qiao, S. Intestinal Microbiota Succession and Immunomodulatory Consequences after Introduction of Lactobacillus reuteri I5007 in Neonatal Piglets. PLoS ONE 2015, 10, e0119505. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Hou, C.; Wang, G.; Jia, H.; Yu, H.; Zeng, X.; Thacker, P.A.; Zhang, G.; Qiao, S. Lactobacillus reuteri I5007 Modulates Intestinal Host Defense Peptide Expression in the Model of IPEC-J2 Cells and Neonatal Piglets. Nutrients 2017, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; Mcdonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Philip, H.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [Green Version]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef] [Green Version]
- Peran, L.; Sierra, S.; Comalada, M.; Lara-Villoslada, F.; Bailón, E.; Nieto, A.; Concha, Á.; Olivares, M.; Zarzuelo, A.; Xaus, J. A comparative study of the preventative effects exerted by two probiotics, Lactobacillus reuteri and Lactobacillus fermentum, in the trinitrobenzenesulfonic acid model of rat colitis. Br. J. Nutr. 2007, 97, 96. [Google Scholar] [CrossRef] [Green Version]
- Madsen, K.L.; Doyle, J.S.; Jewell, L.D.; Tavernini, M.M.; Fedorak, R.N. Lactobacillus species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology 1999, 116, 1107–1114. [Google Scholar] [CrossRef]
- Egger, B.; Bajaj-Elliott, M.; MacDonald, T.T.; Inglin, R.; Eysselein, V.E.; Büchler, M.W. Characterisation of acute murine dextran sodium sulphate colitis: Cytokine profile and dose dependency. Digestion 2000, 62, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Biasi, F.; Astegiano, M.; Maina, M.; Leonarduzzi, G.; Poli, G. Polyphenol supplementation as a complementary medicinal approach to treating inflammatory bowel disease. Curr. Med. Chem. 2011, 18, 4851–4865. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couper, K.N.; Blount, D.G.; Riley, E.M. IL-10: The Master Regulator of Immunity to Infection. J. Immunol. 2008, 180, 5771–5777. [Google Scholar] [CrossRef] [PubMed]
- Petersson, J.; Schreiber, O.; Hansson, G.C.; Gendler, S.J.; Velcich, A.; Lundberg, J.O.; Roos, S.; Holm, L.; Phillipson, M. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G327. [Google Scholar] [CrossRef] [Green Version]
- Dicksved, J.; Schreiber, O.; Willing, B.; Petersson, J.; Rang, S.; Phillipson, M.; Holm, L.; Roos, S. Lactobacillus reuteri maintains a functional mucosal barrier during DSS treatment despite mucus layer dysfunction. PLoS ONE 2012, 7, e46399. [Google Scholar] [CrossRef] [Green Version]
- Johansson, M.E.V.; Phillipson, M.; Petersson, J.; Velcich, A.; Holm, L.; Hansson, G.C. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 15064–15069. [Google Scholar] [CrossRef] [Green Version]
- Gersemann, M.; Becker, S.; Kübler, I.; Koslowski, M.; Wang, G.; Herrlinger, K.R.; Griger, J.; Fritz, P.; Fellermann, K.; Schwab, M.; et al. Differences in goblet cell differentiation between Crohn’s disease and ulcerative colitis. Differentiation 2009, 77, 84–94. [Google Scholar] [CrossRef]
- Yan, F.; Cao, H.; Cover, T.L.; Washington, M.K.; Shi, Y.; Liu, L.; Chaturvedi, R.; Peek, R.M.; Wilson, K.T.; Polk, D.B. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J. Clin. Investig. 2011, 121, 2242–2253. [Google Scholar] [CrossRef] [Green Version]
- Lorenza, P.; Federica, D.C.; Pamela, V.; Michele, C.; Salvatore, C.; Bruno, D.; Group, D.S. Gut Microbiota Dysbiosis as Risk and Premorbid Factors of IBD and IBS Along the Childhood–Adulthood Transition. Inflamm. Bowel Dis. 2015, 22, 487–504. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, B.; Delgado, S.; Blanco-Míguez, A.; Lourenço, A.; Gueimonde, M.; Margolles, A. Probiotics, gut microbiota and their influence on host health and disease. Mol. Nutr. Food Res. 2016. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.-C.; Zhang, F.-C.; Yin, X.; Cheng, B.-J.; Zhao, C.-H.; Wang, Y.-L.; Zhang, Z.-Z.; Hao, H.-W.; Zhang, T.-H.; Ye, H.-Q. Lactobacillus reuteri F-9-35 Prevents DSS-Induced Colitis by Inhibiting Proinflammatory Gene Expression and Restoring the Gut Microbiota in Mice. J. Food Sci. 2018, 83, 2645–2652. [Google Scholar] [CrossRef] [PubMed]
