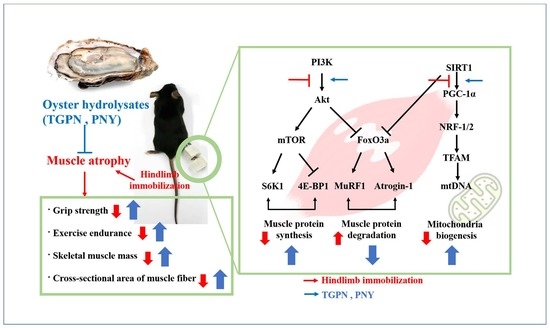
Oyster Hydrolysates Attenuate Muscle Atrophy via Regulating Protein Turnover and Mitochondria Biogenesis in C2C12 Cell and Immobilized Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of TGPN and PNY
2.2. Cell Culture
2.2.1. Cell Culture
2.2.2. Cell Viability Assay
2.3. Treatment with Dexamethasone and TGPN, PNY
2.4. Determination of C2C12 Myotube Diameter
2.5. Animals and Experimental Design
2.6. Muscle Function Test
2.6.1. Grip Strength Test
2.6.2. Treadmill Test
2.7. H&E Staining
2.8. Protein Extract and Western Blot Analysis
2.9. RNA Extract and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
2.10. Quantitative Analysis of Mitochondrial DNA
2.11. Statistical Analysis
3. Results
3.1. TGPN and PNY Protected against Dexamethasone-Induced Muscle Atrophy in C2C12
3.2. TGPN and PNY Ameliorated Muscle Function in Immobilization-Induced Muscle Atrophy Mice
3.3. TGPN and PNY Increased Muscle Mass and Cross-Sectional Area of Muscle Fiber in Immobilization-Induced Muscle Atrophy Mice
3.4. TGPN and PNY Stimulated Muscle Protein Synthesis and Blocked Muscle Protein Degradation via PI3K/Akt Pathway in Immobilization-Induced Muscle Atrophy Mice
3.5. TGPN and PNY Improved Mitochondrial Biogenesis in Immobilization-Induced Muscle Atrophy Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jung, S.; Kim, K. Exercise-induced PGC-1α transcriptional factors in skeletal muscle. Integr. Med. Res. 2014, 3, 155–160. [Google Scholar] [CrossRef] [Green Version]
- Yeung, S.S.; Yeung, S.S.Y.; Reijnierse, E.M.; Pham, V.K.; Trappenburg, M.C.; Lim, W.K.; Meskers, C.G.M.; Maier, A.B. Sarcopenia and its association with falls and fractures in older adults: A systematic review and meta-analysis. J. Cachexia Sarcopenia Muscle 2019, 10, 485–500. [Google Scholar] [CrossRef] [Green Version]
- Fuggle, N.; Shaw, S.; Dennison, E.; Cooper, C. Sarcopenia. Best Pract. Res. Clin. Rheumatol. 2017, 31, 218–242. [Google Scholar] [CrossRef] [Green Version]
- Altun, M.; Grönholdt-Klein, M.; Wang, L.; Ulfhake, B. Cellular degradation machineries in age-related loss of muscle mass (Sarcopenia). In Senescence; IntechOpen: Shanghai, China, 2012. [Google Scholar] [CrossRef] [Green Version]
- Fitts, R.H.; Riley, D.R.; Widrick, J.J. Functional and structural adaptations of skeletal muscle to microgravity. J. Exp. Biol. 2001, 204, 3201–3208. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.A.; Moylan, J.S.; Reid, M.B. Physical inactivity and muscle weakness in the critically ill. Crit. Care Med. 2009, 37, S337–S346. [Google Scholar] [CrossRef]
- Fitts, R.H.; Riley, D.R.; Widrick, J.J. Physiology of a microgravity environment invited review: Microgravity and skeletal muscle. J. Appl. Physiol. 2000, 89, 823–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackman, R.W.; Kandarian, S.C. The molecular basis of skeletal muscle atrophy. Am. J. Physiol. Cell Physiol. 2004, 287, C834–C843. [Google Scholar] [CrossRef] [Green Version]
- Thomason, D.B.; Booth, F.W. Atrophy of the soleus muscle by hindlimb unweighting. J. Appl. Physiol. 1990, 68, 1–12. [Google Scholar] [CrossRef]
- Morley, J.E.; Anker, S.D.; Von Haehling, S. Prevalence, incidence, and clinical impact of sarcopenia: Facts, numbers, and epidemiology—Update 2014. J. Cachexia Sarcopenia Muscle 2014, 5, 253–259. [Google Scholar] [CrossRef]
