Transient Effect of Infant Formula Supplementation on the Intestinal Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects and Sample Collection
2.2. Bacterial DNA Extraction and Sequencing
2.3. Immune-Cell Phenotyping by Flow Cytometry
2.4. Vaccine Responses to Tetanus Toxoid
2.5. Statistical Analysis
3. Results
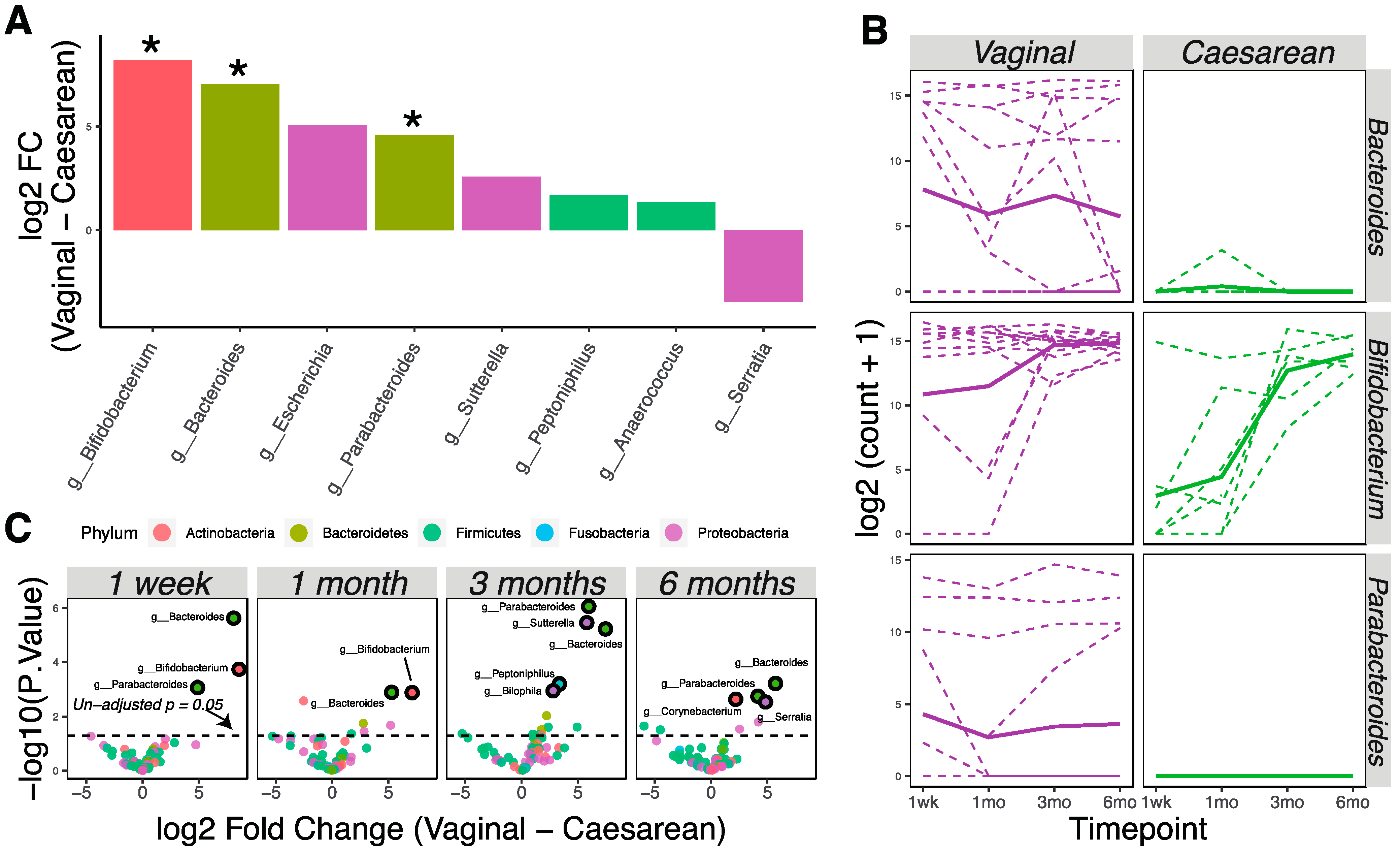
3.1. Different Gut Microbial Communities Associated with Different Ages, Diets, and Delivery Methods
3.2. Delivery Method Influences the Infant Gut Microbiota through 6 Months of Age
3.3. Changes in Microbial Communities Associated with Supplementation by 1 Month of Age
3.4. Characterization of Immune Development and Vaccine Responses of Supplemented and Unsupplemented Infants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eidelman, A.I.; Schanler, R.J. Breastfeeding and the Use of Human Milk. Pediatrics 2012, 129, e827–e841. [Google Scholar] [CrossRef] [Green Version]
- Kramer, M.S.; Kakuma, R. Optimal duration of exclusive breastfeeding. Cochrane Database Syst. Rev. 2012, 2012, CD003517. [Google Scholar] [CrossRef]
- Ladomenou, F.; Moschandreas, J.; Kafatos, A.; Tselentis, Y.; Galanakis, E. Protective effect of exclusive breastfeeding against infections during infancy: A prospective study. Arch. Dis. Child. 2010, 95, 1004–1008. [Google Scholar] [CrossRef] [PubMed]
- Duijts, L.; Jaddoe, V.W.V.; Hofman, A.; Moll, H.A. Prolonged and Exclusive Breastfeeding Reduces the Risk of Infectious Diseases in Infancy. Pediatrics 2010, 126, e18–e25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, C.; Bik, E.M.; DiGiulio, D.B.; Relman, D.A.; Brown, P.O. Development of the Human Infant Intestinal Microbiota. PLoS Biol. 2007, 5, e177. [Google Scholar] [CrossRef] [Green Version]
- Victora, C.G.; Bahl, R.; Barros, A.J.D.; França, G.V.; Horton, S.; Krasevec, J.; Murch, S.; Sankar, M.J.; Walker, N.; Rollins, N.C.; et al. Breastfeeding in the 21st century: Epidemiology, mechanisms, and lifelong effect. Lancet 2016, 387, 475–490. [Google Scholar] [CrossRef] [Green Version]
- Turin, C.G.; Ochoa, T.J. The Role of Maternal Breast Milk in Preventing Infantile Diarrhea in the Developing World. Curr. Trop. Med. Rep. 2014, 1, 97–105. [Google Scholar] [CrossRef] [Green Version]
- Zivkovic, A.M.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc. Natl. Acad. Sci. USA 2010, 108, 4653–4658. [Google Scholar] [CrossRef] [Green Version]
- Houghteling, P.D.; Walker, W.A. Why Is Initial Bacterial Colonization of the Intestine Important to Infants’ and Children’s Health? J. Pediatr. Gastroenterol. Nutr. 2015, 60, 294–307. [Google Scholar] [CrossRef] [Green Version]
