The Structure of Relationships between the Human Exposome and Cardiometabolic Health: The Million Veteran Program
Abstract
1. Background
2. Methods
2.1. Study Population
2.2. Exposome Assessment
2.3. Assessment of Physiological Markers of Cardiometabolic Health
2.4. Assessment of Medication Usage
2.5. Statistical Analysis
3. Results
3.1. Participant Characteristics
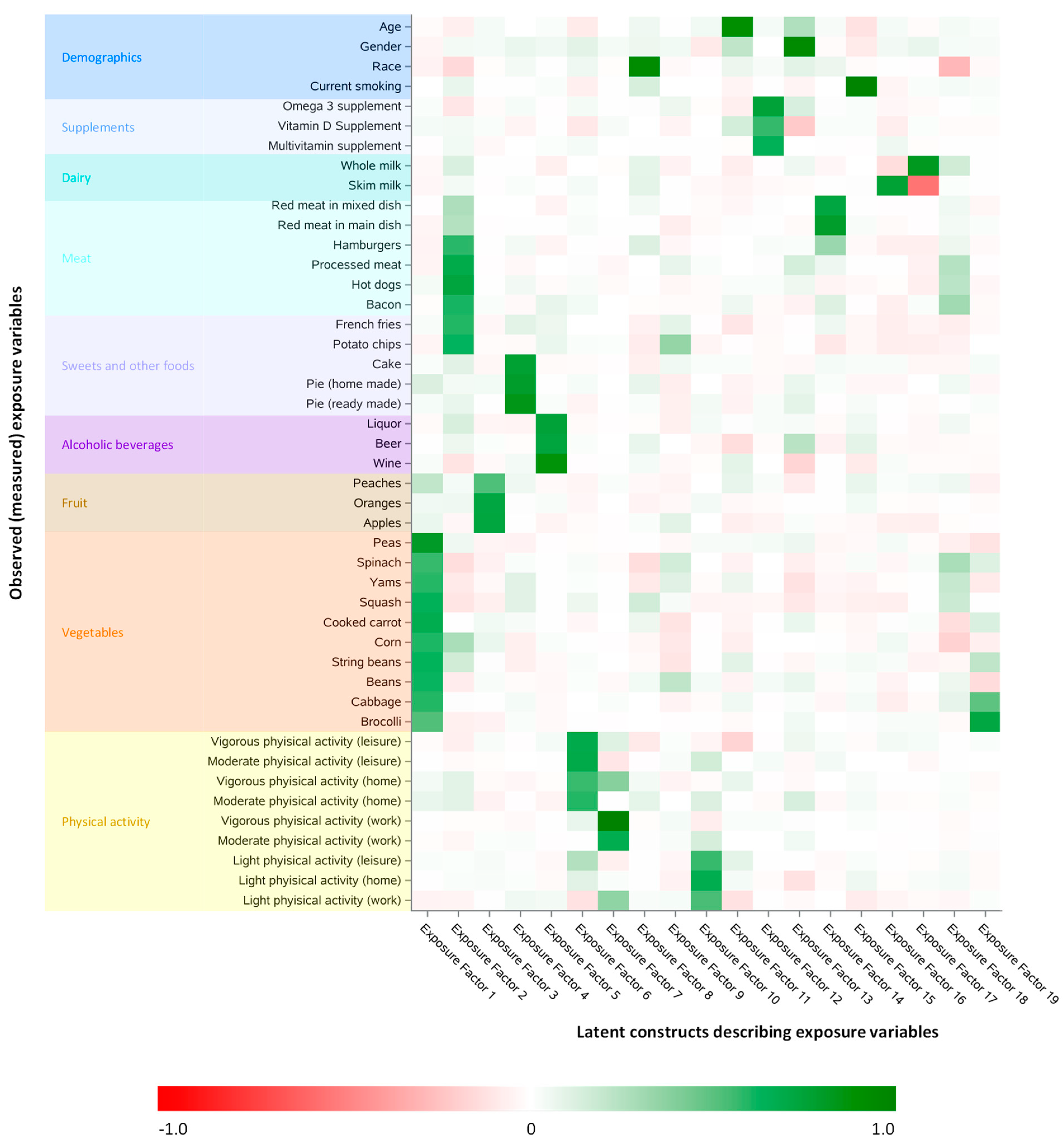
3.2. Latent Constructs Describing