- Tay, L.; Ding, Y.Y.; Leung, B.P.; Ismail, N.H.; Yeo, A.; Yew, S.; Tay, K.S.; Tan, C.H.; Chong, M.S. Sex-specific differences in risk factors for sarcopenia amongst community-dwelling older adults. Age 2015, 37, 121. [Google Scholar] [CrossRef] [Green Version]
- Talbot, J.; Maves, L. Skeletal muscle fiber type: Using insights from muscle developmental biology to dissect targets for susceptibility and resistance to muscle disease. Wiley Interdiscip. Rev. Dev. Biol. 2016, 5, 518–534. [Google Scholar] [CrossRef] [Green Version]
- Röckl, K.S.; Hirshman, M.F.; Brandauer, J.; Fujii, N.; Witters, L.A.; Goodyear, L.J. Skeletal muscle adaptation to exercise training: AMP-activated protein kinase mediates muscle fiber type shift. Diabetes 2007, 56, 2062–2069. [Google Scholar] [CrossRef] [Green Version]
- Schiaffino, S.; Reggiani, C. Myosin isoforms in mammalian skeletal muscle. J. Appl. Physiol. 1994, 77, 493–501. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, Y.; Wu, W.; Hou, L.; Chen, H.; Zuo, B.; Xiong, Y.; Yang, J. Skeletal muscle-specific overexpression of PGC-1α induces fiber-type conversion through enhanced mitochondrial respiration and fatty acid oxidation in mice and pigs. Int. J. Biol. Sci. 2017, 13, 1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, M.A.; Polgar, J.; Weightman, D.; Appleton, D. Data on the distribution of fibre types in thirty-six human muscles: An autopsy study. J. Neurol. Sci. 1973, 18, 111–129. [Google Scholar] [CrossRef]
- Pette, D.; Spamer, C. Metabolie properties of muscle fibers. Fed Proe 1986, 45, 2910–2914. [Google Scholar]
- Arany, Z.; Lebrasseur, N.; Morris, C.; Smith, E.; Yang, W.; Ma, Y.; Chin, S.; Spiegelman, B.M. The transcriptional coactivator PGC-1β drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 2007, 5, 35–46. [Google Scholar] [CrossRef] [Green Version]
- Palus, S.; Springer, J.I.; Doehner, W.; von Haehling, S.; Anker, M.; Anker, S.D.; Springer, J. Models of sarcopenia: Short review. Int. J. Cardiol. 2017, 238, 19–21. [Google Scholar] [CrossRef] [PubMed]
- Fanzani, A.; Conraads, V.M.; Penna, F.; Martinet, W. Molecular and cellular mechanisms of skeletal muscle atrophy: An update. J. Cachexia Sarcopenia Muscle 2012, 3, 163–179. [Google Scholar] [CrossRef]
- Cesarone, G.; Edupuganti, O.P.; Chen, C.-P.; Wickstrom, E. Insulin receptor substrate 1 knockdown in human MCF7 ER+ breast cancer cells by nuclease-resistant IRS1 siRNA conjugated to a disulfide-bridged D-peptide analogue of insulin-like growth factor 1. Bioconjug. Chem. 2007, 18, 1831–1840. [Google Scholar] [CrossRef]
- Zhang, Y.; Aguilar, O.A.; Storey, K.B. Transcriptional activation of muscle atrophy promotes cardiac muscle remodeling during mammalian hibernation. PeerJ 2016, 4, e2317. [Google Scholar] [CrossRef] [Green Version]
- Fanin, M.; Nascimbeni, A.; Angelini, C. Muscle atrophy in L imb G irdle M uscular D ystrophy 2 A: A morphometric and molecular study. Neuropathol. Appl. Neurobiol. 2013, 39, 762–771. [Google Scholar] [CrossRef] [PubMed]
- Sandri, M.; Lin, J.; Handschin, C.; Yang, W.; Arany, Z.P.; Lecker, S.H.; Goldberg, A.L.; Spiegelman, B.M. PGC-1α protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc. Natl. Acad. Sci. USA 2006, 103, 16260–16265. [Google Scholar] [CrossRef] [Green Version]
- Jäger, S.; Handschin, C.; St-Pierre, J.; Spiegelman, B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proc. Natl. Acad. Sci. USA 2007, 104, 12017–12022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rygiel, K.A.; Picard, M.; Turnbull, D.M. The ageing neuromuscular system and sarcopenia: A mitochondrial perspective. J. Physiol. 2016, 594, 4499–4512. [Google Scholar] [CrossRef]
