Effects of Lingonberry (Vaccinium vitis-idaea L.) Supplementation on Hepatic Gene Expression in High-Fat Diet Fed Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Study Design
2.2. RNA Extraction
2.3. Next-Generation Sequencing and Data Analysis
2.4. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
2.5. Statistical Analyses
3. Results
3.1. Body and Liver Weights
3.2. Changes in the Hepatic Gene Expression Caused by High-Fat Diet
3.3. Differences in Hepatic Gene Expression between Lingonberry-Supplemented and Control High-Fat Diet Groups
3.4. Functions and Interactions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Inoue, Y.; Qin, B.; Poti, J.; Sokol, R.; Gordon-Larsen, P. Epidemiology of Obesity in Adults: Latest Trends. Curr. Obes. Rep. 2018, 7, 276–288. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Obesity and Overweight. Available online: https://www.who.int/en/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 18 February 2020).
- Jung, U.J.; Choi, M. Obesity and its Metabolic Complications: The Role of Adipokines and the Relationship between Obesity, Inflammation, Insulin Resistance, Dyslipidemia and Nonalcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2014, 15, 6184–6223. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Lee, I.S.; Choue, R. Obesity, Inflammation and Diet. Pediatr. Gastroenterol. Hepatol. Nutr. 2013, 16, 143–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mraz, M.; Haluzik, M. The Role of Adipose Tissue Immune Cells in Obesity and Low-Grade Inflammation. J. Endocrinol. 2014, 222, 113–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, A.M.; Macedo-de La Concha, L.E.; Pantoja-Meléndez, C.A. Low-Grade Inflammation and its Relation to Obesity and Chronic Degenerative Diseases. Rev. Med. Hosp. Gen. Méx. 2017, 80, 101–105. [Google Scholar] [CrossRef]
- Fazel, Y.; Koenig, A.B.; Sayiner, M.; Goodman, Z.D.; Younossi, Z.M. Epidemiology and Natural History of Non-Alcoholic Fatty Liver Disease. Metab. Clin. Exp. 2016, 65, 1017–1025. [Google Scholar] [CrossRef] [Green Version]
- Polyzos, S.A.; Kountouras, J.; Mantzoros, C.S. Obesity and Nonalcoholic Fatty Liver Disease: From Pathophysiology to Therapeutics. Metabolism 2019, 92, 82–97. [Google Scholar] [CrossRef]
- Papandreou, D.; Andreou, E. Role of Diet on Non-Alcoholic Fatty Liver Disease: An Updated Narrative Review. World J. Hepatol. 2015, 7, 575–582. [Google Scholar] [CrossRef]
- Yu, J.; Marsh, S.; Hu, J.; Feng, W.; Wu, C. The Pathogenesis of Nonalcoholic Fatty Liver Disease: Interplay between Diet, Gut Microbiota, and Genetic Background. Gastroenterol. Res. Pract. 2016, 2016, 2862173. [Google Scholar] [CrossRef] [Green Version]
- Tiniakos, D.G.; Vos, M.B.; Brunt, E.M. Nonalcoholic Fatty Liver Disease: Pathology and Pathogenesis. Annu. Rev. Pathol. Mech. Dis. 2010, 5, 145–171. [Google Scholar] [CrossRef] [Green Version]
- McPherson, S.; Hardy, T.; Henderson, E.; Burt, A.D.; Day, C.P.; Anstee, Q.M. Evidence of NAFLD Progression from Steatosis to Fibrosing-Steatohepatitis using Paired Biopsies: Implications for Prognosis and Clinical Management. J. Hepatol. 2015, 62, 1148–1155. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Baker, R.D.; Bhatia, T.; Zhu, L.; Baker, S.S. Pathogenesis of Nonalcoholic Steatohepatitis. Cell. Mol. Life Sci. 2016, 73, 1969–1987. [Google Scholar] [CrossRef] [PubMed]
- Trovato, F.M.; Castrogiovanni, P.; Malatino, L.; Musumeci, G. Nonalcoholic Fatty Liver Disease (NAFLD) Prevention: Role of Mediterranean Diet and Physical Activity. Hepatobiliary Surg. Nutr. 2019, 8, 167–169. [Google Scholar] [CrossRef]
- Jayarathne, S.; Koboziev, I.; Park, O.; Oldewage-Theron, W.; Shen, C.; Moustaid-Moussa, N. Anti-Inflammatory and Anti-Obesity Properties of Food Bioactive Components: Effects on Adipose Tissue. Prev. Nutr. Food Sci. 2017, 22, 251–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, M.; Lai, C.; Ho, C. Anti-Inflammatory Activity of Natural Dietary Flavonoids. Food Funct. 2010, 1, 15–31. [Google Scholar] [CrossRef]
- Sears, B.; Ricordi, C. Role of Fatty Acids and Polyphenols in Inflammatory Gene Transcription and their Impact on Obesity, Metabolic Syndrome and Diabetes. Eur. Rev. Med. Pharmacol. Sci. 2012, 16, 1137–1154. [Google Scholar]
- Ryan, M.C.; Itsiopoulos, C.; Thodis, T.; Ward, G.; Trost, N.; Hofferberth, S.; O’dea, K.; Desmond, P.V.; Johnson, N.A.; Wilson, A.M. The Mediterranean Diet Improves Hepatic Steatosis and Insulin Sensitivity in Individuals with Non-Alcoholic Fatty Liver Disease. J. Hepatol. 2013, 59, 138–143. [Google Scholar] [CrossRef]
- Anhê, F.F.; Varin, T.V.; Le Barz, M.; Pilon, G.; Dudonné, S.; Trottier, J.; St-Pierre, P.; Harris, C.S.; Lucas, M.; Lemire, M.; et al. Arctic Berry Extracts Target the Gut–liver Axis to Alleviate Metabolic Endotoxaemia, Insulin Resistance and Hepatic Steatosis in Diet-Induced Obese Mice. Diabetologia 2018, 61, 919–931. [Google Scholar] [CrossRef] [Green Version]
- Glisan, S.L.; Ryan, C.; Neilson, A.P.; Lambert, J.D. Cranberry Extract Attenuates Hepatic Inflammation in High-Fat-Fed Obese Mice. J. Nutr. Biochem. 2016, 37, 60–66. [Google Scholar] [CrossRef] [Green Version]
