Vitamin A Transporters in Visual Function: A Mini Review on Membrane Receptors for Dietary Vitamin A Uptake, Storage, and Transport to the Eye
Abstract
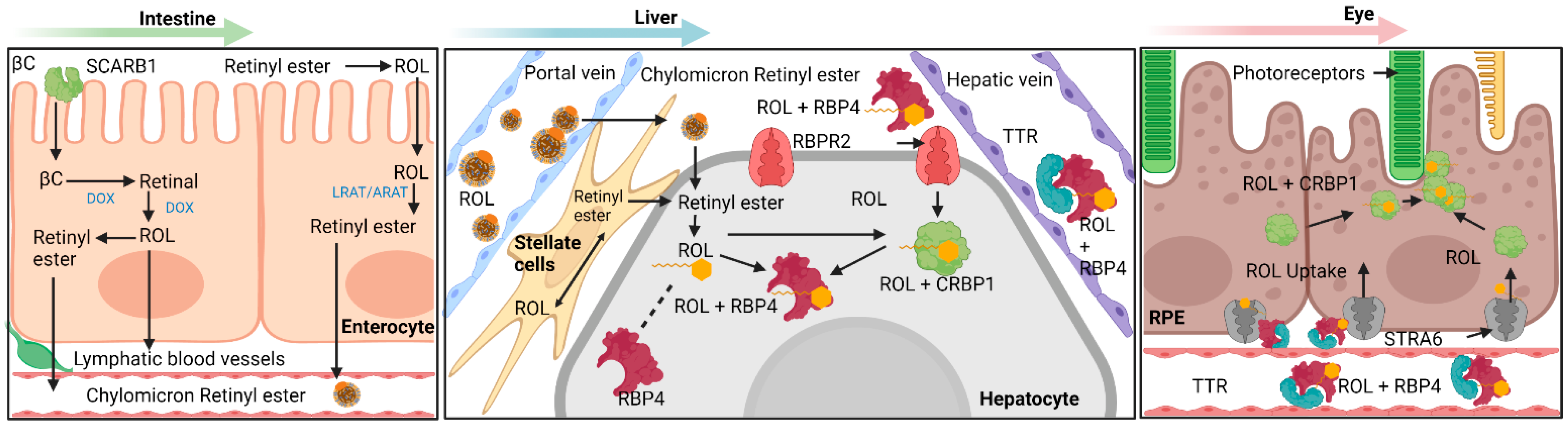
1. Mechanisms Involving Intestinal Absorption of Provitamin A Carotenoids and Preformed Vitamin A
2. Uptake of Carotenoids—SR-B1
2.1. Carotenoid Cleaving Enzymes—BCO1, BCO2
2.2. An Intestinal Transcription Factor Regulates Vitamin A Absorption—ISX
3. Transport of All-Trans Retinol to Extrahepatic Organs—RBP4 and Retinol
4. Known Vitamin A Transporters/RBP4 Receptors
4.1. Stimulated by Retinoic Acid 6 (STRA6)
4.2. Retinol Binding Protein 4 Receptor 2 (RBPR2) in Whole-Body Vitamin A Homeostasis
5. All-Trans Retinoic Acid as a Transcription Factor
6. Vitamin A Metabolites—Retinoids (All-Trans-Retinoic Acid) in Development
7. Vitamin A in Vision
8. Concluding Remarks and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Borel, P.; Desmarchelier, C. Genetic Variations Associated with Vitamin A Status and Vitamin A Bioavailability. Nutrients 2017, 9, 246. [Google Scholar] [CrossRef] [PubMed]
- Kopec, R.E.; Gleize, B.; Borel, P.; Desmarchelier, C.; Caris-Veyrat, C. Are lutein, lycopene, and β-carotene lost through the digestive process? Food Funct. 2017, 8, 1494–1503. [Google Scholar] [CrossRef]
- Bohn, T.; Desmarchelier, C.; El, S.N.; Keijer, J.; Van Schothorst, E.; Rühl, R.; Borel, P. β-Carotene in the human body: Metabolic bioactivation pathways—From digestion to tissue distribution and excretion. Proc. Nutr. Soc. 2019, 78, 68–87. [Google Scholar] [CrossRef] [PubMed]
- Von Lintig, J.; Moon, J.; Lee, J.; Ramkumar, S. Carotenoid metabolism at the intestinal barrier. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2020, 1865, 158580. [Google Scholar] [CrossRef]
- Von Lintig, J.; Moon, J.; Babino, D. Molecular components affecting ocular carotenoid and retinoid homeostasis. Prog. Retin. Eye Res. 2021, 80, 100864. [Google Scholar] [CrossRef]
- Ramkumar, S.; Moon, J.; Golczak, M.; von Lintig, J. LRAT coordinates the negative-feedback regulation of intestinal retinoid biosynthesis from β-carotene. J. Lipid Res. 2021, 62, 100055. [Google Scholar] [CrossRef]
- Blomhoff, R. Transport and Metabolism of Vitamin A. Nutr. Rev. 2009, 52, S13–S23. [Google Scholar] [CrossRef] [PubMed]
- Chelstowska, S.; Widjaja-Adhi, M.A.K.; Silvaroli, J.A.; Golczak, M. Molecular Basis for Vitamin A Uptake and Storage in Vertebrates. Nutrients 2016, 8, 676. [Google Scholar] [CrossRef]
- Chadwick, A.C.; Jensen, D.R.; Hanson, P.J.; Lange, P.T.; Proudfoot, S.C.; Peterson, F.C.; Volkman, B.F.; Sahoo, D. NMR Structure of the C-Terminal Transmembrane Domain of the HDL Receptor, SR-BI, and a Functionally Relevant Leucine Zipper Motif. Structure 2017, 25, 446–457. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Valacchi, G.; Sticozzi, C.; Lim, Y.; Pecorelli, A. Scavenger receptor class B type I: A multifunctional receptor. Ann. N. Y. Acad. Sci. 2011, 1229, E1–E7. [Google Scholar] [CrossRef]
- Widjaja-Adhi, M.A.K.; Lobo, G.P.; Golczak, M.; Von Lintig, J. A genetic dissection of intestinal fat-soluble vitamin and carotenoid absorption. Hum. Mol. Genet. 2015, 24, 3206–3219. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, C.; Sumser, E.; Wernet, M.F.; von Lintig, J. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc. Natl. Acad. Sci. USA 2002, 99, 10581–10586. [Google Scholar] [CrossRef] [PubMed]