- Hwang, D.; Kang, M.-J.; Jo, M.J.; Seo, Y.B.; Park, N.G.; Kim, G.-D. Anti-inflammatory activity of β-thymosin peptide derived from pacific oyster (Crassostrea gigas) on NO and PGE2 production by down-regulating NF-κB in LPS-induced RAW264. 7 macrophage cells. Mar. Drugs 2019, 17, 129. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.-K.; Kim, Y.-S.; Lee, S.-J.; Jeon, Y.-J.; Lee, J.D.; Son, T.-I.; Ahn, C.-B.; Kim, Y.-T.; Moon, S.-H.; Jeon, B.-T.; et al. Anticancer effect of lipids partially purified from Pacific oyster, Crassostrea gigas on PC3 cells. Food Sci. Biotechnol. 2010, 19, 213–217. [Google Scholar] [CrossRef]
- Cheong, K.-L.; Xia, L.-X.; Liu, Y. Isolation and characterization of polysaccharides from oysters (Crassostrea gigas) with anti-tumor activities using an aqueous two-phase system. Mar. Drugs 2017, 15, 338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.-H.; Moon, S.; Xie, C.; Choung, S.-Y. Protective effects of enzymatic oyster hydrolysate on acetaminophen-induced HepG-2 cell damage. J. Korean Soc. Food Sci. Nutr. 2014, 43, 1166–1173. [Google Scholar] [CrossRef]
- Xie, C.-L.; Kang, S.S.; Lu, C.; Choi, Y.J. Quantification of multifunctional dipeptide YA from oyster hydrolysate for quality control and efficacy evaluation. BioMed Res. Int. 2018, 2018, 1–10. [Google Scholar] [CrossRef]
- Han, J.-H.; Bang, J.S.; Choi, Y.J.; Choung, S.-Y. Oral administration of oyster (Crassostrea gigas) hydrolysates protects against wrinkle formation by regulating the MAPK pathway in UVB-irradiated hairless mice. Photochem. Photobiol. Sci. 2019, 18, 1436–1446. [Google Scholar] [CrossRef]
- You, J.-S.; Anderson, G.B.; Dooley, M.S.; Hornberger, T.A. The role of mTOR signaling in the regulation of protein synthesis and muscle mass during immobilization in mice. Dis. Model Mech. 2015, 8, 1059–1069. [Google Scholar] [CrossRef] [Green Version]
- Witteveen, E.; Hoogland, I.C.M.; Wieske, L.; Weber, N.C.; Verhamme, C.; Schultz, M.J.; van Schaik, I.N.; Horn, J. Assessment of intensive care unit-acquired weakness in young and old mice: An E. coli septic peritonitis model. Muscle Nerve 2016, 53, 127–133. [Google Scholar] [CrossRef] [Green Version]
- Buch, A.; Carmeli, E.; Boker, L.K.; Marcus, Y.; Shefer, G.; Kis, O.; Berner, Y.; Stern, N. Muscle function and fat content in relation to sarcopenia, obesity and frailty of old age—An overview. Exp. Gerontol. 2016, 76, 25–32. [Google Scholar] [CrossRef]
- Wall, B.T.; Dirks, M.L.; Snijders, T.; Senden, J.M.G.; Dolmans, J.; van Loon, L.J.C. Substantial skeletal muscle loss occurs during only 5 days of disuse. Acta Physiol. 2014, 210, 600–611. [Google Scholar] [CrossRef] [PubMed]
- Booth, F. Effect of limb immobilization on skeletal muscle. J. Appl. Physiol. 1982, 52, 1113–1118. [Google Scholar] [CrossRef] [PubMed]
- Han, M.J.; Shin, J.E.; Park, S.J.; Choung, S.-Y. Synergetic effect of soluble whey protein hydrolysate and Panax ginseng berry extract on muscle atrophy in hindlimb-immobilized C57BL/6 mice. J. Ginseng Res. 2021, 107, 2411–2502. [Google Scholar] [CrossRef]
- Malavaki, C.; Sakkas, G.J.; Mitrou, G.I.; Kalyva, A.; Stefanidis, I.; Myburgh, K.H.; Karatzaferi, C. Skeletal muscle atrophy: Disease-induced mechanisms may mask disuse atrophy. J. Muscle Res. Cell Motil. 2015, 36, 405–421. [Google Scholar] [CrossRef]