the Exposure Variables in the Training Dataset
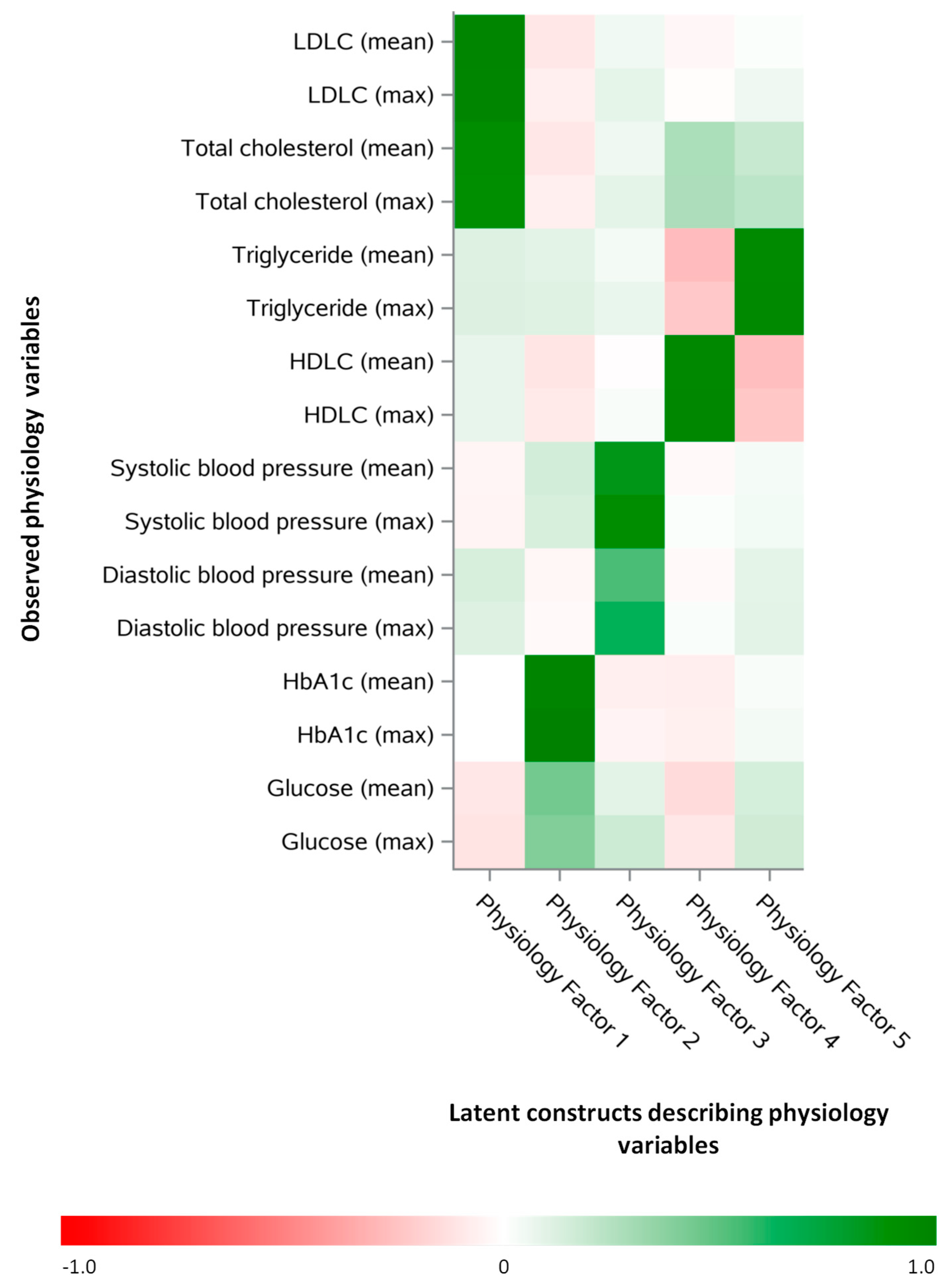
3.3. Latent Constructs Describing the Physiological Variables in the Training Dataset
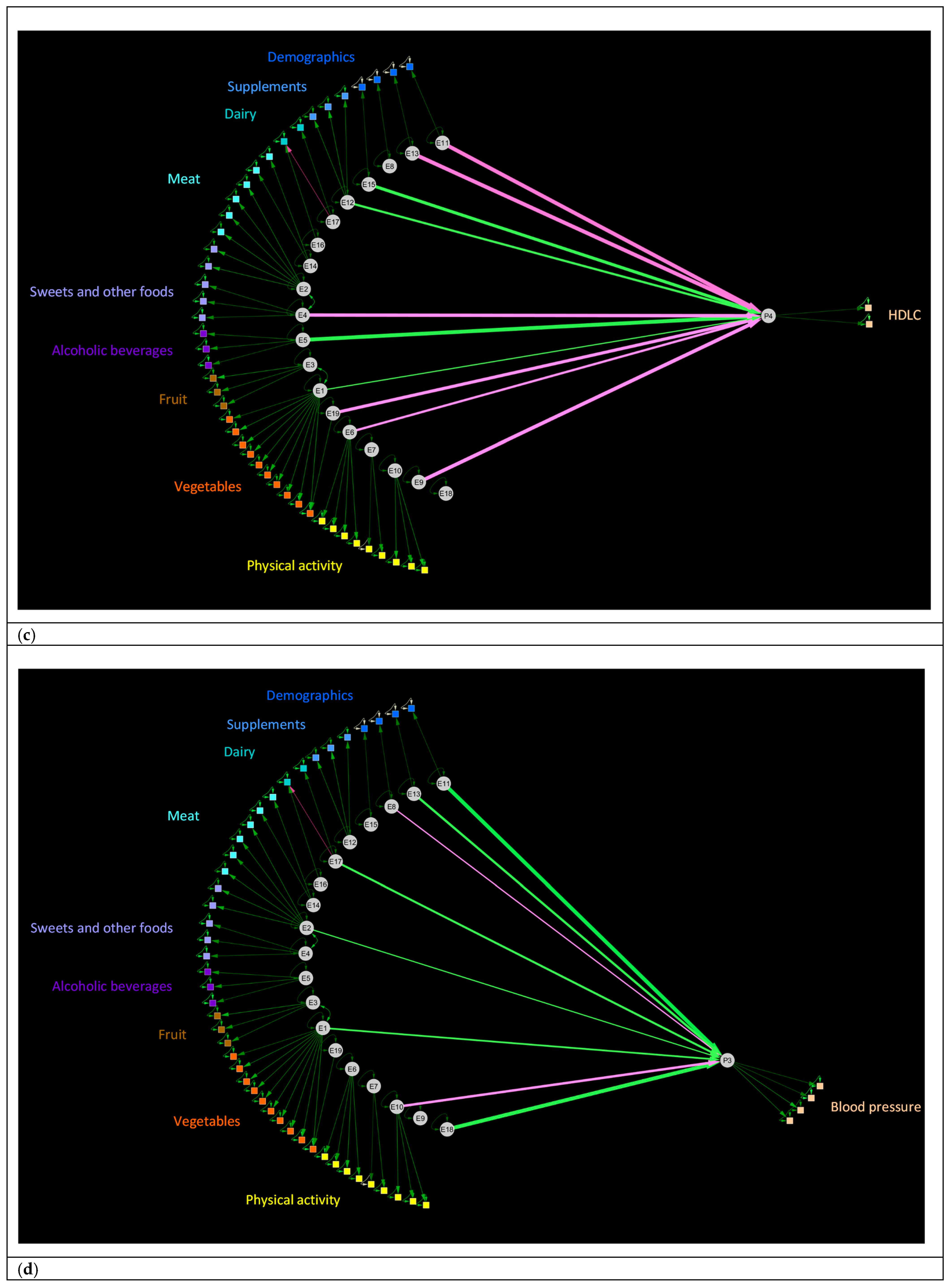