- Lobo, G.P.; Amengual, J.; Palczewski, G.; Babino, D.; von Lintig, J. Mammalian Carotenoid-oxygenases: Key players for carotenoid function and homeostasis. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2012, 1821, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Amengual, J.; Golczak, M.; Palczewski, K.; von Lintig, J. Lecithin:Retinol Acyltransferase Is Critical for Cellular Uptake of Vitamin A from Serum Retinol-binding Protein. J. Biol. Chem. 2012, 287, 24216–24227. [Google Scholar] [CrossRef]
- Von Lintig, J.; Hessel, S.; Isken, A.; Kiefer, C.; Lampert, J.M.; Voolstra, O.; Vogt, K. Towards a better understanding of carotenoid metabolism in animals. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2005, 1740, 122–131. [Google Scholar] [CrossRef]
- Sui, X.; Kiser, P.D.; von Lintig, J.; Palczewski, K. Structural basis of carotenoid cleavage: From bacteria to mammals. Arch. Biochem. Biophys. 2013, 539, 203–213. [Google Scholar] [CrossRef]
- Shete, V.; Quadro, L. Mammalian Metabolism of β-Carotene: Gaps in Knowledge. Nutrients 2013, 5, 4849–4868. [Google Scholar] [CrossRef]
- Lobo, G.P.; Hessel, S.; Eichinger, A.; Noy, N.; Moise, A.R.; Wyss, A.; Palczewski, K.; Von Lintig, J. ISX is a retinoic acid-sensitive gatekeeper that controls intestinal β,β-carotene absorption and vitamin A production. FASEB J. 2010, 24, 1656–1666. [Google Scholar] [CrossRef]
- O’Byrne, S.M.; Blaner, W.S. Retinol and retinyl esters: Biochemistry and physiology. J. Lipid Res. 2013, 54, 1731–1743. [Google Scholar] [CrossRef]
- Blaner, W.S. Vitamin A signaling and homeostasis in obesity, diabetes, and metabolic disorders. Pharmacol. Ther. 2019, 197, 153–178. [Google Scholar] [CrossRef]
- Thompson, S.J.; Sargsyan, A.; Lee, S.-A.; Yuen, J.J.; Cai, J.; Smalling, R.; Ghyselinck, N.; Mark, M.; Blaner, W.S.; Graham, T.E. Hepatocytes Are the Principal Source of Circulating RBP4 in Mice. Diabetes 2016, 66, 58–63. [Google Scholar] [CrossRef]
- Van Bennekum, A.M.; Wei, S.; Gamble, M.V.; Vogel, S.; Piantedosi, R.; Gottesman, M.; Episkopou, V.; Blaner, W.S. Biochemical Basis for Depressed Serum Retinol Levels in Transthyretin-deficient Mice. J. Biol. Chem. 2001, 276, 1107–1113. [Google Scholar] [CrossRef]
- Shirakami, Y.; Lee, S.-A.; Clugston, R.; Blaner, W.S. Hepatic metabolism of retinoids and disease associations. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2012, 1821, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wongsiriroj, N.; Blaner, W.S. The multifaceted nature of retinoid transport and metabolism. Hepatobiliary Surg. Nutr. 2014, 3, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Episkopou, V.; Piantedosi, R.; Maeda, S.; Shimada, K.; Gottesman, M.E.; Blaner, W.S. Studies on the Metabolism of Retinol and Retinol-binding Protein in Transthyretin-deficient Mice Produced by Homologous Recombination. J. Biol. Chem. 1995, 270, 866–870. [Google Scholar] [CrossRef] [PubMed]