- Abdelmoez, A.M.; Puig, L.S.; Smith, J.A.B.; Gabriel, B.M.; Savikj, M.; Dollet, L.; Chibalin, A.V.; Krook, A.; Zierath, J.R.; Pillon, N.J. Comparative profiling of skeletal muscle models reveals heterogeneity of transcriptome and metabolism. Am. J. Physiol.-Cell Physiol. 2020, 318, C615–C626. [Google Scholar] [CrossRef] [Green Version]
- Bhatnagar, S.; Mittal, A.; Gupta, S.K.; Kumar, A. TWEAK causes myotube atrophy through coordinated activation of ubiquitin-proteasome system, autophagy, and caspases. J. Cell. Physiol. 2012, 227, 1042–1051. [Google Scholar] [CrossRef] [Green Version]
- Menconi, M.; Gonnella, P.; Petkova, V.; Lecker, S.; Hasselgren, P.-O. Dexamethasone and corticosterone induce similar, but not identical, muscle wasting responses in cultured L6 and C2C12 myotubes. J. Cell. Biochem. 2008, 105, 353–364. [Google Scholar] [CrossRef] [Green Version]
- Chacon-Cabrera, A.; Lund-Palau, H.; Gea, J.; Barreiro, E. Time-course of muscle mass loss, damage, and proteolysis in gastrocnemius following unloading and reloading: Implications in chronic diseases. PLoS ONE 2016, 11, e0164951. [Google Scholar] [CrossRef]
- Suzuki, H.; Yoshikawab, Y.; Tsujimotoc, H.; Kitaurad, T.; Muraokae, I. Clenbuterol accelerates recovery after immobilization-induced atrophy of rat hindlimb muscle. Acta Histochem. 2020, 122, 151453. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Kim, M.-B.; Hwang, J.-K. Red bean extract inhibits immobilization-induced muscle atrophy in C57BL/6N mice. J. Med. Food 2020, 23, 29–36. [Google Scholar] [CrossRef]
- White, C.; Dixon, K.; Samuel, D.; Stokes, M. Handgrip and quadriceps muscle endurance testing in young adults. Springerplus 2013, 2, 451. [Google Scholar] [CrossRef] [Green Version]
- Bonaldo, P.; Sandri, M. Cellular and molecular mechanisms of muscle atrophy. Dis. Model Mech. 2013, 6, 25–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]








| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| Atrogin-1 | AGAAAGAAAGACATTCAGAACA | GCTCCTTCGTACTTCCTT |
| TFAM | CACCCAGATGCAAAACTTTCAG | CTGCTCTTTATACTTGCTCACAG |
| NRF-1 | CGGTAGCATCACTGGCAGAA | GGATCTGGACCAGGCCATTA |
| NRF-2 | TGAAGTTCGCATTTTGATGGC | CTTTGGTCCTGGCATCTCTAC |
| GAPDH | CGGCCGCATCTTCTTGTG | CCGACCTTCACCATTTTGTCTAC |
| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| mtDNA | CCTATCCTTGCCATCAT | GAGGCTGTTGCTGTGAC |
| nDNA | ATGGAAAGCCTGCCATCATG | TCCTTGTTGTTCAGCATCAC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeon, S.-H.; Choung, S.-Y. Oyster Hydrolysates Attenuate Muscle Atrophy via Regulating Protein Turnover and Mitochondria Biogenesis in C2C12 Cell and Immobilized Mice. Nutrients 2021, 13, 4385. https://doi.org/10.3390/nu13124385
Jeon S-H, Choung S-Y. Oyster Hydrolysates Attenuate Muscle Atrophy via Regulating Protein Turnover and Mitochondria Biogenesis in C2C12 Cell and Immobilized Mice. Nutrients. 2021; 13(12):4385. https://doi.org/10.3390/nu13124385
Chicago/Turabian StyleJeon, So-Hyun, and Se-Young Choung. 2021. "Oyster Hydrolysates Attenuate Muscle Atrophy via Regulating Protein Turnover and Mitochondria Biogenesis in C2C12 Cell and Immobilized Mice" Nutrients 13, no. 12: 4385. https://doi.org/10.3390/nu13124385
APA StyleJeon, S. -H., & Choung, S. -Y. (2021). Oyster Hydrolysates Attenuate Muscle Atrophy via Regulating Protein Turnover and Mitochondria Biogenesis in C2C12 Cell and Immobilized Mice. Nutrients, 13(12), 4385. https://doi.org/10.3390/nu13124385