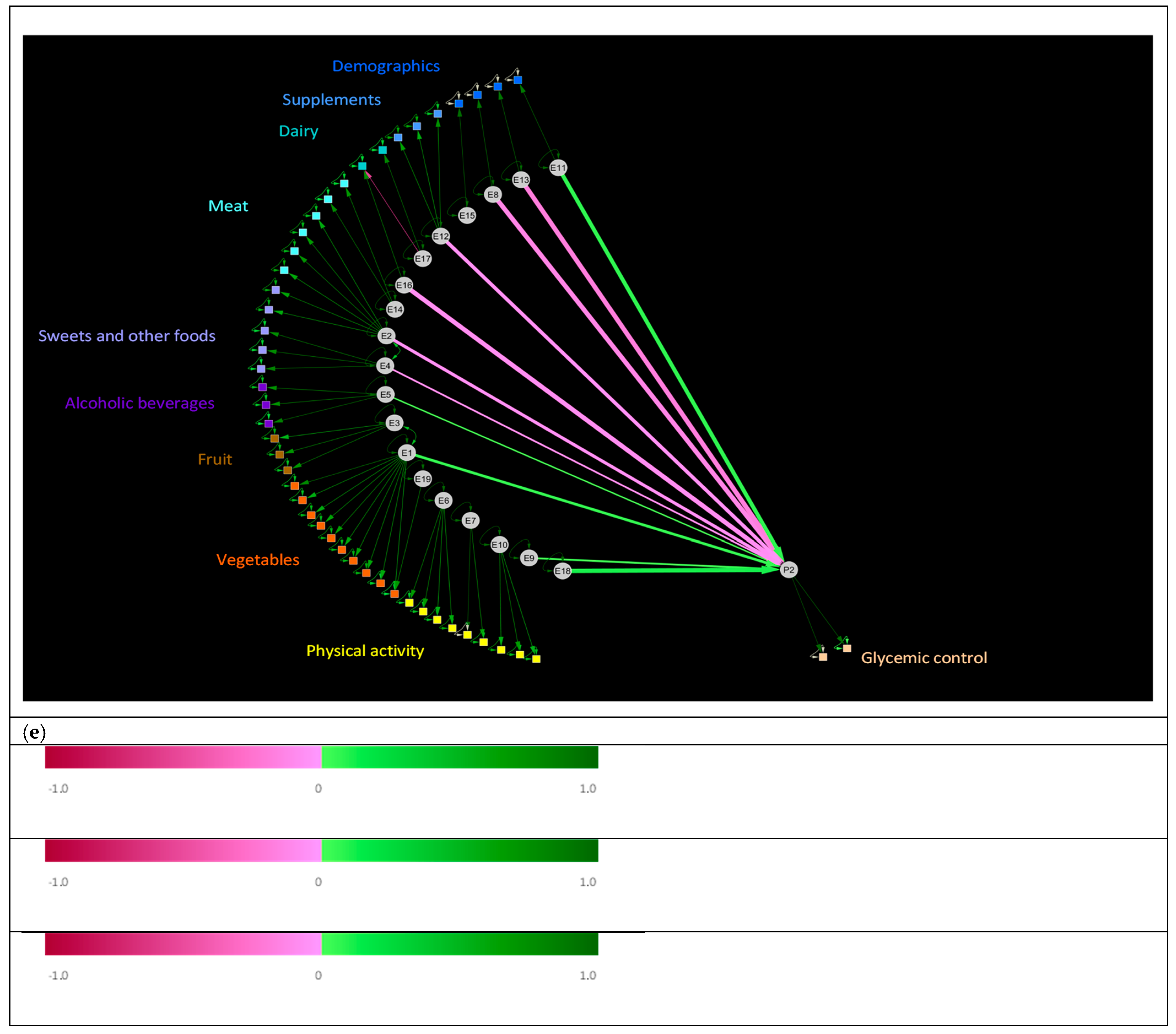
3.4. Relationships between Human Exposures and Physiology in the Validation Dataset
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
- -
- Co-Chair: J. Michael Gaziano, M.D., M.P.H.
