Altered Gut Microbial Metabolites in Amnestic Mild Cognitive Impairment and Alzheimer’s Disease: Signals in Host–Microbe Interplay
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Fecal Metabolomics Analysis
2.2.1. Sample Preparation
2.2.2. Fecal Untargeted Metabolomics Profiling
2.2.3. Multivariate Data Analysis
2.3. Targeted Profiling of Fecal Tryptophan Metabolites
2.4. Targeted Profiling of Fecal Short-Chain Fatty Acids (SCFAs)
2.5. Targeted Profiling of Fecal Bile Acids
2.6. Measurement of Circulating Lipopolysaccharide Level
2.7. Statistical Analysis
3. Results
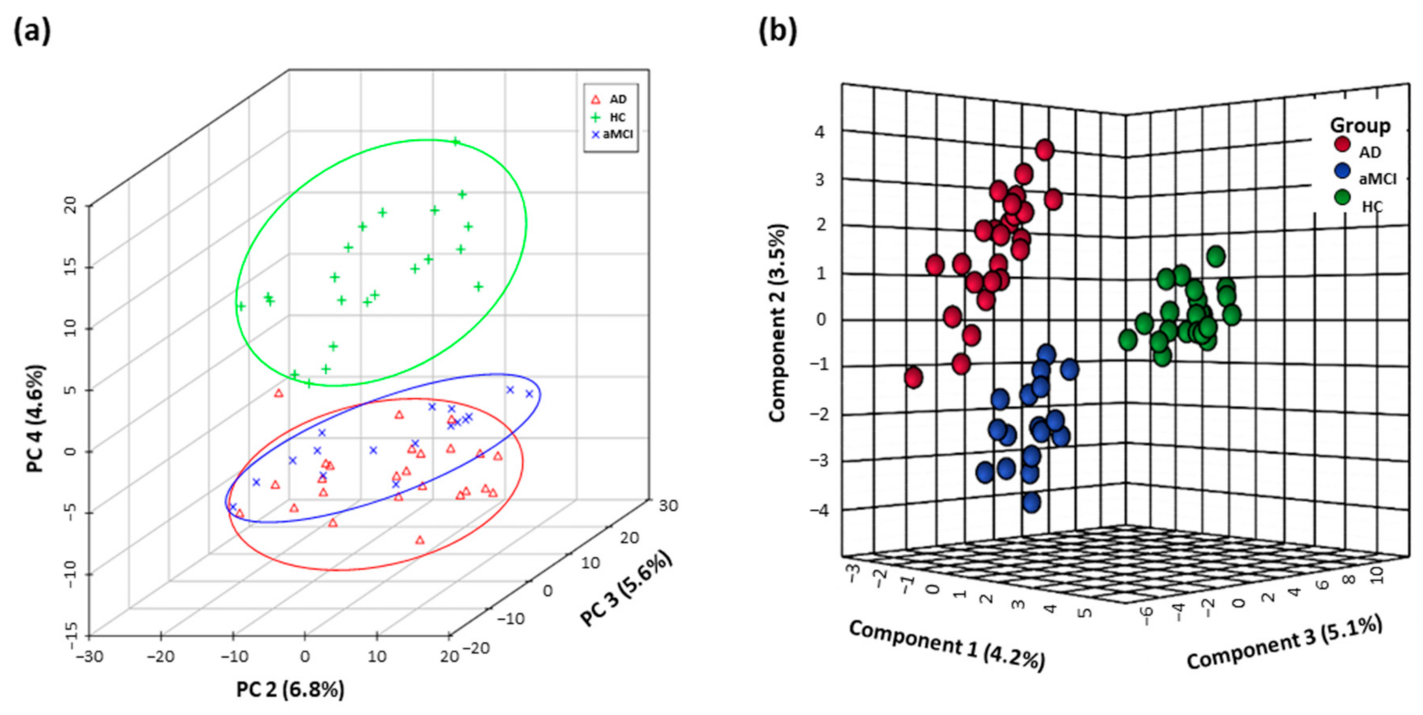
3.1. Distinguishing Metabolomics Profiles among AD, aMCI, and HC
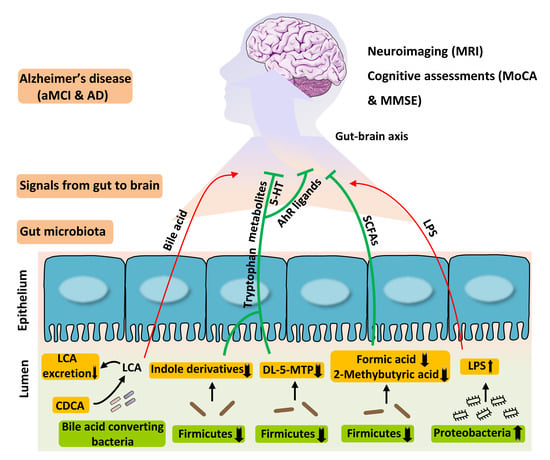
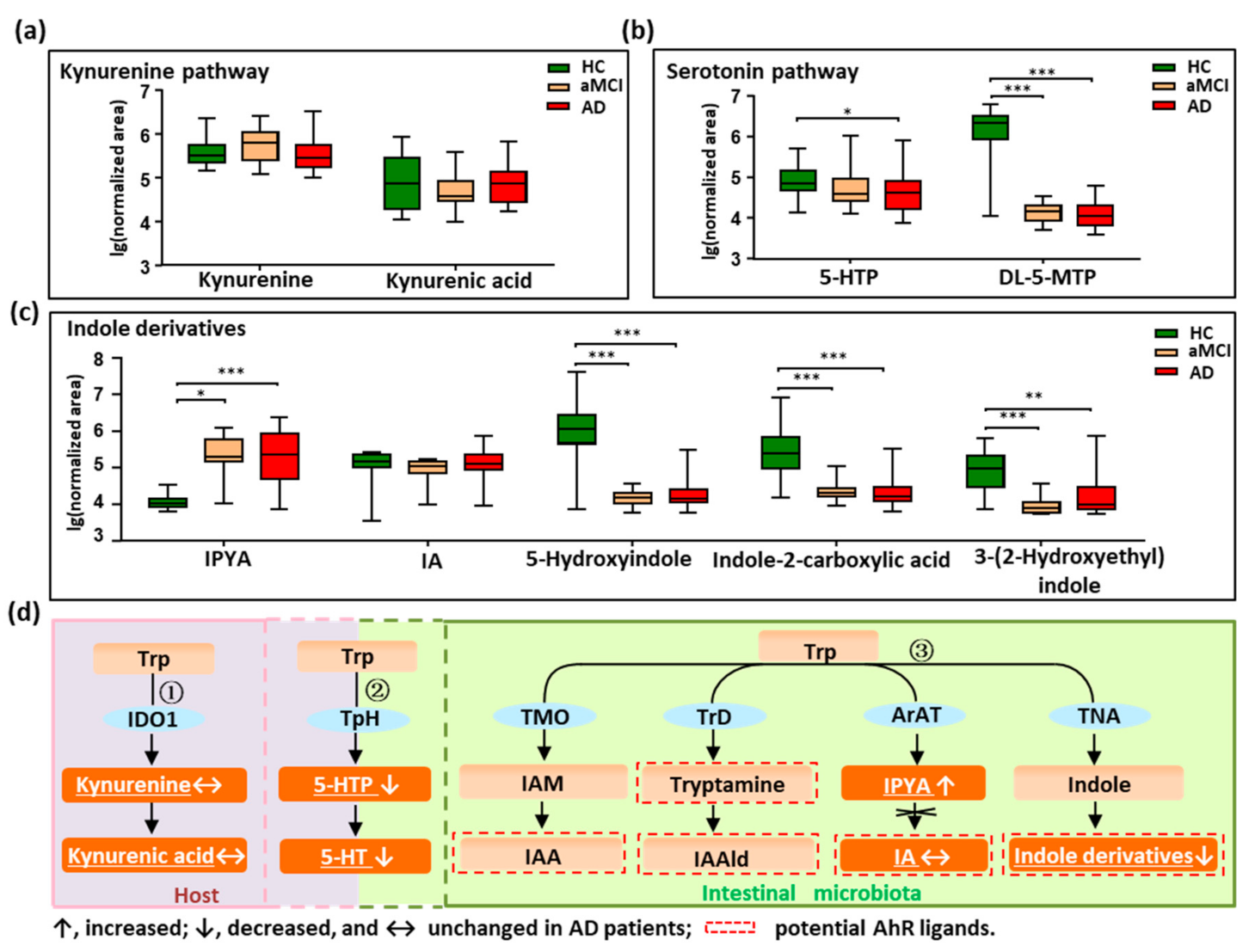
3.2. Disturbed Metabolic Pathways of Tryptophan in Patients with AD
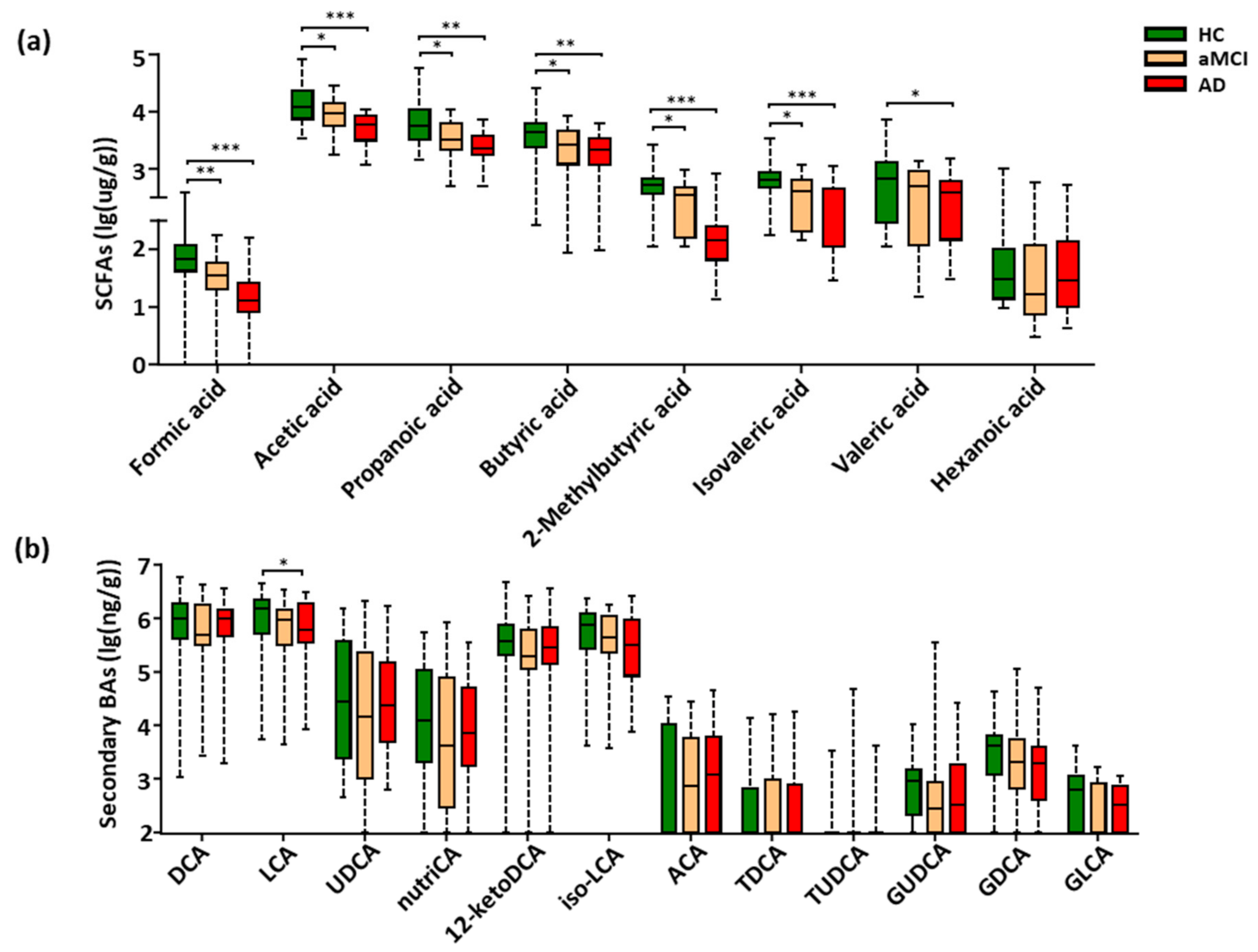
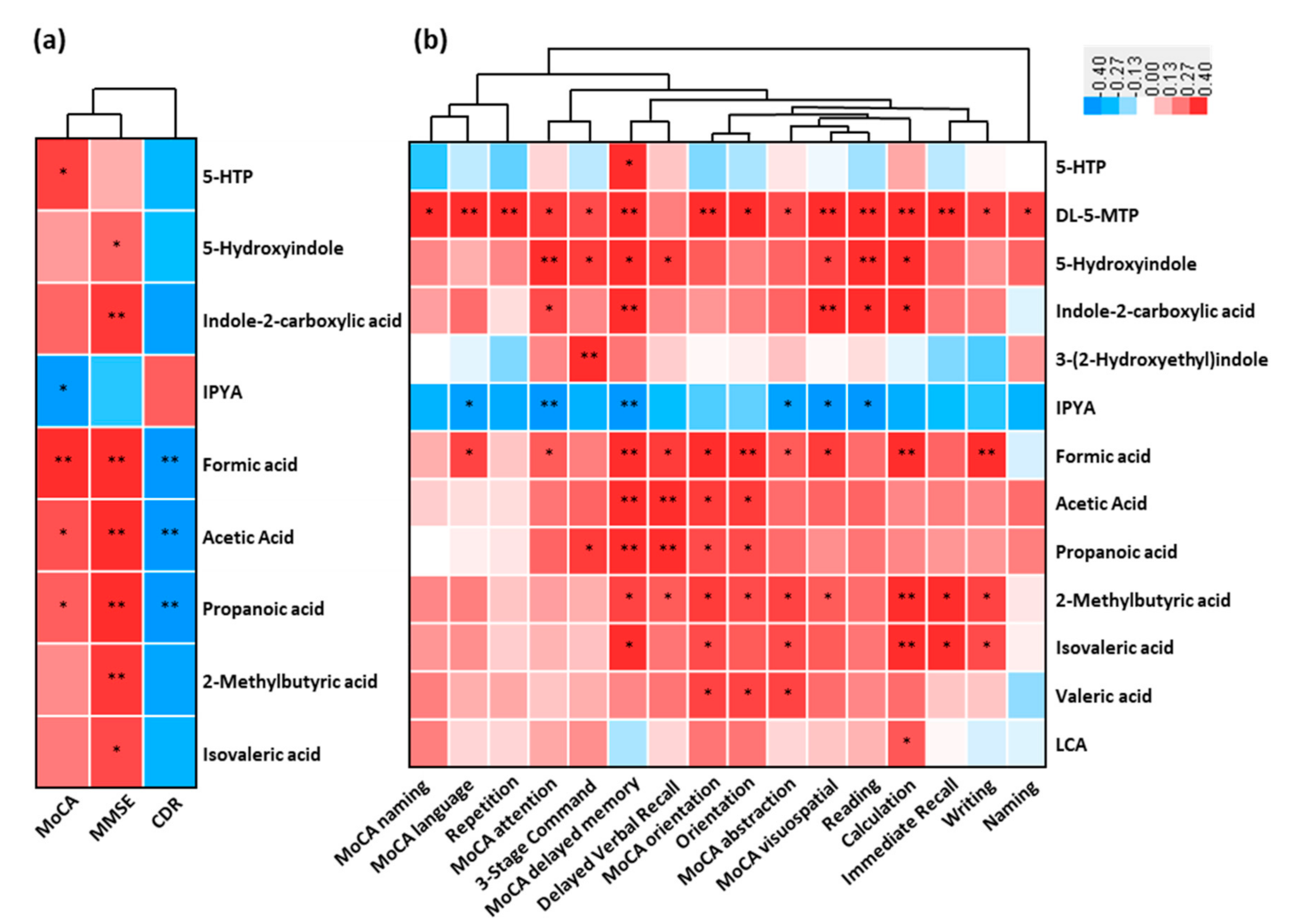
3.3. Fecal SCFAs Associated with Progression from HC to aMCI and AD
3.4. Perturbation of Fecal Bile Acids in AD Patients