- Lehtonen, H.-M.; Suomela, J.-P.; Tahvonen, R.; Vaarno, J.; Venojärvi, M.; Viikari, J.; Kallio, H. Berry Meals and Risk Factors Associated with Metabolic Syndrome. Eur. J. Clin. Nutr. 2010, 64, 614–621. [Google Scholar] [CrossRef]
- Liu, J.; Hefni, M.E.; Witthöft, C.M. Characterization of Flavonoid Compounds in Common Swedish Berry Species. Foods 2020, 9, 358. [Google Scholar] [CrossRef] [Green Version]
- Bujor, O.; Ginies, C.; Popa, V.I.; Dufour, C. Phenolic Compounds and Antioxidant Activity of Lingonberry (Vaccinium Vitis-Idaea L.) Leaf, Stem and Fruit at Different Harvest Periods. Food Chem. 2018, 252, 356–365. [Google Scholar] [CrossRef]
- Rodgers Dinstel, R.; Cascio, J.; Koukel, S. The Antioxidant Level of Alaska’s Wild Berries: High, Higher and Highest. Int. J. Circumpolar Health 2013, 72, 21188. [Google Scholar] [CrossRef]
- Ehala, S.; Vaher, M.; Kaljurand, M. Characterization of Phenolic Profiles of Northern European Berries by Capillary Electrophoresis and Determination of their Antioxidant Activity. J. Agric. Food Chem. 2005, 53, 6484–6490. [Google Scholar] [CrossRef] [PubMed]
- Vilkickyte, G.; Raudone, L.; Petrikaite, V. Phenolic Fractions from Vaccinium Vitis-Idaea L. and their Antioxidant and Anticancer Activities Assessment. Antioxidants 2020, 9, 1261. [Google Scholar] [CrossRef]
- Ryyti, R.; Hämäläinen, M.; Peltola, R.; Moilanen, E. Beneficial Effects of Lingonberry (Vaccinium Vitis-Idaea L.) Supplementation on Metabolic and Inflammatory Adverse Effects Induced by High-Fat Diet in a Mouse Model of Obesity. PLoS ONE 2020, 15, e0232605. [Google Scholar] [CrossRef] [PubMed]
- Heyman, L.; Axling, U.; Blanco, N.; Sterner, O.; Holm, C.; Berger, K. Evaluation of Beneficial Metabolic Effects of Berries in High-Fat Fed C57BL/6J Mice. J. Nutr. Metab. 2014, 2014, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Eid, H.M.; Ouchfoun, M.; Brault, A.; Vallerand, D.; Musallam, L.; Arnason, J.T.; Haddad, P.S. Lingonberry (Vaccinium Vitis-Idaea L.) Exhibits Antidiabetic Activities in a Mouse Model of Diet-Induced Obesity. Evid. Based Complement. Altern. Med. 2014, 2014, 645812. [Google Scholar] [CrossRef] [Green Version]
- Matziouridou, C.; Marungruang, N.; Nguyen, T.D.; Nyman, M.; Fåk, F. Lingonberries Reduce Atherosclerosis in Apoe-/- Mice in Association with Altered Gut Microbiota Composition and Improved Lipid Profile. Mol. Nutr. Food Res. 2016, 60, 1150–1160. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.A. Quality Control Tool for High Throughput Sequence Data (Fast QC). Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 25 January 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics Enrichment Tools: Paths Toward the Comprehensive Functional Analysis of Large Gene Lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherman, B.T.; Lempicki, R.A.; Huang, D.W. Systematic and Integrative Analysis of Large Gene Lists using DAVID Bioinformatics Resources. Nat. Protoc. 2008, 4, 44–57. [Google Scholar]
- Expansion of the Gene Ontology Knowledgebase and Resources. Nucleic Acids Res. 2017, 45, D331–D338. [CrossRef] [PubMed] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. the Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-Protein Interaction Networks, Integrated Over the Tree of Life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- NCBI (National Center for Biotechnology Information). Available online: https://www.ncbi.nlm.nih.gov/gene/13112 (accessed on 19 September 2020).
- UniProt Consortium Knowledgebase. Available online: https://www.uniprot.org/uniprot/Q64459 (accessed on 19 September 2020).
- Bechmann, L.P.; Hannivoort, R.A.; Gerken, G.; Hotamisligil, G.S.; Trauner, M.; Canbay, A. The Interaction of Hepatic Lipid and Glucose Metabolism in Liver Diseases. J. Hepatol. 2012, 56, 952–964. [Google Scholar] [CrossRef] [Green Version]
- Radonjic, M.; de Haan, J.R.; van Erk, M.J.; van Dijk, K.W.; van den Berg, S.A.A.; de Groot, P.J.; Müller, M.; van Ommen, B. Genome-Wide mRNA Expression Analysis of Hepatic Adaptation to High-Fat Diets Reveals Switch from an Inflammatory to Steatotic Transcriptional Program. PLoS ONE 2009, 4, e6646. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Ye, R.D. Serum Amyloid A1: Structure, Function and Gene Polymorphism. Gene 2016, 583, 48–57. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Tao, Q.; Wang, X.; Wang, X.; Zhang, X. Impact of High-Fat Diet on Liver Genes Expression Profiles in Mice Model of Nonalcoholic Fatty Liver Disease. Environ. Toxicol. Pharmacol. 2016, 45, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Rui, L. Leptin Signaling and Leptin Resistance. Front. Med. 2013, 7, 207–222. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Guo, J.; Su, Z. Advances in Understanding the Interrelations between Leptin Resistance and Obesity. Physiol. Behav. 2014, 130, 157–169. [Google Scholar] [CrossRef]
- Rensen, S.S.; Slaats, Y.; Driessen, A.; Peutz-Kootstra, C.; Nijhuis, J.; Steffensen, R.; Greve, J.W.; Buurman, W.A. Activation of the Complement System in Human Nonalcoholic Fatty Liver Disease. Hepatology 2009, 50, 1809–1817. [Google Scholar] [CrossRef]