- VA Boston Healthcare System, 150 S. Huntington Avenue, Boston, MA 02130, USA
- -
- Co-Chair: Sumitra Muralidhar, Ph.D.
- US Department of Veterans Affairs, 810 Vermont Avenue NW, Washington, DC 20420, USA
- -
- Rachel Ramoni, D.M.D., Sc.D., Chief VA Research and Development Officer
- US Department of Veterans Affairs, 810 Vermont Avenue NW, Washington, DC 20420, USA
- -
- Jean Beckham, Ph.D.
- Durham VA Medical Center, 508 Fulton Street, Durham, NC 27705, USA
- -
- Kyong-Mi Chang, M.D.
- Philadelphia VA Medical Center, 3900 Woodland Avenue, Philadelphia, PA 19104, USA
- -
- Christopher J. O’Donnell, M.D., M.P.H.
- VA Boston Healthcare System, 150 S. Huntington Avenue, Boston, MA 02130, USA
- -
- Philip S. Tsao, Ph.D.
- VA Palo Alto Health Care System, 3801 Miranda Avenue, Palo Alto, CA 94304, USA
- -
- James Breeling, M.D., Ex-Officio
- US Department of Veterans Affairs, 810 Vermont Avenue NW, Washington, DC 20420, USA
- -
- Grant Huang, Ph.D., Ex-Officio
- US Department of Veterans Affairs, 810 Vermont Avenue NW, Washington, DC 20420, USA
- -
- Juan P. Casas, M.D., Ph.D., Ex-Officio
- VA Boston Healthcare System, 150 S. Huntington Avenue, Boston, MA 02130, USA
- -
- Sumitra Muralidhar, Ph.D.
- ∘
- US Department of Veterans Affairs, 810 Vermont Avenue NW, Washington, DC 20420, USA
- -
- Jennifer Moser, Ph.D.
- ∘
- US Department of Veterans Affairs, 810 Vermont Avenue NW, Washington, DC 20420, USA
- -
- Recruitment/Enrollment Director/Deputy Director, Boston—Stacey B. Whitbourne, Ph.D.; Jessica V. Brewer, M.P.H.
- VA Boston Healthcare System, 150 S. Huntington Avenue, Boston, MA 02130, USA
- -
- MVP Coordinating Centers
- Clinical Epidemiology Research Center (CERC), West Haven—Mihaela Aslan, Ph.D.
- West Haven VA Medical Center, 950 Campbell Avenue, West Haven, CT 06516, USA
- Cooperative Studies Program Clinical Research Pharmacy Coordinating Center, Albuquerque—Todd Connor, Pharm.D.; Dean P. Argyres, B.S., M.S.
- New Mexico VA Health Care System, 1501 San Pedro Drive SE, Albuquerque, NM 87108, USA
- Genomics Coordinating Center, Palo Alto—Philip S. Tsao, Ph.D.
- VA Palo Alto Health Care System, 3801 Miranda Avenue, Palo Alto, CA 94304, USA
- MVP Boston Coordinating Center, Boston—J. Michael Gaziano, M.D., M.P.H.
- VA Boston Healthcare System, 150 S. Huntington Avenue, Boston, MA 02130, USA
- MVP Information Center, Canandaigua—Brady Stephens, M.S.