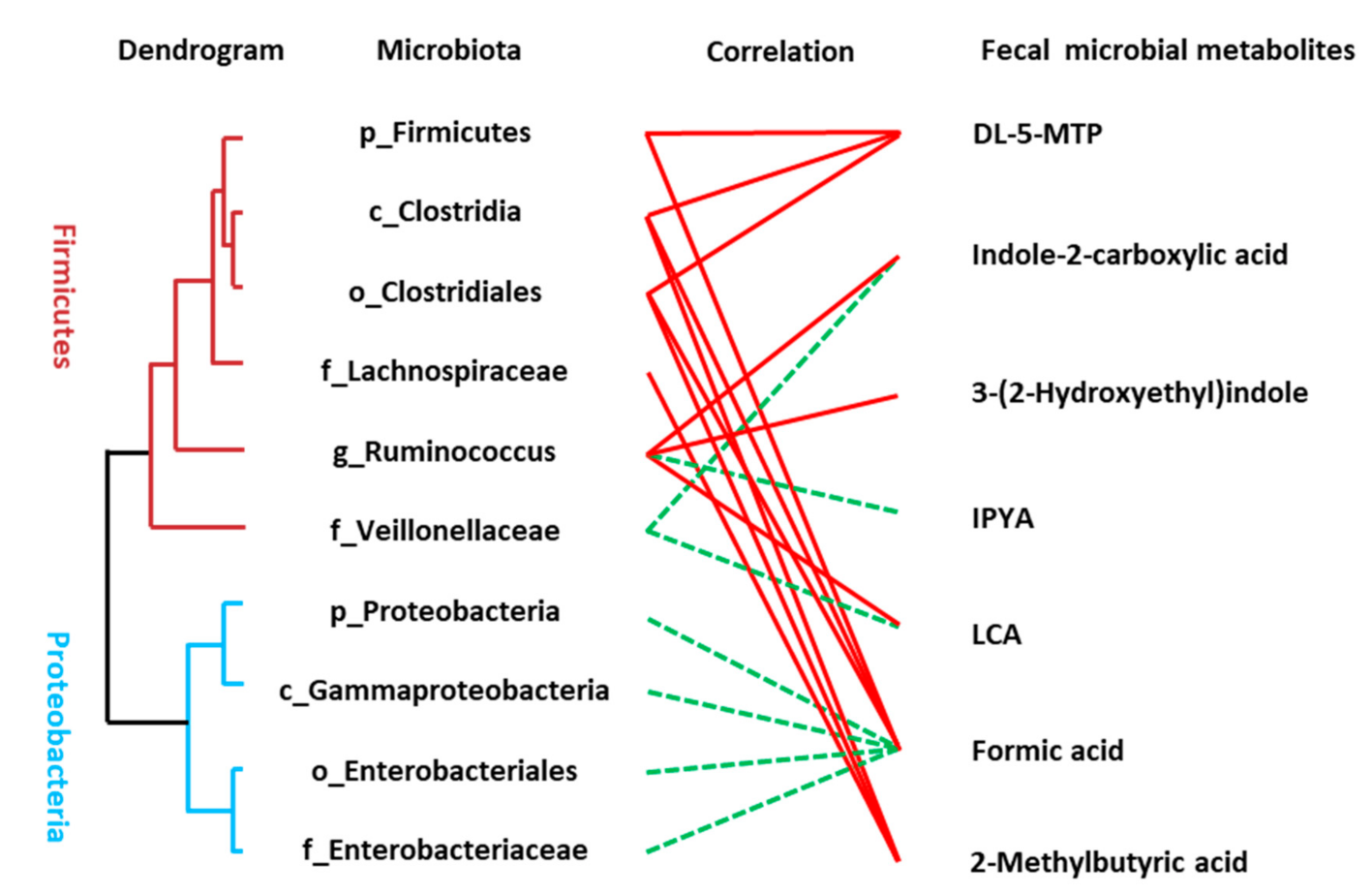
3.5. Association between the Disturbed Microbes of AD and Fecal Metabolites
3.6. Serum Levels of Endotoxin Prone to Increase in AD
3.7. Association between the Altered Metabolites of AD and Cognitive Impairment
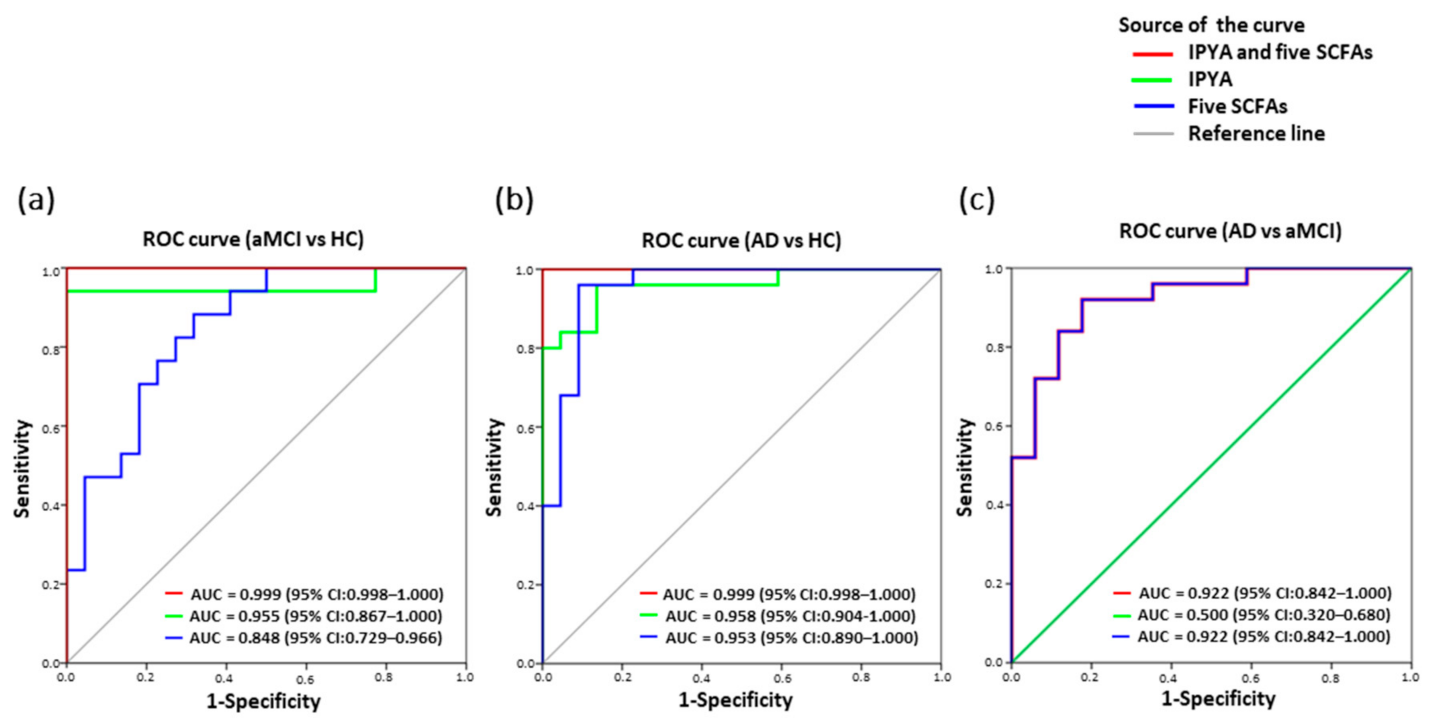
3.8. Classification of AD from aMCI and HC by Fecal Microbial Signatures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sperling, R.A.; Aisen, P.S.; Beckett, L.A.; Bennett, D.A.; Craft, S.; Fagan, A.M.; Iwatsubo, T.; Jack, C.R., Jr.; Kaye, J.; Montine, T.J.; et al. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2011, 7, 280–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Australia, D.; Baker, S.; Banerjee, S. Alzheimer’s Disease International. World Alzheimer Report 2019: Attitudes to Dementia; Alzheimer’s Disease International: London, UK, 2019. [Google Scholar]
- Alzheimer’s Association. 2019 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2019, 15, 321–387. [CrossRef]