- Quadro, L.; Blaner, W.S.; Salchow, D.J.; Vogel, S.; Piantedosi, R.; Gouras, P.; Freeman, S.; Cosma, M.P.; Colantuoni, V.; Gottesman, M.E. Impaired retinal function and vitamin A availability in mice lacking retinol-binding protein. EMBO J. 1999, 18, 4633–4644. [Google Scholar] [CrossRef]
- Blaner, W.S. STRA6, a Cell-Surface Receptor for Retinol-Binding Protein: The Plot Thickens. Cell Metab. 2007, 5, 164–166. [Google Scholar] [CrossRef]
- Kawaguchi, R.; Yu, J.; Honda, J.; Hu, J.; Whitelegge, J.; Ping, P.; Wiita, P.; Bok, D.; Sun, H. A Membrane Receptor for Retinol Binding Protein Mediates Cellular Uptake of Vitamin A. Science 2007, 315, 820–825. [Google Scholar] [CrossRef]
- Kelly, M.; von Lintig, J. STRA6: Role in Cellular Retinol Uptake and Efflux. Hepatobiliary Surg. Nutr. 2015, 4, 229. [Google Scholar]
- Bouillet, P.; Sapin, V.; Chazaud, C.; Messaddeq, N.; Décimo, D.; Dollé, P.; Chambon, P. Developmental expression pattern of Stra6, a retinoic acid-responsive gene encoding a new type of membrane protein. Mech. Dev. 1997, 63, 173–186. [Google Scholar] [CrossRef]
- Kawaguchi, R.; Zhong, M.; Kassai, M.; Ter-Stepanian, M.; Sun, H. STRA6-Catalyzed Vitamin A Influx, Efflux, and Exchange. J. Membr. Biol. 2012, 245, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Clarke, O.B.; Kim, J.; Stowe, S.; Kim, Y.-K.; Assur, Z.; Cavalier, M.; Godoy-Ruiz, R.; von Alpen, D.C.; Manzini, C.; et al. Structure of the STRA6 receptor for retinol uptake. Science 2016, 353, 353. [Google Scholar] [CrossRef] [PubMed]
- Pasutto, F.; Sticht, H.; Hammersen, G.; Gillessen-Kaesbach, G.; FitzPatrick, D.R.; Nürnberg, G.; Brasch, F.; Schirmer-Zimmermann, H.; Tolmie, J.L.; Chitayat, D.; et al. Mutations in STRA6 Cause a Broad Spectrum of Malformations Including Anophthalmia, Congenital Heart Defects, Diaphragmatic Hernia, Alveolar Capillary Dysplasia, Lung Hypoplasia, and Mental Retardation. Am. J. Hum. Genet. 2007, 80, 550–560. [Google Scholar] [CrossRef]
- Golzio, C.; Martinovic-Bouriel, J.; Thomas, S.; Mougou-Zrelli, S.; Grattagliano-Bessières, B.; Bonnière, M.; Delahaye, S.; Munnich, A.; Encha-Razavi, F.; Lyonnet, S.; et al. Matthew-Wood Syndrome Is Caused by Truncating Mutations in the Retinol-Binding Protein Receptor Gene STRA6. Am. J. Hum. Genet. 2007, 80, 1179–1187. [Google Scholar] [CrossRef]
- Isken, A.; Golczak, M.; Oberhauser, V.; Hunzelmann, S.; Driever, W.; Imanishi, Y.; Palczewski, K.; von Lintig, J. RBP4 Disrupts Vitamin A Uptake Homeostasis in a STRA6-Deficient Animal Model for Matthew-Wood Syndrome. Cell Metab. 2008, 7, 258–268. [Google Scholar] [CrossRef]
- Ruiz, A.; Mark, M.; Jacobs, H.; Klopfenstein, M.; Hu, J.; Lloyd, M.; Habib, S.; Tosha, C.; A Radu, R.; Ghyselinck, N.B.; et al. Retinoid Content, Visual Responses, and Ocular Morphology Are Compromised in the Retinas of Mice Lacking the Retinol-Binding Protein Receptor, STRA6. Investig. Opthalmol. Vis. Sci. 2012, 53, 3027–3039. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.C.; Jacobs, H.; Marwarha, G.; Gely-Pernot, A.; O’Byrne, S.M.; DeSantis, D.; Klopfenstein, M.; Feret, B.; Dennefeld, C.; Blaner, W.S.; et al. The STRA6 Receptor Is Essential for Retinol-binding Protein-induced Insulin Resistance but Not for Maintaining Vitamin A Homeostasis in Tissues Other Than the Eye. J. Biol. Chem. 2013, 288, 24528–24539. [Google Scholar] [CrossRef]