- Imhann, F.; Vich Vila, A.; Jan Bonder, M. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2016, 67, 108–119. [Google Scholar] [CrossRef]
- Zhang, Q.; Wu, Y.; Wang, J.; Wu, G.; Long, W.; Xue, Z.; Wang, L.; Zhang, X.; Pang, X.; Zhao, Y.; et al. Accelerated dysbiosis of gut microbiota during aggravation of DSS-induced colitis by a butyrate-producing bacterium. Sci. Rep. 2016, 6, 27572. [Google Scholar] [CrossRef]
- Jacobs, J.P.; Goudarzi, M.; Singh, N.; Tong, M.; Mchardy, I.H.; Ruegger, P.; Asadourian, M.; Moon, B.H.; Ayson, A.; Borneman, J. A Disease-Associated Microbial and Metabolomics State in Relatives of Pediatric Inflammatory Bowel Disease Patients. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 750–766. [Google Scholar] [CrossRef] [Green Version]
- Sokol, H.; Seksik, P. The intestinal microbiota in inflammatory bowel diseases: Time to connect with the host. Curr. Opin. Gastroenterol. 2010, 26, 327–331. [Google Scholar] [CrossRef]
- Lupp, C.; Robertson, M.L.; Wickham, M.E.; Sekirov, I.; Champion, O.L.; Gaynor, E.C.; Finlay, B.B. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2007, 2, 204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokol, H.; Lepage, P.; Seksik, P.; Doré, J.; Marteau, P. Temperature Gradient Gel Electrophoresis of Fecal 16S rRNA Reveals Active Escherichia coli in the Microbiota of Patients with Ulcerative Colitis. J. Clin. Microbiol. 2006, 44, 3172–3177. [Google Scholar] [CrossRef] [Green Version]
- Darfeuille-Michaud, A.; Boudeau, J.; Bulois, P.; Neut, C.; Glasser, A.-L.; Barnich, N.; Bringer, M.-A.; Swidsinski, A.; Beaugerie, L.; Colombel, J.-F. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 2004, 127, 412–421. [Google Scholar] [CrossRef]
- Frank, D.N.; Amand, A.L., St.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia Vilela, E.; De Lourdes De Abreu Ferrari, M.; Oswaldo Da Gama Torres, H.; Guerra Pinto, A.; Carolina Carneiro Aguirre, A.; Paiva Martins, F.; Marcos Andrade Goulart, E.; Sales Da Cunha, A. Influence of Saccharomyces boulardii on the intestinal permeability of patients with Crohn’s disease in remission. Scand. J. Gastroenterol. 2008, 43, 842–848. [Google Scholar] [CrossRef] [PubMed]
- Ljungh, A.; Wadström, T. Lactic acid bacteria as probiotics. Curr. Issues Intest. Microbiol. 2006, 7, 73–89. [Google Scholar] [PubMed]
- Kristensen, N.B.; Bryrup, T.; Allin, K.H.; Nielsen, T.; Hansen, T.H.; Pedersen, O. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: A systematic review of randomized controlled trials. Genome Med. 2016, 8, 52. [Google Scholar] [CrossRef] [Green Version]
- Oz, H.S.; Chen, T.S.; Nagasawa, H. Comparative efficacies of 2 cysteine prodrugs and a glutathione delivery agent in a colitis model. Transl. Res. 2007, 150, 122–129. [Google Scholar] [CrossRef] [Green Version]
- Son, M.; Ko, J.I.; Kim, W.B.; Kang, H.K.; Kim, B.K. Taurine Can Ameliorate Inflammatory Bowel Disease in Rats. Adv Exp Med Biol. 1998, 442, 291–298. [Google Scholar] [CrossRef]
- Ni, J.; Shen, T.C.D.; Chen, E.Z.; Bittinger, K.; Bailey, A.; Roggiani, M.; Sirota-Madi, A.; Friedman, E.S.; Chau, L.; Lin, A. A role for bacterial urease in gut dysbiosis and Crohn’s disease. Sci. Transl. Med. 2017, 9, eaah6888. [Google Scholar] [CrossRef] [Green Version]
- Sugihara, K.; Morhardt, T.L.; Kamada, N. The Role of Dietary Nutrients in Inflammatory Bowel Disease. Front. Immunol. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef] [Green Version]
- Kolho, K.-L.; Pessia, A.; Jaakkola, T.; de Vos, W.M.; Velagapudi, V. Faecal and Serum Metabolomics in Paediatric Inflammatory Bowel Disease. J. Crohns Colitis 2017, 11, 321–334. [Google Scholar] [CrossRef]