- Kobori, M.; Masumoto, S.; Akimoto, Y.; Oike, H. Chronic Dietary Intake of Quercetin Alleviates Hepatic Fat Accumulation Associated with Consumption of a Western-Style Diet in C57/BL6J Mice. Mol. Nutr. Food Res. 2011, 55, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Skat-Rørdam, J.; Højland Ipsen, D.; Lykkesfeldt, J.; Tveden-Nyborg, P. A Role of Peroxisome Proliferator-activated Receptor Γ in Non-alcoholic Fatty Liver Disease. Basic Clin. Pharmacol. Toxicol. 2019, 124, 528–537. [Google Scholar] [CrossRef]
- Choudhary, N.S.; Kumar, N.; Duseja, A. Peroxisome Proliferator-Activated Receptors and their Agonists in Nonalcoholic Fatty Liver Disease. J. Clin. Exp. Hepatol. 2019, 9, 731–739. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.K.; Park, J.E.; Lee, M.; Hardwick, J.P. Hepatic Lipid Homeostasis by Peroxisome Proliferator-Activated Receptor Gamma 2. Liver Res. 2018, 2, 209–215. [Google Scholar] [CrossRef]
- Pan, W.W.; Myers, M.G. Leptin and the Maintenance of Elevated Body Weight. Nat. Rev. Neurosci. 2018, 19, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Paz-Filho, G.; Mastronardi, C.; Franco, C.B.; Wang, K.B.; Wong, M.; Licinio, J. Leptin: Molecular Mechanisms, Systemic Pro-Inflammatory Effects, and Clinical Implications. Arq. Bras. Endocrinol. Metabol. 2012, 56, 597–607. [Google Scholar] [CrossRef] [Green Version]
- Feng, H.; Zheng, L.; Feng, Z.; Zhao, Y.; Zhang, N. The Role of Leptin in Obesity and the Potential for Leptin Replacement Therapy. Endocrine 2012, 44, 33–39. [Google Scholar] [CrossRef]
- St-Pierre, J.; Tremblay, M.L. Modulation of Leptin Resistance by Protein Tyrosine Phosphatases. Cell Metab. 2012, 15, 292–297. [Google Scholar] [CrossRef] [Green Version]
- Koskinen-Kolasa, A.; Vuolteenaho, K.; Korhonen, R.; Moilanen, T.; Moilanen, E. Catabolic and Proinflammatory Effects of Leptin in Chondrocytes are Regulated by Suppressor of Cytokine Signaling-3. Arthritis Res. Ther. 2016, 18, 215. [Google Scholar] [CrossRef] [Green Version]
- Howard, J.K.; Flier, J.S. Attenuation of Leptin and Insulin Signaling by SOCS Proteins. Trends Endocrinol. Metab. 2006, 17, 365–371. [Google Scholar] [CrossRef]
- Rufino, A.T.; Costa, V.M.; Carvalho, F.; Fernandes, E. Flavonoids as Antiobesity Agents: A Review. Med. Res. Rev. 2021, 41, 556–585. [Google Scholar] [CrossRef] [PubMed]
- Adriouch, S.; Lampuré, A.; Nechba, A.; Baudry, J.; Assmann, K.; Kesse-Guyot, E.; Hercberg, S.; Scalbert, A.; Touvier, M.; Fezeu, L.K. Prospective Association between Total and Specific Dietary Polyphenol Intakes and Cardiovascular Disease Risk in the Nutrinet-Santé French Cohort. Nutrients 2018, 10, 1587. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Moustaid-Moussa, N.; Chen, L.; Mo, H.; Shastri, A.; Su, R.; Bapat, P.; Kwun, I.; Shen, C. Novel Insights of Dietary Polyphenols and Obesity. J. Nutr. Biochem. 2014, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Susara, M.H.; Prashar, S.; Karmin, O.; Siow, Y.L. Lingonberry Improves Non-Alcoholic Fatty Liver Disease by Reducing Hepatic Lipid Accumulation, Oxidative Stress and Inflammatory Response. Antioxidants 2021, 10, 565. [Google Scholar]
- Heyman-Lindén, L.; Seki, Y.; Storm, P.; Jones, H.A.; Charron, M.J.; Berger, K.; Holm, C. Berry Intake Changes Hepatic Gene Expression and DNA Methylation Patterns Associated with High-Fat Diet. J. Nutr. Biochem. 2016, 27, 79–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, A.; Whitehead, A.S. Mapping of the Mouse Serum Amyloid A Gene Cluster by Long-Range Polymerase Chain Reaction. Immunogenetics 1996, 44, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Rajala, M.W.; Berger, J.P.; Moller, D.E.; Barzilai, N.; Scherer, P.E. Hyperglycemia-Induced Production of Acute Phase Reactants in Adipose Tissue. J. Biol. Chem. 2001, 276, 42077–42083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sack, G.H. Serum Amyloid A—A Review. Mol. Med. 2018, 24, 46. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Chatterjee, M.; Schmid, H.; Beck, S.; Gawaz, M. CXCL14 as an Emerging Immune and Inflammatory Modulator. J. Inflamm. 2016, 13, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Shuai, C.; Gao, S.; Jiang, J.; Luan, J.; Lv, X. Chemokine CXCL14 Acts as a Potential Genetic Target for Liver Fibrosis. Int. Immunopharmacol. 2020, 89, 107067. [Google Scholar] [CrossRef]
- Li, J.; Gao, J.; Yan, D.; Yuan, Y.; Sah, S.; Satyal, U.; Liu, M.; Han, W.; Yu, Y. Neutralization of Chemokine CXCL14 (BRAK) Expression Reduces CCl4 Induced Liver Injury and Steatosis in Mice. Eur. J. Pharmacol. 2011, 671, 120–127. [Google Scholar] [CrossRef]
- Wang, Y. Small Lipid-Binding Proteins in Regulating Endothelial and Vascular Functions: Focusing on Adipocyte Fatty Acid Binding Protein and Lipocalin-2. Br. J. Pharmacol. 2012, 165, 603–621. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Jin, D.; Zhang, Y.; Wright, W.; Bazuine, M.; Brockman, D.A.; Bernlohr, D.A.; Chen, X. Lipocalin-2 Deficiency Impairs Thermogenesis and Potentiates Diet-Induced Insulin Resistance in Mice. Diabetes 2010, 59, 1376–1385. [Google Scholar] [CrossRef] [Green Version]