- Amengual, J.; Zhang, N.; Kemerer, M.; Maeda, T.; Palczewski, K.; von Lintig, J. STRA6 is critical for cellular vitamin A uptake and homeostasis. Hum. Mol. Genet. 2014, 23, 5402–5417. [Google Scholar] [CrossRef]
- Alapatt, P.; Guo, F.; Komanetsky, S.M.; Wang, S.; Cai, J.; Sargsyan, A.; Diaz, E.; Bacon, B.; Aryal, P.; Graham, T.E. Liver Retinol Transporter and Receptor for Serum Retinol-binding Protein (RBP4). J. Biol. Chem. 2013, 288, 1250–1265. [Google Scholar] [CrossRef]
- Shi, Y.; Obert, E.; Rahman, B.; Rohrer, B.; Lobo, G.P. The Retinol Binding Protein Receptor 2 (Rbpr2) is required for Photoreceptor Outer Segment Morphogenesis and Visual Function in Zebrafish. Sci. Rep. 2017, 7, 16207. [Google Scholar] [CrossRef] [PubMed]
- During, A.; Harrison, E.H. Mechanisms of provitamin A (carotenoid) and vitamin A (retinol) transport into and out of intestinal Caco-2 cells. J. Lipid Res. 2007, 48, 2283–2294. [Google Scholar] [CrossRef] [PubMed]
- Solanki, A.K.; Kondkar, A.A.; Fogerty, J.; Su, Y.; Kim, S.-H.; Lipschutz, J.H.; Nihalani, D.; Perkins, B.D.; Lobo, G.P. A Functional Binding Domain in the Rbpr2 Receptor Is Required for Vitamin A Transport, Ocular Retinoid Homeostasis, and Photoreceptor Cell Survival in Zebrafish. Cells 2020, 9, 1099. [Google Scholar] [CrossRef]
- Costabile, B.K.; Kim, Y.-K.; Chen, Y.; Clarke, O.B.; Quadro, L.; Mancia, F. Sample preparation for structural and functional analyses of the STRA6 receptor for retinol-binding protein. In Methods in Enzymology; Pohl, E., Ed.; Academic Press: Cambridge, MA, USA, 2020; Volume 637, pp. 95–117. [Google Scholar]
- Le Maire, A.; Teyssier, C.; Balaguer, P.; Bourguet, W.; Germain, P. Regulation of RXR-RAR Heterodimers by RXR- and RAR-Specific Ligands and Their Combinations. Cells 2019, 8, 1392. [Google Scholar] [CrossRef] [PubMed]
- Das, B.C.; Thapa, P.; Verma, A.; Ray, S.K.; Evans, T.; Karki, R.; Das, S.; Mahapatra, S.; Liu, T.-C.; Torregroza, I.; et al. Retinoic acid signaling pathways in development and diseases. Bioorg. Med. Chem. 2014, 22, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, R.E.; Putzke, A.P.; Myers, J.P.; Margaretha, L.; Moens, C.B. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development 2007, 134, 177–187. [Google Scholar] [CrossRef]
- White, J.A.; Guo, Y.-D.; Baetz, K.; Beckett-Jones, B.; Bonasoro, J.; Hsu, K.E.; Dilworth, F.J.; Jones, G.; Petkovich, M. Identification of the Retinoic Acid-inducible All-trans-retinoic Acid 4-Hydroxylase. J. Biol. Chem. 1996, 271, 29922–29927. [Google Scholar] [CrossRef] [PubMed]
- Niederreither, K.; Dollé, P. Retinoic acid in development: Towards an integrated view. Nat. Rev. Genet. 2008, 9, 541–553. [Google Scholar] [CrossRef]
- Niederreither, K.; Abu-Abed, S.; Schuhbaur, B.; Petkovich, M.; Chambon, P.; Dollé, P. Genetic evidence that oxidative derivatives of retinoic acid are not involved in retinoid signaling during mouse development. Nat. Genet. 2002, 31, 84–88. [Google Scholar] [CrossRef]
- Cvekl, A.; Ashery-Padan, R. The cellular and molecular mechanisms of vertebrate lens development. Development 2014, 141, 4432–4447. [Google Scholar] [CrossRef]