- Underwood, M.A.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Bifidobacterium longum subspecies infantis: Champion colonizer of the infant gut. Pediatr. Res. 2015, 77, 229–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pacheco, A.R.; Barile, D.; Underwood, M.A.; Mills, D.A. The impact of the milk glycobiome on the neonate gut microbiota. Annu. Rev. Anim. Biosci. 2015, 3, 419–445. [Google Scholar] [CrossRef] [Green Version]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huda, M.N.; Lewis, Z.; Kalanetra, K.M.; Rashid, M.; Ahmad, S.M.; Raqib, R.; Qadri, F.; Underwood, M.A.; Mills, D.A.; Stephensen, C.B. Stool Microbiota and Vaccine Responses of Infants. Pediatrics 2014, 134, e362–e372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asarat, M.; Apostolopoulos, V.; Vasiljevic, T.; Donkor, O. Short-Chain Fatty Acids Regulate Cytokines and Th17/Treg Cells in Human Peripheral Blood Mononuclear Cellsin vitro. Immunol. Investig. 2016, 45, 205–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, S.; Friedberg, I.; Ivanov, I.V.; Davidson, L.A.; Goldsby, J.S.; Dahl, D.B.; Herman, D.; Wang, M.; Donovan, S.M.; Chapkin, R.S. A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in immune response. Genome Biol. 2012, 13, R32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henrick, B.M.; Chew, S.; Casaburi, G.; Brown, H.K.; Frese, S.A.; Zhou, Y.; Underwood, M.A.; Smilowitz, J.T. Colonization by B. infantis EVC001 modulates enteric inflammation in exclusively breastfed infants. Pediatr. Res. 2019, 86, 749–757. [Google Scholar] [CrossRef]
- Escribano, J.; Ferré, N.; Gispert-Llaurado, M.; Luque, V.; Rubio-Torrents, C.; Zaragoza-Jordana, M.; Polanco, I.; Codoñer, F.M.; Chenoll, E.; Morera, M.; et al. Bifidobacterium longum subsp infantis CECT7210-supplemented formula reduces diarrhea in healthy infants: A randomized controlled trial. Pediatr. Res. 2018, 83, 1120–1128. [Google Scholar] [CrossRef] [PubMed]
- Del Giudice, M.M.; Indolfi, C.; Capasso, M.; Maiello, N.; Decimo, F.; Ciprandi, G. Bifidobacterium mixture (B longum BB536, B infantis M-63, B breve M-16V) treatment in children with seasonal allergic rhinitis and intermittent asthma. Ital. J. Pediatr. 2017, 43, 25. [Google Scholar] [CrossRef] [Green Version]
- Centers for Disease Control and Prevention Breastfeeding Report Card. Available online: https://www.cdc.gov/breastfeeding/pdf/2018breastfeedingreportcard.pdf (accessed on 19 February 2020).
- Kuzniewicz, M.W.; Escobar, G.J.; Wi, S.; Liljestrand, P.; McCulloch, C.; Newman, T.B. Risk Factors for Severe Hyperbilirubinemia among Infants with Borderline Bilirubin Levels: A Nested Case-Control Study. J. Pediatr. 2008, 153, 234–240. [Google Scholar] [CrossRef] [Green Version]
- Lain, S.J.; Roberts, C.L.; Bowen, J.R.; Nassar, N. Early Discharge of Infants and Risk of Readmission for Jaundice. Pediatrics 2015, 135, 314–321. [Google Scholar] [CrossRef] [Green Version]
- Escobar, G.J.; Gonzales, V.M.; Armstrong, M.A.; Folck, B.F.; Xiong, B.; Newman, T.B. Rehospitalization for Neonatal Dehydration. Arch. Pediatr. Adolesc. Med. 2002, 156, 155–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flaherman, V.J.; Narayan, N.R.; Hartigan-O’Connor, D.; Cabana, M.D.; McCulloch, C.E.; Paul, I.M. The Effect of Early Limited Formula on Breastfeeding, Readmission, and Intestinal Microbiota: A Randomized Clinical Trial. J. Pediatr. 2018, 196, 84–90.e1. [Google Scholar] [CrossRef]