- Bosch, S.; Struys, E.A.; van Gaal, N.; Bakkali, A.; Jansen, E.W.; Diederen, K.; Benninga, M.A.; Mulder, C.J.; de Boer, N.K.H.; de Meij, T.G.J. Fecal Amino Acid Analysis Can Discriminate De Novo Treatment-Naïve Pediatric Inflammatory Bowel Disease From Controls. J. Pediatr. Gastroenterol. Nutr. 2018, 66, 773–778. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, X.; Hu, C.-A.A. Therapeutic Potential of Amino Acids in Inflammatory Bowel Disease. Nutrients 2017, 9, 920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorburn, A.; Macia, L.; Mackay, C. Diet, Metabolites, and “Western-Lifestyle” Inflammatory Diseases. Immunity 2014, 40, 833–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.; Manicassamy, S.; Munn, D. Activation of Gpr109a, Receptor for Niacin and the Commensal Metabolite Butyrate, Suppresses Colonic Inflammation and Carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [Green Version]
- Tao, J.H.; Duan, J.A.; Wei, Z.; Shu, J.; Guo, J.M.; Wei, D.D. Polysaccharides From Chrysanthemum morifolium Ramat Ameliorate Colitis Rats via Regulation of the Metabolic Profiling and NF-κ B/TLR4 and IL-6/JAK2/STAT3 Signaling Pathways. Front. Pharmacol. 2018, 9, 746. [Google Scholar] [CrossRef]
- Langmann, T.; Moehle, C.; Mauerer, R.; Scharl, M.; Liebisch, G.; Zahn, A.; Stremmel, W.; Schmitz, G. Loss of detoxification in inflammatory bowel disease: Dysregulation of pregnane X receptor target genes. Gastroenterology 2004, 127, 26–40. [Google Scholar] [CrossRef]




| Target Genes | Forward (F)/Reverse (R) | Sequence | Size (bp) |
|---|---|---|---|
| M-GAPDH | F | AACTTTGGCATTGTGGAAGG | 115 |
| R | ACACATTGGGGGTAGGAACA | ||
| M-IL-6 | F | AGACTTCCATCCAGTTGCCT | 175 |
| R | CATTTCCACGATTTCCCAGAGA | ||
| M-IL-1β | F | TCAGGCAGGCAGTATCACTC | 250 |
| R | AGCTCATATGGGTCCGACAG | ||
| M-IFN-γ | F | ACTGGCAAAAGGATGGTGAC | 237 |
| R | TGAGCTCATTGAATGCTTGG | ||
| M-TNF-α | F | ACCCTCACACTCACAAACCA | 246 |
| R | GGCAGAGAGGAGGTTGACTT | ||
| M-IL-17A | F | GACTACCTCAACCGTTCCAC | 118 |
| R | CCTCCGCATTGACACAGC | ||
| M-Muc-2 | F | GACTGCCGAGACTCCTACAA | 129 |
| R | CTTGTGGGTGAGGTAGATGG | ||
| H-IL-8 | F | TGGCTCTCTTGGCAGTC | 238 |
| R | TGCACCCAGTTTTCCTTGGG | ||
| H-TNF-α | F | AGCCCATGTTGTAGCAAACC | 134 |
| R | TGAGGTACAGGCCCTCTGAT | ||
| H-IL-1β | F | ATGATGGCTTATTACAGTGGCAA | 132 |
| R | GTCGGAGATTCGTAGCTGGA | ||
| H-IL-10 | F | AAAGAAGGCATGCACAGCTC | 132 |
| R | AAGCATGTTAGGCAGGTTGC | ||
| H-GAPDH | F | CAACGACCACTTTGTCAAGC | 140 |
| R | TTCCTCTTGTGCTCTTGCTG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Huang, S.; Cai, S.; Yu, H.; Wang, Y.; Zeng, X.; Qiao, S. Lactobacillus reuteri Ameliorates Intestinal Inflammation and Modulates Gut Microbiota and Metabolic Disorders in Dextran Sulfate Sodium-Induced Colitis in Mice. Nutrients 2020, 12, 2298. https://doi.org/10.3390/nu12082298
Wang G, Huang S, Cai S, Yu H, Wang Y, Zeng X, Qiao S. Lactobacillus reuteri Ameliorates Intestinal Inflammation and Modulates Gut Microbiota and Metabolic Disorders in Dextran Sulfate Sodium-Induced Colitis in Mice. Nutrients. 2020; 12(8):2298. https://doi.org/10.3390/nu12082298
Chicago/Turabian StyleWang, Gang, Shuo Huang, Shuang Cai, Haitao Yu, Yuming Wang, Xiangfang Zeng, and Shiyan Qiao. 2020. "Lactobacillus reuteri Ameliorates Intestinal Inflammation and Modulates Gut Microbiota and Metabolic Disorders in Dextran Sulfate Sodium-Induced Colitis in Mice" Nutrients 12, no. 8: 2298. https://doi.org/10.3390/nu12082298
APA StyleWang, G., Huang, S., Cai, S., Yu, H., Wang, Y., Zeng, X., & Qiao, S. (2020). Lactobacillus reuteri Ameliorates Intestinal Inflammation and Modulates Gut Microbiota and Metabolic Disorders in Dextran Sulfate Sodium-Induced Colitis in Mice. Nutrients, 12(8), 2298. https://doi.org/10.3390/nu12082298