- Yan, Q.-W.; Yang, Q.; Mody, N.; Graham, T.E.; Hsu, C.-H.; Xu, Z.; Houstis, N.E.; Kahn, B.B.; Rosen, E.D. The Adipokine Lipocalin 2 is Regulated by Obesity and Promotes Insulin Resistance. Diabetes 2007, 56, 2533–2540. [Google Scholar] [CrossRef] [Green Version]
- Deis, J.A.; Guo, H.; Wu, Y.; Liu, C.; Bernlohr, D.A.; Chen, X. Lipocalin 2 Regulates Retinoic Acid-Induced Activation of Beige Adipocytes. J. Mol. Endocrinol. 2018, 61, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Foncea, R.; O’Byrne, S.M.; Jiang, H.; Zhang, Y.; Deis, J.A.; Blaner, W.S.; Bernlohr, D.A.; Chen, X. Lipocalin 2, a Regulator of Retinoid Homeostasis and Retinoid-Mediated Thermogenic Activation in Adipose Tissue. J. Biol. Chem. 2016, 291, 11216–11229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, A.M.; Soufi, N.; Chambers, K.T.; Chen, Z.; Schweitzer, G.G.; McCommis, K.S.; Erion, D.M.; Graham, M.J.; Su, X.; Finck, B.N. Abrogating Monoacylglycerol Acyltransferase Activity in Liver Improves Glucose Tolerance and Hepatic Insulin Signaling in Obese Mice. Diabetes 2014, 63, 2284–2296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, A.K.; Tunison, K.; Dalal, J.S.; Yen, C.E.; Farese, J.; Robert, V.; Horton, J.D.; Garg, A. Mogat1 Deletion does Not Ameliorate Hepatic Steatosis in Lipodystrophic (Agpat2-/-) Or Obese (Ob/Ob) Mice. J. Lipid Res. 2016, 57, 616–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, Y.; Suemitsu, E.; Kajimoto, K.; Sato, Y.; Akhter, A.; Sakurai, Y.; Hatakeyama, H.; Hyodo, M.; Kaji, N.; Baba, Y.; et al. Hepatic Monoacylglycerol O-Acyltransferase 1 as a Promising Therapeutic Target for Steatosis, Obesity, and Type 2 Diabetes. Mol. Ther. Nucleic Acids 2014, 3, e154. [Google Scholar] [CrossRef]
- Carr, R.M.; Ahima, R.S. Pathophysiology of Lipid Droplet Proteins in Liver Diseases. Exp. Cell Res. 2016, 340, 187–192. [Google Scholar] [CrossRef] [Green Version]
- Okumura, T. Role of Lipid Droplet Proteins in Liver Steatosis. J. Physiol. Biochem. 2011, 67, 629–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, S.; Matsusue, K.; Kashireddy, P.; Cao, W.; Yeldandi, V.; Yeldandi, A.V.; Rao, M.S.; Gonzalez, F.J.; Reddy, J.K. Adipocyte-Specific Gene Expression and Adipogenic Steatosis in the Mouse Liver due to Peroxisome Proliferator-Activated Receptor Gamma1 (PPARgamma1) Overexpression. J. Biol. Chem. 2003, 278, 498–505. [Google Scholar] [CrossRef] [Green Version]
- Haywood, N.J.; Slater, T.A.; Matthews, C.J.; Wheatcroft, S.B. The Insulin Like Growth Factor and Binding Protein Family: Novel Therapeutic Targets in Obesity & Diabetes. Mol. Metab. 2019, 19, 86–96. [Google Scholar] [PubMed]
- Ko, J.M.; Park, H.K.; Yang, S.; Hwang, I.T. Influence of Catch-Up Growth on IGFBP-2 Levels and Association between IGFBP-2 and Cardiovascular Risk Factors in Korean Children Born SGA. Endocr. J. 2012, 59, 725–733. [Google Scholar] [CrossRef] [Green Version]
- Heald, A.H.; Kaushal, K.; Siddals, K.W.; Rudenski, A.S.; Anderson, S.G.; Gibson, J.M. Insulin-Like Growth Factor Binding Protein-2 (IGFBP-2) is a Marker for the Metabolic Syndrome. Exp. Clin. Endocrinol. Diabetes 2006, 114, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Rajpathak, S.N.; He, M.; Sun, Q.; Kaplan, R.C.; Muzumdar, R.; Rohan, T.E.; Gunter, M.J.; Pollak, M.; Kim, M.; Pessin, J.E.; et al. Insulin-Like Growth Factor Axis and Risk of Type 2 Diabetes in Women. Diabetes 2012, 61, 2248–2254. [Google Scholar] [CrossRef] [Green Version]
- Wheatcroft, S.B.; Kearney, M.T.; Shah, A.M.; Ezzat, V.A.; Miell, J.R.; Modo, M.; Williams, S.C.R.; Cawthorn, W.P.; Medina-Gomez, G.; Vidal-Puig, A.; et al. IGF-Binding Protein-2 Protects Against the Development of Obesity and Insulin Resistance. Diabetes 2007, 56, 285–294. [Google Scholar] [CrossRef] [Green Version]
- Zanger, U.M.; Schwab, M. Cytochrome P450 Enzymes in Drug Metabolism: Regulation of Gene Expression, Enzyme Activities, and Impact of Genetic Variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef] [PubMed]
- Hannemann, F.; Bichet, A.; Ewen, K.M.; Bernhardt, R. Cytochrome P450 Systems—biological Variations of Electron Transport Chains. Biochim. Biophys. Acta 2007, 1770, 330–344. [Google Scholar] [CrossRef] [PubMed]
- Yoshinari, K.; Takagi, S.; Yoshimasa, T.; Sugatani, J.; Miwa, M. Hepatic CYP3A Expression is Attenuated in Obese Mice Fed a High-Fat Diet. Pharm. Res. 2006, 23, 1188–1200. [Google Scholar] [CrossRef]
- Maximos, S.; Chamoun, M.; Gravel, S.; Turgeon, J.; Michaud, V. Tissue Specific Modulation of Cyp2c and Cyp3a mRNA Levels and Activities by Diet-Induced Obesity in Mice: The Impact of Type 2 Diabetes on Drug Metabolizing Enzymes in Liver and Extra-Hepatic Tissues. Pharmaceutics 2017, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Tomankova, V.; Anzenbacher, P.; Anzenbacherova, E. Effects of Obesity on Liver Cytochromes P450 in various Animal Models. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub 2017, 161, 144–151. [Google Scholar] [CrossRef] [Green Version]