- Taft, D.; Ho, S.; Tancredi, D.; Stephensen, C.; Hinde, K.; Von Mutius, E.; Kirjavainen, P.; Dalphin, J.-C.; Lauener, R.; Riedler, J.; et al. Population Duration of Breastfeeding and Prevalence of Bifidobacterium Longum Subspecies Infantis (OR01-01-19). Curr. Dev. Nutr. 2019, 3, 93185. [Google Scholar] [CrossRef]
- Chung, H.; Pamp, S.J.; Hill, J.A.; Surana, N.K.; Edelman, S.M.; Troy, E.B.; Reading, N.C.; Villablanca, E.J.; Wang, S.; Mora, J.R.; et al. Gut Immune Maturation Depends on Colonization with a Host-Specific Microbiota. Cell 2012, 149, 1578–1593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamada, N.; Seo, S.-U.; Chen, G.Y.; Núñez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013, 13, 321–335. [Google Scholar] [CrossRef]
- Sjögren, Y.M.; Tomicic, S.; Lundberg, A.; Fagerås-Böttcher, M.; Björkstén, B.; Sverremark-Ekström, E.; Jenmalm, M.C. Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin. Exp. Allergy 2009, 39, 1842–1851. [Google Scholar] [CrossRef] [Green Version]
- Lundell, A.-C.; Björnsson, V.; Ljung, A.; Ceder, M.; Johansen, S.; Lindhagen, G.; Törnhage, C.-J.; Adlerberth, I.; Wold, A.E.; Rudin, A. Infant B Cell Memory Differentiation and Early Gut Bacterial Colonization. J. Immunol. 2012, 188, 4315–4322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gronlund, M.-M.; Arvilommi, H.; Kero, P.; Lehtonen, O.-P.; Isolauri, E. Importance of intestinal colonisation in the maturation of humoral immunity in early infancy: A prospective follow up study of healthy infants aged 0-6 months. Arch. Dis. Child. Fetal Neonatal Ed. 2000, 83, 186F–192F. [Google Scholar] [CrossRef] [Green Version]
- Huda, M.N.; Ahmad, S.M.; Alam, M.J.; Khanam, A.; Kalanetra, K.M.; Taft, D.H.; Raqib, R.; Underwood, M.A.; Mills, D.A.; Stephensen, C.B. Bifidobacterium Abundance in Early Infancy and Vaccine Response at 2 Years of Age. Pediatrics 2019, 143, e20181489. [Google Scholar] [CrossRef] [Green Version]
- Centers for Disease Control and Prevention (CDC). Updated Recommendations for Use of Tetanus Toxoid, Reduced Diphtheria Toxoid, and Acellular Pertussis Vaccine (Tdap) in Pregnant Women-Advisory Committee on Immunization Practices (ACIP), 2012. MMWR. Morb. Mortal. Wkly. Rep. 2013, 62, 131–135. [Google Scholar]
- Simon, A. FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinform. 2019. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 29 April 2019).
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing 2019. Available online: https://www.R-project.org/ (accessed on 29 April 2019).