- Levey, A.I.; Lah, J.; Goldstein, F.; Steenland, K.; Bliwise, D. Mild cognitive impairment: An opportunity to identify patients at high risk for progression to Alzheimer’s disease. Clin. Ther. 2006, 28, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Canteli, M.; Iadecola, C. Alzheimer’s Disease and Vascular Aging: JACC Focus Seminar. J. Am. Coll. Cardiol. 2020, 75, 942–951. [Google Scholar] [CrossRef]
- Busche, M.A.; Hyman, B.T. Synergy between amyloid-β and tau in Alzheimer’s disease. Nat. Neurosci. 2020, 23, 1183–1193. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef] [Green Version]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef]
- Vogt, N.M.; Kerby, R.L.; Dill-McFarland, K.A. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 2017, 7, 13537. [Google Scholar] [CrossRef]
- Liu, P.; Wu, L.; Peng, G.; Han, Y.; Tang, R.; Ge, J.; Zhang, L.; Jia, L.; Yue, S.; Zhou, K.; et al. Altered microbiomes distinguish Alzheimer’s disease from amnestic mild cognitive impairment and health in a Chinese cohort. Brain Behav. Immun. 2019, 80, 633–643. [Google Scholar] [CrossRef]
- Harach, T.; Marungruang, N.; Duthilleul, N.; Cheatham, V.; Mc Coy, K.D.; Frisoni, G.; Neher, J.; Fåk, F.; Jucker, M.; Lasser, T.; et al. Reduction of Abeta amyloid pathology in APPPS1 transgenic mice in the absence of gut microbiota. Sci. Rep. 2017, 7, 41802. [Google Scholar] [CrossRef]
- Zierer, J.; Jackson, M.A. The fecal metabolome as a functional readout of the gut microbiome. Nat. Genet. 2018, 50, 790–795. [Google Scholar] [CrossRef]
- Mahmoudiandehkordi, S.; Arnold, M.; Nho, K.; Ahmad, S.; Jia, W.; Xie, G.; Louie, G.; Kueider-Paisley, A.; Moseley, M.A.; Thompson, J.W.; et al. Altered bile acid profile associates with cognitive impairment in Alzheimer’s disease-An emerging role for gut microbiome. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2019, 15, 76–92. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Liu, X.; Wang, M.; Liu, L.; Sun, X.; Ma, L.; Xie, W.; Wang, C.; Tang, S.; Wang, D.; et al. Lysophosphatidylcholine and amide as metabolites for detecting alzheimer disease using ultrahigh-performance liquid chromatography-quadrupole time-of-flight mass spectrometry-based metabonomics. J. Neuropathol. Exp. Neurol. 2014, 73, 954–963. [Google Scholar] [CrossRef] [Green Version]
- Varma, V.R.; Oommen, A.; Varma, S.; Casanova, R.; An, Y.; Andrews, R.M.; O’Brien, R.; Pletnikova, O.; Troncoso, J.C.; Toledo, J.B.; et al. Brain and blood metabolite signatures of pathology and progression in Alzheimer disease: A targeted metabolomics study. PLoS Med. 2018, 15, e1002482. [Google Scholar] [CrossRef]
- Kaddurah-Daouk, R.; Rozen, S.; Matson, W.; Han, X.; Hulette, C.M.; Burke, J.R.; Doraiswamy, P.M.; Welsh-Bohmer, K.A. Metabolomic changes in autopsy-confirmed Alzheimer’s disease. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2011, 7, 309–317. [Google Scholar] [CrossRef] [Green Version]
- Diagnostic and Statistical Manual of Mental Disorders, 4th ed.; American Psychiatric Association: Washington, DC, USA, 1994; p. 886.