- Nelson, D.R.; Zeldin, D.C.; Hoffman, S.M.G.; Maltais, L.J.; Wain, H.M.; Nebert, D.W. Comparison of Cytochrome P450 (CYP) Genes from the Mouse and Human Genomes, Including Nomenclature Recommendations for Genes, Pseudogenes and Alternative-Splice Variants. Pharmacogenetics 2004, 14, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Tang, Q.; Zhao, L.; Zhang, Q.; Wu, Y.; Hu, H.; Liu, L.; Liu, X.; Zhu, Y.; Guo, A.; et al. Time Serial Transcriptome Reveals Cyp2c29 as a Key Gene in Hepatocellular Carcinoma Development. Cancer Biol. Med. 2020, 17, 401–417. [Google Scholar] [CrossRef]
- Han, M.; Piorońska, W.; Wang, S.; Nwosu, Z.C.; Sticht, C.; Wang, S.; Gao, Y.; Ebert, M.P.; Dooley, S.; Meyer, C. Hepatocyte Caveolin-1 Modulates Metabolic Gene Profiles and Functions in Non-Alcoholic Fatty Liver Disease. Cell Death Dis. 2020, 11, 104. [Google Scholar] [CrossRef]
- Barretto, S.A.; Lasserre, F.; Fougerat, A.; Smith, L.; Fougeray, T.; Lukowicz, C.; Polizzi, A.; Smati, S.; Régnier, M.; Naylies, C.; et al. Gene Expression Profiling Reveals that PXR Activation Inhibits Hepatic PPARα Activity and Decreases FGF21 Secretion in Male C57Bl6/J Mice. Int. J. Mol. Sci. 2019, 20, 3767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konno, Y.; Kamino, H.; Moore, R.; Lih, F.; Tomer, K.B.; Zeldin, D.C.; Goldstein, J.A.; Negishi, M. The Nuclear Receptors Constitutive Active/Androstane Receptor and Pregnane X Receptor Activate the Cyp2c55 Gene in Mouse Liver. Drug Metab. Dispos. 2010, 38, 1177–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karcheva-Bahchevanska, D.; Lukova, P.; Nikolova, M.; Mladenov, R.; Iliev, I. Inhibition Effect of Bulgarian Lingonberry (Vaccinium Vitis-Idaea L.) Extracts on A-Amylase Activity. C. R. de L’Acad. Bulg. des Sci. 2019, 72, 212–218. [Google Scholar]
- Rimando, A.M.; Kalt, W.; Magee, J.B.; Dewey, J.; Ballington, J.R. Resveratrol, Pterostilbene, and Piceatannol in Vaccinium Berries. J. Agric. Food Chem. 2004, 52, 4713–4719. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, J.K.; Thomas, S.; Nanjan, M.J. Resveratrol Supplementation Improves Glycemic Control in Type 2 Diabetes Mellitus. Nutr. Res. 2012, 32, 537–541. [Google Scholar] [CrossRef]
- Laavola, M.; Nieminen, R.; Leppänen, T.; Eckerman, C.; Holmbom, B.; Moilanen, E. Pinosylvin and Monomethylpinosylvin, Constituents of an Extract from the Knot of Pinus Sylvestris, Reduce Inflammatory Gene Expression and Inflammatory Responses In Vivo. J. Agric. Food Chem. 2015, 63, 3445–3453. [Google Scholar] [CrossRef]
- Wahab, A.; Gao, K.; Jia, C.; Zhang, F.; Tian, G.; Murtaza, G.; Chen, J. Significance of Resveratrol in Clinical Management of Chronic Diseases. Molecules 2017, 22, 1329. [Google Scholar] [CrossRef] [Green Version]
- Eräsalo, H.; Hämäläinen, M.; Leppänen, T.; Mäki-Opas, I.; Laavola, M.; Haavikko, R.; Yli-Kauhaluoma, J.; Moilanen, E. Natural Stilbenoids have Anti-Inflammatory Properties in Vivo and Down-Regulate the Production of Inflammatory Mediators NO, IL6, and MCP1 Possibly in a PI3K/Akt-Dependent Manner. J. Nat. Prod. 2018, 81, 1131–1142. [Google Scholar] [CrossRef]
- Laavola, M.; Leppänen, T.; Hämäläinen, M.; Vuolteenaho, K.; Moilanen, T.; Nieminen, R.; Moilanen, E. IL-6 in Osteoarthritis: Effects of Pine Stilbenoids. Molecules 2018, 24, 109. [Google Scholar] [CrossRef] [Green Version]
- Kivimäki, K.; Leppänen, T.; Hämäläinen, M.; Vuolteenaho, K.; Moilanen, E. Pinosylvin Shifts Macrophage Polarization to Support Resolution of Inflammation. Molecules 2021, 26, 2772. [Google Scholar] [CrossRef] [PubMed]
- Anhê, F.F.; Nachbar, R.T.; Varin, T.V.; Vilela, V.; Dudonné, S.; Pilon, G.; Fournier, M.; Lecours, M.; Desjardins, Y.; Roy, D.; et al. A Polyphenol-Rich Cranberry Extract Reverses Insulin Resistance and Hepatic Steatosis Independently of Body Weight Loss. Mol. Metab. 2017, 6, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, K.; Ono, M.; Imoto, A.; Nagayama, H.; Tetsumura, N.; Terada, T.; Tomita, K.; Nishinaka, T. Cranberry Attenuates Progression of Non-Alcoholic Fatty Liver Disease Induced by High-Fat Diet in Mice. Biol. Pharm. Bull 2019, 42, 1295–1302. [Google Scholar] [CrossRef] [Green Version]
- Hormoznejad, R.; Mohammad Shahi, M.; Rahim, F.; Helli, B.; Alavinejad, P.; Sharhani, A. Combined Cranberry Supplementation and Weight Loss Diet in Non-Alcoholic Fatty Liver Disease: A Double-Blind Placebo-Controlled Randomized Clinical Trial. Int. J. Food Sci. Nutr. 2020, 71, 991–1000. [Google Scholar] [CrossRef] [PubMed]
- Snyder, S.; Zhao, B.; Luo, T.; Kaiser, C.; Cavender, G.; Hamilton-Reeves, J.; Sullivan, D.; Shay, N. Consumption of Quercetin and Quercetin- Containing Apple and Cherry Extracts Affects Blood Glucose Concentration, Hepatic Metabolism, and Gene Expression Patterns in Obese C57BL/6J High Fat-Fed Mice 1–4. J. Nutr. 2016, 146, 1001–1007. [Google Scholar] [CrossRef] [PubMed]



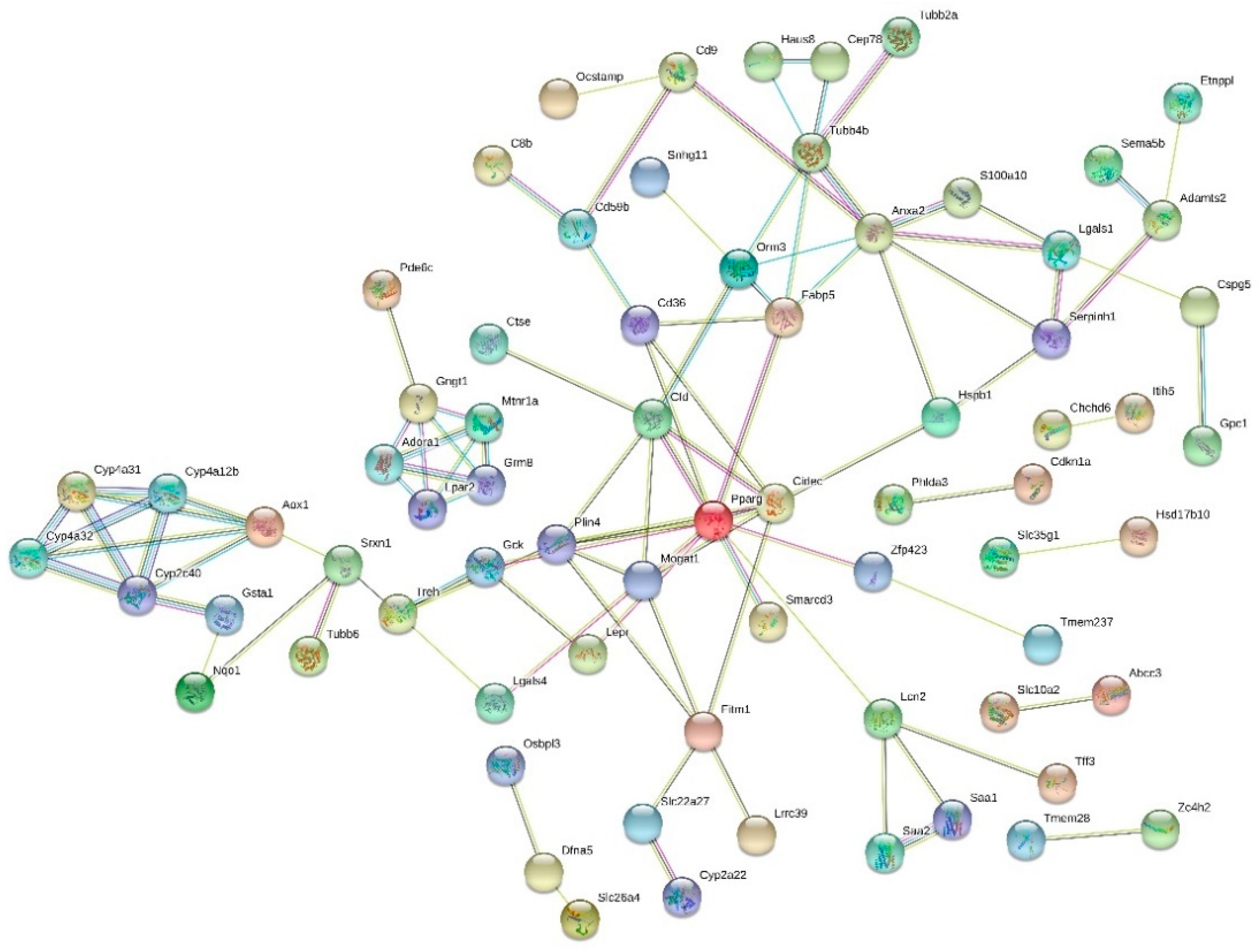
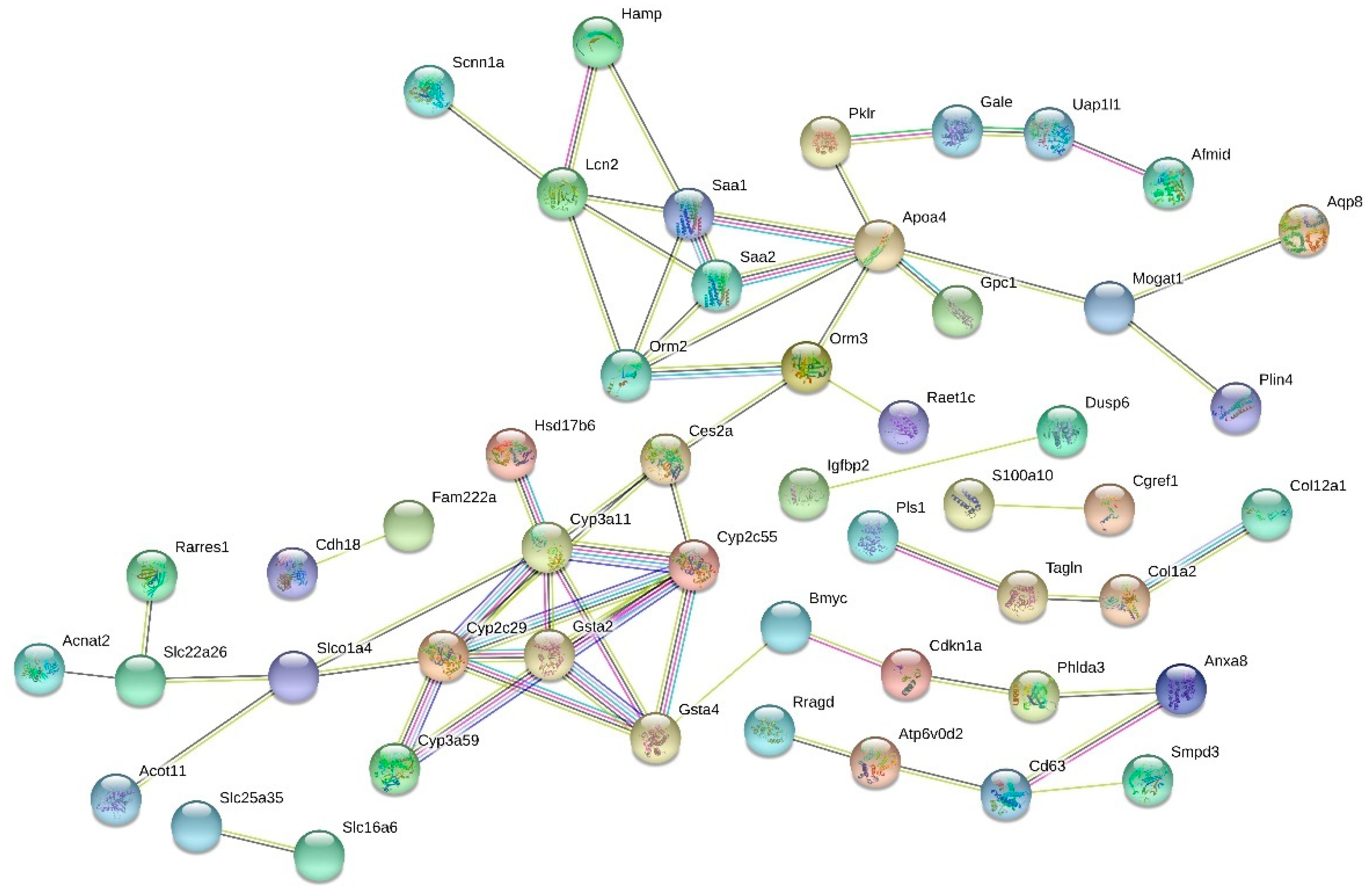
| Gene | Name | Functions in Mouse | Mean (LF) | Mean (HF) | Fold Change | p-Value (FDR adj.) |
|---|---|---|---|---|---|---|
| Themis | Thymocyte selection associated | T cell receptor signaling pathway, immune response | 36.9 | 186.5 | 2.69 | <0.0001 |
| Mogat1 | Monoacylglycerol O-acyltransferase 1 | Lipid metabolic process | 19.5 | 66.4 | 2.51 | <0.0001 |
| Kbtbd11 | Kelch repeat and BTB (POZ) domain containing 11 | 11.9 | 46.4 | 2.36 | <0.0001 | |