- Lewis, Z.T.; Bokulich, N.A.; Kalanetra, K.M.; Ruiz-Moyano, S.; Underwood, M.A.; Mills, D.A. Use of bifidobacterial specific terminal restriction fragment length polymorphisms to complement next generation sequence profiling of infant gut communities. Anaerobe 2013, 19, 62–69. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLOS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, G.F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package 2019. Available online: https://github.com/vegandevs/vegan/ (accessed on 29 April 2019).
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Nieuwdorp, M.; Groen, A.K.; Zwinderman, A.H. Statistical evaluation of diet-microbe associations. BMC Microbiol. 2019, 19, 90. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef] [Green Version]
- Storey, J.D.; Bass, A.J.; Dabney, A.; Robinson, D. Qvalue: Q-Value Estimation for False Discovery Rate Control 2019. Available online: http://github.com/jdstorey/qvalue (accessed on 29 April 2019).
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stiemsma, L.T.; Michels, K.B. The Role of the Microbiome in the Developmental Origins of Health and Disease. Pediatrics 2018, 141, e20172437. [Google Scholar] [CrossRef] [Green Version]
- Ho, N.T.; Li, F.; Lee-Sarwar, K.A.; Tun, H.M.; Brown, B.P.; Pannaraj, P.S.; Bender, J.M.; Azad, M.B.; Thompson, A.L.; Weiss, S.T.; et al. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Penders, J.; Thijs, C.; Vink, C.; Stelma, F.F.; Snijders, B.; Kummeling, I.; Van den Brandt, P.A.; Stobberingh, E.E. Factors Influencing the Composition of the Intestinal Microbiota in Early Infancy. Pediatrics 2006, 118, 511–521. [Google Scholar] [CrossRef] [Green Version]
- Biasucci, G.; Rubini, M.; Riboni, S.; Morelli, L.; Bessi, E.; Retetangos, C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum. Dev. 2010, 86, 13–15. [Google Scholar] [CrossRef]
- Azad, M.B.; Konya, T.; Maughan, H.; Guttman, D.S.; Field, C.J.; Chari, R.S.; Sears, M.R.; Becker, A.B.; Scott, J.A.; Kozyrskyj, A.L.; et al. Gut microbiota of healthy Canadian infants: Profiles by mode of delivery and infant diet at 4 months. Can. Med. Assoc. J. 2013, 185, 385–394. [Google Scholar] [CrossRef] [Green Version]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Forster, S.C.; Tsaliki, E.; Vervier, K.; Strang, A.; Simpson, N.; Kumar, N.; Stares, M.D.; Rodger, A.; Brocklehurst, P.; et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nat. Cell Biol. 2019, 574, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Dogra, S.; Sakwinska, O.; Soh, S.-E.; Ngom-Bru, C.; Brück, W.M.; Berger, B.; Brüssow, H.; Lee, Y.S.; Yap, F.; Chong, Y.-S.; et al. Dynamics of Infant Gut Microbiota Are Influenced by Delivery Mode and Gestational Duration and Are Associated with Subsequent Adiposity. mBio 2015, 6, e02419-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arrieta, M.-C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015, 7, 307ra152. [Google Scholar] [CrossRef] [PubMed]