- Dubois, B.; Feldman, H.H.; Jacova, C.; DeKosky, S.T.; Barberger-Gateau, P.; Cummings, J.L.; Delacourte, A.; Galasko, D.; Gauthier, S.; Jicha, G.A.; et al. Research criteria for the diagnosis of Alzheimer’s disease: Revising the NINCDS-ADRDA criteria. Lancet Neurol. 2007, 6, 734–746. [Google Scholar] [CrossRef]
- Petersen, R.C. Mild cognitive impairment as a diagnostic entity. J. Intern. Med. 2004, 256, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, B.; Zhang, M.; Rantalainen, M.; Wang, S.; Zhou, H.; Zhang, Y.; Shen, J.; Pang, X.; Zhang, M.; et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl. Acad. Sci. USA 2008, 105, 2117–2122. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Chen, D.; Chen, Y.; Hu, Z.; Cao, M.; Xie, Q.; Chen, Y.; Xu, J.; Zheng, S.; Li, L. Metabonomic Profiles Discriminate Hepatocellular Carcinoma from Liver Cirrhosis by Ultraperformance Liquid Chromatography–Mass Spectrometry. J. Proteome Res. 2012, 11, 1217–1227. [Google Scholar] [CrossRef]
- Cao, H.; Huang, H.; Xu, W.; Chen, D.; Yu, J.; Li, J.; Li, L. Fecal metabolome profiling of liver cirrhosis and hepatocellular carcinoma patients by ultra performance liquid chromatography-mass spectrometry. Anal. Chim. Acta 2011, 691, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Simonato, M.; Fochi, I.; Vedovelli, L.; Giambelluca, S.; Carollo, C.; Padalino, M.; Carnielli, V.P.; Cogo, P. Urinary metabolomics reveals kynurenine pathway perturbation in newborns with transposition of great arteries after surgical repair. Metab. Off. J. Metab. Soc. 2019, 15, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Wang, W.; Lv, S.; Yin, P.; Zhao, X.; Lu, X.; Zhang, F.; Xu, G. Metabonomics study of liver cancer based on ultra performance liquid chromatography coupled to mass spectrometry with HILIC and RPLC separations. Anal. Chim. Acta 2009, 650, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Furuhashi, T.; Sugitate, K.; Nakai, T.; Jikumaru, Y.; Ishihara, G. Rapid profiling method for mammalian feces short chain fatty acids by GC-MS. Anal. Biochem. 2018, 543, 51–54. [Google Scholar] [CrossRef]
- Zheng, X.; Qiu, Y.; Zhong, W.; Baxter, S.; Su, M.; Li, Q.; Xie, G.; Ore, B.M.; Qiao, S.; Spencer, M.D.; et al. A targeted metabolomic protocol for short-chain fatty acids and branched-chain amino acids. Metab. Off. J. Metab. Soc. 2013, 9, 818–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, T.W.; Chen, H.-C.; Chen, C.-Y.; Yen, C.Y.T.; Lin, C.-J.; Prajnamitra, R.P.; Chen, L.-L.; Ruan, S.-C.; Lin, J.-H.; Lin, P.-J.; et al. Loss of Gut Microbiota Alters Immune System Composition and Cripples Postinfarction Cardiac Repair. Circulation 2019, 139, 647–659. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; An, Y.; Tang, H.; Wang, Y. Alterations of Bile Acids and Gut Microbiota in Obesity Induced by High Fat Diet in Rat Model. J. Agric. Food Chem. 2019, 67, 3624–3632. [Google Scholar] [CrossRef]
- Penno, C.A.; Arsenijevic, D.; Da Cunha, T.; Kullak-Ublick, G.A.; Montani, J.-P.; Odermatt, A. Quantification of multiple bile acids in uninephrectomized rats using ultra-performance liquid chromatography-tandem mass spectrometry. Anal. Methods 2013, 5, 1155–1164. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.Y.; Gillilland, M., 3rd; Wu, X.; Leelasinjaroen, P.; Zhang, G.; Zhou, H.; Ye, B.; Lu, Y.; Owyang, C. FODMAP diet modulates visceral nociception by lipopolysaccharide-mediated intestinal inflammation and barrier dysfunction. J. Clin. Investig. 2018, 128, 267–280. [Google Scholar] [CrossRef]
- De Hoon, M.J.L.; Imoto, S.; Nolan, J.; Miyano, S. Open source clustering software. Bioinformatics 2004, 20, 1453–1454. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Li, F.; Wang, L.; Zhao, W.; Li, D.; Xian, Y.; Jiang, X. A risk prediction model of sperm retrieval failure with fine needle aspiration in males with non-obstructive azoospermia. Hum. Reprod. 2019, 34, 200–208. [Google Scholar] [CrossRef] [Green Version]