| Aatk | Apoptosis-associated tyrosine kinase | Apoptosis | 63.4 | 201.3 | 2.35 | <0.0001 |
| Tpm2 | Tropomyosin 2, beta | Actin filament stabilization | 59.4 | 216.7 | 2.31 | <0.0001 |
| Cfd | Complement factor D, adipsin | Complement activation and inflammation | 6.8 | 125.8 | 2.23 | <0.0001 |
| Lgals1 | Lectin, galactose binding, soluble 1 | Cell adhesion, regulation of apoptosis | 272.4 | 838.3 | 2.20 | <0.0001 |
| Adgrv1 | Adhesion G protein-coupled receptor V1 | Cell adhesion | 62.2 | 157.3 | 2.14 | <0.0001 |
| Lrrc14b | Leucine rich repeat containing 14B | 6.2 | 21.2 | 2.14 | <0.0001 | |
| Tmem28 | Transmembrane protein 28 | Calcium ion transport | 27.4 | 81.9 | 2.13 | <0.0001 |
| Slc22a29 | Solute carrier family 22. member 29 | Organic anion transport | 8.7 | 39.9 | 2.11 | <0.0001 |
| Clstn3 | Calsyntenin 3 | Cell adhesion | 285.4 | 713.2 | 2.07 | <0.0001 |
| Hspb1 | Heat shock protein 1 | Negative regulation of apoptosis, positive regulation of interleukin-1 beta production | 45.8 | 109.4 | 2.06 | <0.0001 |
| Tafa2 | TAFA chemokine-like family member 2 | Receptor ligand activity | 4.3 | 16.9 | 2.04 | <0.0001 |
| Treh | Trehalase (brush-border membrane glycoprotein) | Metabolism | 22.0 | 54.0 | 1.97 | <0.0001 |
| Sema5b | Sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B | Cell differentiation, positive regulation of cell migration | 40.6 | 102.8 | 1.95 | <0.0001 |
| Osbpl3 | Oxysterol binding protein-like 3 | Lipid transport | 52.0 | 181.6 | 1.93 | <0.0001 |
| Fitm1 | Fat storage-inducing transmembrane protein 1 | Lipid droplet organization, phospholipid biosynthetic process | 409.0 | 939.7 | 1.92 | <0.0001 |
| Anxa2 | Annexin A2 | Regulation of cholesterol metabolism | 141.8 | 311.8 | 1.89 | <0.0001 |
| Hectd2os | Hectd2, opposite strand | 1342.7 | 3302.2 | 1.89 | <0.0001 |
| Gene | Name | Functions in Mouse | Mean (LF) | Mean (HF) | Fold Change | p-Value (FDR adj.) |
|---|---|---|---|---|---|---|
| Lepr | Leptin receptor | Regulation of metabolism | 296.0 | 43.2 | −3.48 | <0.0001 |
| Adgrf1 | Adhesion G protein-coupled receptor F1 | G-protein coupled receptor activity | 122.3 | 40.0 | −2.06 | <0.0001 |
| Igfbp2 | Insulin-like growth factor binding protein 2 | Glucose metabolism, insulin sensitivity | 7680.0 | 3614.7 | −1.95 | <0.0001 |
| Fabp5 | Fatty acid binding protein 5, epidermal | Glucose and lipid metabolism | 1273.4 | 156.8 | −1.93 | <0.0001 |
| Grm8 | Glutamate receptor, metabotropic 8 | Glutamate receptor activity | 21.7 | 8.1 | −1.91 | <0.0001 |
| Adam11 | A disintegrin and metallopeptidase domain 11 | Metalloendopeptidase activity | 134.1 | 53.2 | −1.88 | <0.0001 |
| Srgap3 | Insulin-like growth factor binding protein 2 | Negative regulation of cell migration | 106.8 | 43.8 | −1.82 | <0.0001 |
| Cyp2c40 | Cytochrome P450, family 2, subfamily c, polypeptide 40 | Arachidonic acid epoxygenase activity, metal ion binding | 21.3 | 5.8 | −1.73 | 0.0003 |
| Slc35g1 | Solute carrier family 35, member G1 | Regulation of cytosolic calcium ion concentration | 399.4 | 215.0 | −1.72 | <0.0001 |
| Lpar2 | Lysophosphatidic acid receptor 2 | Activation of MAPK activity | 55.6 | 29.3 | −1.65 | <0.0001 |
| Pde6c | Phosphodiesterase 6C, cGMP specific, cone, alpha prime | 3′,5′-cyclic-GMP phosphodiesterase activity, metal ion and nucleotide binding | 33.5 | 16.1 | −1.64 | 0.0008 |
| St3gal5 | ST3 beta-galactoside alpha−2,3-sialyltransferase 5 | Protein glycosylation | 2464.8 | 1193.9 | −1.62 | 0.0005 |
| Sds | Serine hydratase | L-serine ammonia-lyase activity | 3961.3 | 2026.8 | −1.62 | 0.0007 |
| Sox12 | SRY (sex determining region Y)-box 12 | Cell differentiation | 113.2 | 64.0 | −1.61 | <0.0001 |
| Tff3 | Trefoil factor 3, intestinal | Regulation of glucose metabolism | 37.1 | 15.4 | −1.61 | 0.0020 |
| Cadm4 | Cell adhesion molecule 4 | Regulation of cell proliferation | 138.7 | 83.8 | −1.59 | <0.0001 |
| Rnf145 | Ring finger protein 145 | Metal ion binding, transferase activity | 581.0 | 326.8 | −1.58 | <0.0001 |
| Lgals4 | Lectin, galactose binding, soluble 4 | Cell adhesion | 130.7 | 72.8 | −1.58 | 0.0001 |
| Cspg5 | Chondroitin sulfate proteoglycan 5 | Cell differentiation | 21.8 | 9.1 | −1.56 | 0.0065 |
| Cd9 | CD9 antigen | Cell adhesion | 428.2 | 243.1 | −1.55 | 0.0002 |
| Gene | Name | Functions in Mouse | Mean (HF) | Mean (HF + LGB) | Fold Change | p-Value (FDR adj.) |
|---|---|---|---|---|---|---|
| Wfdc2 | WAP four-disulfide core domain 2 | Endopeptidase inhibitor activity | 185.0 | 54.2 | −2.28 | <0.0001 |
| Apoa4 | Apolipoprotein A-IV | Antioxidant activity, cholesterol and lipid homeostasis | 11,871.9 | 3002.7 | −2.13 | <0.0001 |
| Gpc1 | Glypican 1 | Cell migration | 542.7 | 211.2 | −2.04 | <0.0001 |
| Slc35f2 | Solute carrier family 35, member F2 | Transmembrane transporter activity | 34.4 | 10.5 | −2.04 | <0.0001 |
| Ifi27l2b | Interferon, alpha-inducible protein 27 like 2B | Immune system process, intrinsic apoptotic signaling pathway | 102.6 | 31.2 | −2.04 | <0.0001 |
| Rad51b | RAD51 paralog B | DNA recombination and repair, positive regulation of cell proliferation | 90.9 | 23.3 | −2.03 | <0.0001 |
| Lcn2 | Lipocalin 2 | Apoptotic process, inflammation | 174.8 | 42.2 | −1.99 | <0.0001 |