- Stokholm, J.; Blaser, M.J.; Thorsen, J.; Rasmussen, M.A.; Waage, J.; Vinding, R.K.; Schoos, A.-M.M.; Kunøe, A.; Fink, N.R.; Chawes, B.L.; et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat. Commun. 2018, 9, 141. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; LeVan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W.; et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016, 22, 1187–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Gao, J.; Du, M.; Mao, X. Milk fat globule membrane supplementation modulates the gut microbiota and attenuates metabolic endotoxemia in high-fat diet-fed mice. J. Funct. Foods 2018, 47, 56–65. [Google Scholar] [CrossRef]
- Eboix-Amorós, A.; Ecollado, M.C.; Emira, A. Relationship between Milk Microbiota, Bacterial Load, Macronutrients, and Human Cells during Lactation. Front. Microbiol. 2016, 7, 492. [Google Scholar] [CrossRef] [Green Version]
- Holgerson, P.L.; Vestman, N.R.; Claesson, R.; Öhman, C.; Domellöf, M.; Tanner, A.C.; Hernell, O.; Johansson, I. Oral Microbial Profile Discriminates Breast-fed From Formula-fed Infants. J. Pediatr. Gastroenterol. Nutr. 2013, 56, 127–136. [Google Scholar] [CrossRef] [Green Version]
- Ardeshir, A.; Narayan, N.R.; Méndez-Lagares, G.; Lu, D.; Rauch, M.; Huang, Y.; Van Rompay, K.K.A.; Lynch, S.V.; Hartigan-O’Connor, D.J. Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci. Transl. Med. 2014, 6, 252ra120. [Google Scholar] [CrossRef] [Green Version]
- Timmerman, H.M.; Rutten, N.B.M.M.; Boekhorst, J.; Saulnier, D.M.; Kortman, G.A.M.; Contractor, N.; Kullen, M.; Floris, E.; Harmsen, H.J.M.; Vlieger, A.M.; et al. Intestinal colonisation patterns in breastfed and formula-fed infants during the first 12 weeks of life reveal sequential microbiota signatures. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Jones, D.; Collins, M.D. Taxonomic studies on some human cutaneous coryneform bacteria: Description of Dermabacter hominisgen.nov., sp.nov. FEMS Microbiol. Lett. 1988, 51, 51–55. [Google Scholar] [CrossRef]
- Ruiz-Palacios, G.M.; Calva, J.J.; Pickering, L.K.; Lopez-Vidal, Y.; Volkow, P.; Pezzarossi, H.; West, M.S. Protection of breast-fed infants against Campylobacter diarrhea by antibodies in human milk. J. Pediatr. 1990, 116, 707–713. [Google Scholar] [CrossRef]
- Morrow, A.L.; Ruiz-Palacios, G.M.; Altaye, M.; Jiang, X.; Guerrero, M.L.; Meinzen-Derr, J.K.; Farkas, T.; Chaturvedi, P.; Pickering, L.K.; Newburg, D.S. Human milk oligosaccharides are associated with protection against diarrhea in breast-fed infants. J. Pediatr. 2004, 145, 297–303. [Google Scholar] [CrossRef]
- Bian, X.; Garber, J.M.; Cooper, K.K.; Huynh, S.; Jones, J.; Mills, M.K.; Rafala, D.; Nasrin, D.; Kotloff, K.L.; Parker, C.T.; et al. Campylobacter Abundance in Breastfed Infants and Identification of a New Species in the Global Enterics Multicenter Study. mSphere 2020, 5, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, Y.; Hammarström, M.-L.; Lönnerdal, B.; Graverholt, G.; Fält, H.; Hernell, O. Formula Feeding Skews Immune Cell Composition toward Adaptive Immunity Compared to Breastfeeding. J. Immunol. 2009, 183, 4322–4328. [Google Scholar] [CrossRef] [Green Version]