- Tripodi, V.; Contin, M.; Fernández, M.A.; Lemberg, A. Bile acids content in brain of common duct ligated rats. Ann. Hepatol. 2012, 11, 930–934. [Google Scholar] [CrossRef]
- Van Dyke, R.W.; Stephens, J.E.; Scharschmidt, B.F. Bile acid transport in cultured rat hepatocytes. Am. J. Physiol. 1982, 243, G484–G492. [Google Scholar] [CrossRef] [PubMed]
- Krähenbühl, S.; Talos, C.; Fischer, S.; Reichen, J. Toxicity of bile acids on the electron transport chain of isolated rat liver mitochondria. Hepatology 1994, 19, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Chi, H. Metabolic control of regulatory T cell development and function. Trends Immunol. 2015, 36, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Alkasir, R.; Li, J.; Li, X.; Jin, M.; Zhu, B. Human gut microbiota: The links with dementia development. Protein Cell 2017, 8, 90–102. [Google Scholar] [CrossRef] [Green Version]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [Green Version]
- Sparkman, N.L.; Buchanan, J.B.; Heyen, J.R.R.; Chen, J.; Beverly, J.L.; Johnson, R.W. Interleukin-6 facilitates lipopolysaccharide-induced disruption in working memory and expression of other proinflammatory cytokines in hippocampal neuronal cell layers. J. Neurosci. Off. J. Soc. Neurosci. 2006, 26, 10709–10716. [Google Scholar] [CrossRef] [Green Version]
- de J.R. De-Paula, V.; Forlenza, A.S.; Forlenza, O.V. Relevance of gutmicrobiota in cognition, behaviour and Alzheimer’s disease. Pharmacol. Res. 2018, 136, 29–34. [Google Scholar] [CrossRef]
- Xu, Y.; Yan, J.; Zhou, P.; Li, J.; Gao, H.; Xia, Y.; Wang, Q. Neurotransmitter receptors and cognitive dysfunction in Alzheimer’s disease and Parkinson’s disease. Prog. Neurobiol. 2012, 97, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Ma, N.; He, T.; Johnston, L.J.; Ma, X. Host-microbiome interactions: The aryl hydrocarbon receptor as a critical node in tryptophan metabolites to brain signaling. Gut Microbes 2020, 11, 1203–1219. [Google Scholar] [CrossRef]
- Rothhammer, V.; Mascanfroni, I.D.; Bunse, L.; Takenaka, M.C.; Kenison, J.E.; Mayo, L.; Chao, C.-C.; Patel, B.; Yan, R.; Blain, M.; et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016, 22, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Braniste, V.; Al-Asmakh, M.; Kowal, C.; Anuar, F.; Abbaspour, A.; Tóth, M.; Korecka, A.; Bakocevic, N.; Ng, L.G.; Kundu, P.; et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 2014, 6, 263ra158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoyles, L.; Snelling, T.; Umlai, U.-K.; Nicholson, J.K.; Carding, S.R.; Glen, R.C.; McArthur, S. Microbiome-host systems interactions: Protective effects of propionate upon the blood-brain barrier. Microbiome 2018, 6, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huuskonen, J.; Suuronen, T.; Nuutinen, T.; Kyrylenko, S.; Salminen, A. Regulation of microglial inflammatory response by sodium butyrate and short-chain fatty acids. Br. J. Pharmacol. 2004, 141, 874–880. [Google Scholar] [CrossRef]
- Nagpal, R.; Neth, B.J.; Wang, S.; Craft, S.; Yadav, H. Modified Mediterranean-ketogenic diet modulates gut microbiome and short-chain fatty acids in association with Alzheimer’s disease markers in subjects with mild cognitive impairment. EBioMedicine 2019, 47, 529–542. [Google Scholar] [CrossRef] [Green Version]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef] [Green Version]
- Erny, D.; De Angelis, A.L.H.; Jaitin, D.; Wieghofer, P.; Staszewski, O.; David, E.; Keren-Shaul, H.; Mahlakoiv, T.; Jakobshagen, K.; Buch, T.; et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015, 18, 965–977. [Google Scholar] [CrossRef]
- Govindarajan, N.; Agis-Balboa, R.C.; Walter, J.; Sananbenesi, F.; Fischer, A. Sodium butyrate improves memory function in an Alzheimer’s disease mouse model when administered at an advanced stage of disease progression. J. Alzheimer’ s Dis. JAD 2011, 26, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Olazarán, J.; Gil-De-Gómez, L.; Rodríguez-Martín, A.; Valentí-Soler, M.; Frades-Payo, B.; Marín-Muñoz, J.; Antúnez, C.; Frank-García, A.; Acedo-Jiménez, C.; Morlán-Gracia, L.; et al. A blood-based, 7-metabolite signature for the early diagnosis of Alzheimer’s disease. J. Alzheimer’ s Dis. JAD 2015, 45, 1157–1173. [Google Scholar]