| Morc4 | Microrchidia 4 | Metal ion and zinc ion binding | 72.5 | 30.8 | −1.95 | <0.0001 |
| Rarres1 | Retinoic acid receptor responder (tazarotene induced) 1 | Metalloendopeptidase inhibitor activity | 1168.6 | 425.4 | −1.95 | <0.0001 |
| Fam129b | Family with sequence similarity 129, member B | Negative regulation of DNA biosynthetic process and cell proliferation | 304.6 | 130.0 | −1.87 | <0.0001 |
| Bmyc | Brain expressed myelocytomatosis oncogene | Regulation of DNA transcription | 89.6 | 38.7 | −1.83 | <0.0001 |
| Smpd3 | Sphingomyelin phosphodiesterase 3, neutral | Extracellular matrix assembly, regulation of cell proliferation | 106.8 | 36.2 | −1.83 | <0.0001 |
| Saa2 | Serum amyloid A 2 | Acute-phase response, inflammation | 762.9 | 222.7 | −1.83 | <0.0001 |
| Aqp8 | Aquaporin 8 | Canalicular bile acid transport, water transport | 5539.7 | 2377.6 | −1.82 | <0.0001 |
| Cyp46a1 | Cytochrome P450, family 46, subfamily a, polypeptide 1 | Cholesterol catabolic process, iron ion binding | 92.2 | 32.3 | −1.82 | <0.0001 |
| Ly6d | Lymphocyte antigen 6 complex, locus D | Response to stilbenoid | 45.0 | 11.1 | −1.80 | <0.0001 |
| Phlda3 | Pleckstrin homology-like domain, family A, member 3 | Phosphatidylinositol-phosphates binding; apoptotic process positive regulation | 35.0 | 13.8 | −1.78 | <0.0001 |
| Tsc22d1 | TSC22 domain family, member 1 | Regulation of apoptosis, cell proliferation | 1940.3 | 947.4 | −1.77 | <0.0001 |
| Extl1 | Exostoses (multiple)-like 1 | Glycosaminoglycan biosynthesis | 324.5 | 153.5 | −1.77 | <0.0001 |
| Saa1 | Serum amyloid A 1 | Acute-phase response, inflammation, cholesterol metabolic process | 1277.5 | 471.0 | −1.75 | <0.0001 |
| Gene | Name | Functions in Mouse | Mean (HF) | Mean (HF + LGB) | Fold Change | p-Value (FDR adj.) |
|---|---|---|---|---|---|---|
| Cyp3a11 | Cytochrome P450, family 3, subfamily a, polypeptide 11 | Oxidation and reduction, steroid metabolism | 6628.2 | 27,365.0 | 2.85 | <0.0001 |
| Cyp2c55 | Cytochrome P450, family 2, subfamily c, polypeptide 55 | Fatty acid metabolism | 27.3 | 84.5 | 2.22 | <0.0001 |
| Adgrf1 | Adhesion G protein-coupled receptor F1 | G protein receptor activity | 36.2 | 143.8 | 1.91 | <0.0001 |
| Emp2 | Epithelial membrane protein 2 | Cell adhesion, regulation of angiogenesis | 125.1 | 253.5 | 1.79 | <0.0001 |
| Cyp2c29 | Cytochrome P450, family 2, subfamily c, polypeptide 29 | Fatty acid metabolism | 8826.3 | 16,460.9 | 1.75 | <0.0001 |
| Grid1 | Glutamate receptor, ionotropic, delta 1 | Glutamate receptor activity, ion transport | 17.4 | 42.0 | 1.75 | <0.0001 |
| Hsd17b6 | Hydroxysteroid (17-beta) dehydrogenase 6 | Estradiol dehydrogenase activity, lipid and steroid metabolic process | 1545.8 | 3033.3 | 1.74 | <0.0001 |
| Ces2a | Carboxylesterase 2A | Carboxylic ester hydrolase activity, protein glycosylation | 1987.2 | 3581.2 | 1.73 | <0.0001 |
| Fam222a | Family with sequence similarity 222, member A | 16.2 | 38.3 | 1.72 | 0.0001 | |
| Igfbp2 | Insulin-like growth factor binding protein 2 | Glucose metabolism, insulin sensitivity | 3388.8 | 6263.7 | 1.71 | <0.0001 |
| Asap3 | ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 | Cell migration | 77.5 | 140.1 | 1.69 | <0.0001 |
| Neb | Nebulin | Actin filament and protein binding | 161.9 | 337.1 | 1.68 | <0.0001 |
| Slc7a2 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | Amino acid import across plasma membrane, regulation of inflammation | 7225.2 | 13,116.6 | 1.65 | <0.0001 |
| Scnn1a | Sodium channel, nonvoltage-gated 1 alpha | Sodium ion homeostasis | 302.0 | 511.2 | 1.59 | <0.0001 |
| Sorbs3 | Sorbin and SH3 domain containing 3 | Actin filament organization, cell adhesion | 333.5 | 564.2 | 1.59 | <0.0001 |
| Slco1a4 | Solute carrier organic anion transporter family, member 1a4 | Bile acid and bile salt transport | 489.7 | 951.0 | 1.58 | 0.0008 |
| Gsta2 | Glutathione S-transferase, alpha 2 (Yc2) | Glutathione metabolic process, response to bacterium and stilbenoid, xenobiotic metabolic process | 176.3 | 444.3 | 1.58 | 0.0026 |
| Csad | Cysteine sulfinic acid decarboxylase | Amino acid metabolism | 1955.8 | 4247.9 | 1.57 | 0.0029 |
| Enho | Energy homeostasis associated | Negative regulation of lipid biosynthetic process | 84.9 | 234.9 | 1.57 | 0.0030 |
| Gsta4 | Glutathione S-transferase, alpha 4 | Drug binding, glutathione metabolic process | 619.4 | 1065.4 | 1.56 | <0.0001 |
| Gene | Name | Functions in Mouse | Mean (LF) | Mean (HF) * | Mean (HF + LGB) * | FC (HF vs. LF) | p-Value (HF vs. LF) | FC (HF + LGB vs. HF) | p-Value (HF + LGB vs. HF) |
|---|---|---|---|---|---|---|---|---|---|
| Mogat1 | Monoacylglycerol O-acyltransferase 1 | Lipid metabolic process | 19.5 | 68.4 | 27.1 | 2.51 | <0.0001 | −1.69 | 0.0003 |
| Tmem2 | Transmembrane protein 28 | Calcium ion transport | 27.4 | 79.5 | 28.5 | 2.13 | <0.0001 | −1.73 | 0.0001 |
| Ifi27l2b | Interferon, alpha-inducible protein 27-like 2B | Regulation of growth | 44.1 | 97.9 | 31.2 | 1.83 | <0.0001 | −2.04 | <0.0001 |
| Gpc1 | Glypican 1 | Cell migration | 293.2 | 535.1 | 211.2 | 1.78 | <0.0001 | −2.04 | <0.0001 |