- Fouda, G.G.; Martinez, D.R.; Swamy, G.K.; Permar, S.R. The Impact of IgG Transplacental Transfer on Early Life Immunity. ImmunoHorizons 2018, 2, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.M. Maternal antibodies and infant immune responses to vaccines. Vaccine 2015, 33, 6469–6472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henrick, B.M.; Hutton, A.A.; Palumbo, M.C.; Casaburi, G.; Mitchell, R.D.; Underwood, M.A.; Smilowitz, J.T.; Frese, S.A. Elevated Fecal pH Indicates a Profound Change in the Breastfed Infant Gut Microbiome Due to Reduction of Bifidobacterium over the Past Century. mSphere 2018, 3, e00041-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Variable | n = 24 |
|---|---|
| Mother primiparous | 15 (63%) |
| Race | |
| White non-Hispanic | 17 (71%) |
| Hispanic | 1 (4%) |
| Southeast Asian | 3 (13%) |
| West Asian | 3 (13%) |
| Delivery method | |
| Vaginal | 16 (67%) |
| Cesarean | 8 (33%) |
| Sex | |
| Female | 10 (42%) |
| Male | 14 (58%) |
| Diet | |
| Any breastfeeding at 1 week | 24 (100%) |
| Breastfeeding without formula at 1 week | 19 (79%) |
| Any breastfeeding at 1 month | 24 (100%) |
| Breastfeeding without formula 1 month | 13 (54%) |
| Any breastfeeding at 3 months | 23 (96%) |
| Breastfeeding without formula at 3 months | 13 (54%) |
| Any breastfeeding at 6 months | 22 (92%) |
| Breastfeeding without formula 6 months | 3 (21%) |
| Semi-solid or solid foods at 6 months | 17 (81% of 21; information missing for 3) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chin, N.; Méndez-Lagares, G.; Taft, D.H.; Laleau, V.; Kieu, H.; Narayan, N.R.; Roberts, S.B.; Mills, D.A.; Hartigan-O’Connor, D.J.; Flaherman, V.J. Transient Effect of Infant Formula Supplementation on the Intestinal Microbiota. Nutrients 2021, 13, 807. https://doi.org/10.3390/nu13030807
Chin N, Méndez-Lagares G, Taft DH, Laleau V, Kieu H, Narayan NR, Roberts SB, Mills DA, Hartigan-O’Connor DJ, Flaherman VJ. Transient Effect of Infant Formula Supplementation on the Intestinal Microbiota. Nutrients. 2021; 13(3):807. https://doi.org/10.3390/nu13030807
Chicago/Turabian StyleChin, Ning, Gema Méndez-Lagares, Diana H. Taft, Victoria Laleau, Hung Kieu, Nicole R. Narayan, Susan B. Roberts, David A. Mills, Dennis J. Hartigan-O’Connor, and Valerie J. Flaherman. 2021. "Transient Effect of Infant Formula Supplementation on the Intestinal Microbiota" Nutrients 13, no. 3: 807. https://doi.org/10.3390/nu13030807
APA StyleChin, N., Méndez-Lagares, G., Taft, D. H., Laleau, V., Kieu, H., Narayan, N. R., Roberts, S. B., Mills, D. A., Hartigan-O’Connor, D. J., & Flaherman, V. J. (2021). Transient Effect of Infant Formula Supplementation on the Intestinal Microbiota. Nutrients, 13(3), 807. https://doi.org/10.3390/nu13030807