- Marksteiner, J.; Blasko, I.; Kemmler, G.; Koal, T.; Humpel, C. Bile acid quantification of 20 plasma metabolites identifies lithocholic acid as a putative biomarker in Alzheimer’s disease. Metab. Off. J. Metab. Soc. 2018, 14, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, X.; Elliott, C.T.; McGuinness, B.; Passmore, P.; Kehoe, P.G.; Holscher, C.; McClean, P.L.; Graham, S.F.; Green, B.D. Metabolomic Profiling of Bile Acids in Clinical and Experimental Samples of Alzheimer’s Disease. Metabolites 2017, 7, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, M.; Jiang, Y.; Zhao, Q.; Zhu, Z.; Liang, X.; Zhang, K.; Wu, W.; Dong, Q.; An, Y.; Tang, H.; et al. Metabolomics and incident dementia in older Chinese adults: The Shanghai Aging Study. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2020, 16, 779–788. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | HC | aMCI | AD |
|---|---|---|---|
| (n = 28) | (n = 22) | (n = 27) | |
| Age (years) | 74.25 ± 9.03 | 70.00 ± 11.33 | 74.15 ± 11.16 |
| Sex (male/female) | 14/14 | 9/13 | 15/12 |
| Education (years) | 9 (6–12) | 9 (9–12) | 9 (6–9) |
| BMI (kg/m2) | 22.29 ± 2.18 | 22.68 ± 2.17 | 21.87 ± 1.15 |
| Diabetes (%) | 0 (0%) | 2 (9.1%) | 5 (18.7%) |
| Hypertension (%) | 9 (32.1%) | 9 (40.9%) | 11 (40.7%) |
| Fasting glucose (mmol/L) | 5.23 ± 0.69 | 6.37 ± 1.93 | 5.02 ± 0.54 |
| Hemoglobin (g/L) | 138.42 ± 17.85 | 142.19 ± 11.23 | 136.79 ± 11.38 |
| Folic acid (ng/mL) | 11.08 ± 5.16 | 8.97 ± 2.90 | 8.75 ± 3.66 |
| Vitamin B12 (pg/mL) | 484.00 (435.50–716.00) | 510.00 (379.00–685.00) | 383.50 (341.00–506.25) |
| TT4 (nmol/L) | 107.05 ± 22.31 | 108.44 ± 19.74 | 103.50 ± 35.06 |
| TT3 (nmol/L) | 1.53 (1.43–1.92) | 1.72 (1.44–1.89) | 1.55 (1.38–1.75) |
| MMSE | 29.00 (26.00–29.50) | 27.00 (26.00–28.00) | 18.00 (13.50–23.00) ***### |
| MoCA | 26.00 (24.50–27.00) | 22.00 (18.00–24.00) * | 17.00 (14.50–19.00) ***# |
| MoCA Sub Item | HC | aMCI | AD |
|---|---|---|---|
| (n = 28) | (n = 22) | (n = 27) | |
| Visuospatial | 5 (4.75–5) | 4 (3–5) | 3 (1.25–3.75) ***# |
| Naming | 3 (3–3) | 3 (2–3) | 2 (2–3) ** |
| Attention | 3 (3–3) | 3 (2–3) | 1.5 (1–2.75) **# |
| Language | 2.5 (2–3) | 2 (1–2) | 1 (0–1) ***# |
| Abstraction | 1.5 (1–2) | 0 (0–1) | 0 (0–0) *** |
| Delayed recall | 3.5 (3–5) | 1 (0–3) ** | 0.5 (0–1) *** |
| Orientation | 6 (5.75–6) | 6 (5–6) | 3 (3–5.75) **## |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, L.; Han, Y.; Zheng, Z.; Peng, G.; Liu, P.; Yue, S.; Zhu, S.; Chen, J.; Lv, H.; Shao, L.; et al. Altered Gut Microbial Metabolites in Amnestic Mild Cognitive Impairment and Alzheimer’s Disease: Signals in Host–Microbe Interplay. Nutrients 2021, 13, 228. https://doi.org/10.3390/nu13010228
Wu L, Han Y, Zheng Z, Peng G, Liu P, Yue S, Zhu S, Chen J, Lv H, Shao L, et al. Altered Gut Microbial Metabolites in Amnestic Mild Cognitive Impairment and Alzheimer’s Disease: Signals in Host–Microbe Interplay. Nutrients. 2021; 13(1):228. https://doi.org/10.3390/nu13010228
Chicago/Turabian StyleWu, Li, Yuqiu Han, Zhipeng Zheng, Guoping Peng, Ping Liu, Siqing Yue, Shuai Zhu, Jun Chen, Hanying Lv, Lifang Shao, and et al. 2021. "Altered Gut Microbial Metabolites in Amnestic Mild Cognitive Impairment and Alzheimer’s Disease: Signals in Host–Microbe Interplay" Nutrients 13, no. 1: 228. https://doi.org/10.3390/nu13010228
APA StyleWu, L., Han, Y., Zheng, Z., Peng, G., Liu, P., Yue, S., Zhu, S., Chen, J., Lv, H., Shao, L., Sheng, Y., Wang, Y., Li, L., Li, L., & Wang, B. (2021). Altered Gut Microbial Metabolites in Amnestic Mild Cognitive Impairment and Alzheimer’s Disease: Signals in Host–Microbe Interplay. Nutrients, 13(1), 228. https://doi.org/10.3390/nu13010228