| Cdkn1a | Cyclin-dependent kinase inhibitor 1A (P21) | Regulation of cell cycle | 46.7 | 108.6 | 49.8 | 1.73 | 0.0002 | −1.69 | <0.0001 |
| Tubb6 | Tubulin, beta 6 class V | Cell cycle | 42.2 | 85.3 | 44.7 | 1.68 | 0.0002 | −1.54 | 0.0033 |
| Pdlim2 | PDZ and LIM domain 2 | Actin cytoskeleton organization | 17.8 | 37.3 | 20.0 | 1.68 | 0.0005 | −1.56 | 0.0017 |
| Wfdc2 | WAP four-disulfide core domain 2 | Endopeptidase inhibitor activity | 92.0 | 179.5 | 54.2 | 1.66 | 0.0005 | −2.28 | <0.0001 |
| Lcn2 | Lipocalin 2 | Apoptotic process, inflammation | 54.7 | 165.9 | 42.2 | 1.66 | 0.0012 | −1.99 | <0.0001 |
| Saa1 | Serum amyloid A 1 | Acute-phase response, inflammation, cholesterol metabolic process | 513.5 | 1245.2 | 471.0 | 1.65 | 0.0014 | −1.75 | <0.0001 |
| Plin4 | Perilipin 4 | Lipid droplet-associated protein | 74.5 | 201.9 | 105.4 | 1.65 | 0.0016 | −1.55 | 0.0030 |
| Cxcl14 | Chemokine (C-X-C motif) ligand 14 | Immune response, inflammation | 13.2 | 25.9 | 8.4 | 1.60 | 0.0016 | −1.74 | 0.0001 |
| Saa2 | Serum amyloid A 2 | Acute-phase response, inflammation | 285.4 | 738.1 | 222.7 | 1.60 | 0.0030 | −1.83 | <0.0001 |
| Tceal8 | Transcription elongation factor A (SII)-like 8 | WW domain binding | 322.3 | 546.1 | 275.7 | 1.59 | <0.0001 | −1.74 | <0.0001 |
| Tubb2a | Tubulin, beta 2A class IIA | Cell cycle | 293.9 | 1038.1 | 274.6 | 1.59 | 0.0026 | −1.52 | 0.0038 |
| Orm3 | Orosomucoid 3 | 9.7 | 22.3 | 8.5 | 1.58 | 0.0046 | −1.52 | 0.0078 | |
| Phlda3 | Pleckstrin homology-like domain, family A, member 3 | Phosphatidylinositol-phosphates binding; apoptotic process positive regulation | 17.9 | 34.6 | 13.8 | 1.57 | 0.0037 | −1.78 | <0.0001 |
| Slc25a35 | Solute carrier family 25, member 35 | Mitochondrial inner membrane | 11.6 | 21.6 | 11.2 | 1.56 | 0.0024 | −1.53 | 0.0030 |
| S100a10 | S100 calcium binding protein A10 (calpactin) | Regulation of cell migration, inflammation | 1264.2 | 2097.6 | 1116.5 | 1.55 | <0.0001 | −1.66 | <0.0001 |
| Rad51b | RAD51 paralog B | DNA recombination and repair, positive regulation of cell proliferation | 31.9 | 87.1 | 23.3 | 1.54 | 0.0067 | −2.03 | <0.0001 |
| Gale | Galactose-4-epimerase, UDP | UDP-N-acetylglucosamine 4-epimerase activity, identical protein binding | 232.0 | 444.4 | 215.9 | 1.53 | 0.0067 | −1.57 | 0.0020 |
| Adgrf1 | Adhesion G protein-coupled receptor F1 | G-protein coupled receptor activity | 122.3 | 38.1 | 84.5 | −2.06 | <0.0001 | 2.22 | <0.0001 |
| Igfbp2 | Insulin-like growth factor binding protein 2 | Glucose metabolism, insulin sensitivity | 7680.0 | 3501.8 | 6263.7 | −1.95 | <0.0002 | 1.71 | <0.0001 |
| GO Term | Description | p-Value (FDR adj.) |
|---|---|---|
| HF vs. LF | ||
| GO:0006629 | Lipid metabolic process | 0.0005 |
| GO:0035634 | Response to stilbenoid | 0.0012 |
| GO:0050727 | Regulation of inflammatory response | 0.0222 |
| GO:0071404 | Cellular response to low-density lipoprotein particle stimulus | 0.0324 |
| GO:0044255 | Cellular lipid metabolic process | 0.0367 |
| HF vs. HF + LGB | ||
| GO:0006629 | Lipid metabolic process | 4.17 × 10−5 |
| GO:0072330 | Monocarboxylic acid biosynthetic process | 0.0030 |
| GO:0035634 | Response to stilbenoid | 0.0042 |
| GO:0005975 | Carbohydrate metabolic process | 0.0042 |
| GO:0033559 | Unsaturated fatty acid metabolic process | 0.0073 |
| GO:0017144 | Drug metabolic process | 0.0086 |
| GO:0042866 | Pyruvate biosynthetic process | 0.0128 |
| GO:0055114 | Oxidation-reduction process | 0.0157 |
| GO:0006690 | Eicosanoid metabolic process | 0.0168 |
| GO:0046890 | Regulation of lipid biosynthetic process | 0.0190 |
| GO:0044262 | Cellular carbohydrate metabolic process | 0.0202 |
| GO:0051156 | Glucose 6-phosphate metabolic process | 0.0241 |
| GO:0032429 | Regulation of phospholipase A2 activity | 0.0255 |
| GO:1901135 | Carbohydrate derivative metabolic process | 0.0246 |
| GO:0008202 | Steroid metabolic process | 0.0288 |
| GO:0019216 | Regulation of lipid metabolic process | 0.0294 |
| GO:0006637 | Acyl-CoA metabolic process | 0.0355 |
| GO:0006953 | Acute-phase response | 0.0419 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ryyti, R.; Pemmari, A.; Peltola, R.; Hämäläinen, M.; Moilanen, E. Effects of Lingonberry (Vaccinium vitis-idaea L.) Supplementation on Hepatic Gene Expression in High-Fat Diet Fed Mice. Nutrients 2021, 13, 3693. https://doi.org/10.3390/nu13113693
Ryyti R, Pemmari A, Peltola R, Hämäläinen M, Moilanen E. Effects of Lingonberry (Vaccinium vitis-idaea L.) Supplementation on Hepatic Gene Expression in High-Fat Diet Fed Mice. Nutrients. 2021; 13(11):3693. https://doi.org/10.3390/nu13113693
Chicago/Turabian StyleRyyti, Riitta, Antti Pemmari, Rainer Peltola, Mari Hämäläinen, and Eeva Moilanen. 2021. "Effects of Lingonberry (Vaccinium vitis-idaea L.) Supplementation on Hepatic Gene Expression in High-Fat Diet Fed Mice" Nutrients 13, no. 11: 3693. https://doi.org/10.3390/nu13113693
APA StyleRyyti, R., Pemmari, A., Peltola, R., Hämäläinen, M., & Moilanen, E. (2021). Effects of Lingonberry (Vaccinium vitis-idaea L.) Supplementation on Hepatic Gene Expression in High-Fat Diet Fed Mice. Nutrients, 13(11), 3693. https://doi.org/10.3390/nu13113693