- LE, H.-G.T.; Dowling, J.E.; Cameron, D.J. Early retinoic acid deprivation in developing zebrafish results in microphthalmia. Vis. Neurosci. 2012, 29, 219–228. [Google Scholar] [CrossRef]
- Kiser, P.D.; Palczewski, K. Retinoids and Retinal Diseases. Annu. Rev. Vis. Sci. 2016, 2, 197–234. [Google Scholar] [CrossRef]
- Kiser, P.D.; Golczak, M.; Maeda, A.; Palczewski, K. Key enzymes of the retinoid (visual) cycle in vertebrate retina. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2012, 1821, 137–151. [Google Scholar] [CrossRef]
- Shichida, Y.; Morizumi, T. Mechanism of G-protein Activation by Rhodopsin. Photochem. Photobiol. 2006, 83, 70–75. [Google Scholar] [CrossRef]
- Maeda, A.; Maeda, T.; Imanishi, Y.; Kuksa, V.; Alekseev, A.; Bronson, J.D.; Zhang, H.; Zhu, L.; Sun, W.; Saperstein, D.A.; et al. Role of Photoreceptor-specific Retinol Dehydrogenase in the Retinoid Cycle in Vivo. J. Biol. Chem. 2005, 280, 18822–18832. [Google Scholar] [CrossRef] [PubMed]
- Mata, N.L.; Radu, R.A.; Clemmons, R.S.; Travis, G.H. Isomerization and Oxidation of Vitamin A in Cone-Dominant Retinas: A Novel Pathway for Visual-Pigment Regeneration in Daylight. Neuron 2002, 36, 69–80. [Google Scholar] [CrossRef]
- Kiser, P.D.; Golczak, M.; Palczewski, K. Chemistry of the Retinoid (Visual) Cycle. Chem. Rev. 2014, 114, 194–232. [Google Scholar] [CrossRef] [PubMed]
- Bereta, G.; Kiser, P.D.; Golczak, M.; Sun, W.; Heon, E.; Saperstein, D.A.; Palczewski, K. Impact of Retinal Disease-Associated RPE65 Mutations on Retinoid Isomerization. Biochemistry 2008, 47, 9856–9865. [Google Scholar] [CrossRef]
- Kiser, P.D.; Golczak, M.; Lodowski, D.T.; Chance, M.R.; Palczewski, K. Crystal structure of native RPE65, the retinoid isomerase of the visual cycle. Proc. Natl. Acad. Sci. USA 2009, 106, 17325–17330. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Jastrzebska, B.; Golczak, M.; Gulati, S.; Tang, H.; Seibel, W.; Li, X.; Jin, H.; Han, Y.; et al. A novel small molecule chaperone of rod opsin and its potential therapy for retinal degeneration. Nat. Commun. 2018, 9, 1976. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin Ask, N.; Leung, M.; Radhakrishnan, R.; Lobo, G.P. Vitamin A Transporters in Visual Function: A Mini Review on Membrane Receptors for Dietary Vitamin A Uptake, Storage, and Transport to the Eye. Nutrients 2021, 13, 3987. https://doi.org/10.3390/nu13113987
Martin Ask N, Leung M, Radhakrishnan R, Lobo GP. Vitamin A Transporters in Visual Function: A Mini Review on Membrane Receptors for Dietary Vitamin A Uptake, Storage, and Transport to the Eye. Nutrients. 2021; 13(11):3987. https://doi.org/10.3390/nu13113987
Chicago/Turabian StyleMartin Ask, Nicasio, Matthias Leung, Rakesh Radhakrishnan, and Glenn P. Lobo. 2021. "Vitamin A Transporters in Visual Function: A Mini Review on Membrane Receptors for Dietary Vitamin A Uptake, Storage, and Transport to the Eye" Nutrients 13, no. 11: 3987. https://doi.org/10.3390/nu13113987
APA StyleMartin Ask, N., Leung, M., Radhakrishnan, R., & Lobo, G. P. (2021). Vitamin A Transporters in Visual Function: A Mini Review on Membrane Receptors for Dietary Vitamin A Uptake, Storage, and Transport to the Eye. Nutrients, 13(11), 3987. https://doi.org/10.3390/nu13113987