- Canandaigua VA Medical Center, 400 Fort Hill Avenue, Canandaigua, NY 14424, USA
- -
- VA Central Biorepository, Boston—Mary T. Brophy M.D., M.P.H.; Donald E. Humphries, Ph.D.; Luis E. Selva, Ph.D.
- VA Boston Healthcare System, 150 S. Huntington Avenue, Boston, MA 02130, USA
- -
- MVP Informatics, Boston—Nhan Do, M.D.; Shahpoor (Alex) Shayan, M.S.
- VA Boston Healthcare System, 150 S. Huntington Avenue, Boston, MA 02130, USA
- -
- MVP Data Operations/Analytics, Boston—Kelly Cho, M.P.H., Ph.D.
- VA Boston Healthcare System, 150 S. Huntington Avenue, Boston, MA 02130, USA
- -
- Director of Regulatory Affairs—Lori Churby, B.S.
- VA Palo Alto Health Care System, 3801 Miranda Avenue, Palo Alto, CA 94304, USA
- -
- Science Operations—Christopher J. O’Donnell, M.D., M.P.H.
- -
- VA Boston Healthcare System, 150 S. Huntington Avenue, Boston, MA 02130, USA
- -
- Genomics Core—Christopher J. O’Donnell, M.D., M.P.H.; Saiju Pyarajan Ph.D.
- -
- VA Boston Healthcare System, 150 S. Huntington Avenue, Boston, MA 02130, USA
- -
- Philip S. Tsao, Ph.D.
- -
- VA Palo Alto Health Care System, 3801 Miranda Avenue, Palo Alto, CA 94304, USA
- -
- Data Core—Kelly Cho, M.P.H, Ph.D.
- -
- VA Boston Healthcare System, 150 S. Huntington Avenue, Boston, MA 02130, USA
- -
- VA Informatics and Computing Infrastructure (VINCI)—Scott L. DuVall, Ph.D.
- -
- VA Salt Lake City Health Care System, 500 Foothill Drive, Salt Lake City, UT 84148, USA
- -
- Data and Computational Sciences—Saiju Pyarajan, Ph.D.
- -
- VA Boston Healthcare System, 150 S. Huntington Avenue, Boston, MA 02130, USA
- -
- Statistical Genetics—Elizabeth Hauser, Ph.D.
- -
- Durham VA Medical Center, 508 Fulton Street, Durham, NC 27705, USA
- -
- Yan Sun, Ph.D.
- -
- Atlanta VA Medical Center, 1670 Clairmont Road, Decatur, GA 30033, USA
- -
- Hongyu Zhao, Ph.D.
- -
- West Haven VA Medical Center, 950 Campbell Avenue, West Haven, CT 06516, USA
- -
- Atlanta VA Medical Center (Peter Wilson, M.D.)
- ∘
- 1670 Clairmont Road, Decatur, GA 30033, USA
- -
- Bay Pines VA Healthcare System (Rachel McArdle, Ph.D.)
- ∘
- 10,000 Bay Pines Blvd Bay Pines, FL 33744, USA
- -
- Birmingham VA Medical Center (Louis Dellitalia, M.D.)
- ∘
- 700 S. 19th Street, Birmingham, AL 35233, USA
- -
- Central Western Massachusetts Healthcare System (Kristin Mattocks, Ph.D., M.P.H.)
- ∘
- 421 North Main Street, Leeds, MA 01053, USA
- -
- Cincinnati VA Medical Center (John Harley, M.D., Ph.D.)
- ∘
- 3200 Vine Street, Cincinnati, OH 45220, USA
- -
- Clement J. Zablocki VA Medical Center (Jeffrey Whittle, M.D., M.P.H.)
- ∘
- 5000 West National Avenue, Milwaukee, WI 53295, USA
- -
- VA Northeast Ohio Healthcare System (Frank Jacono, M.D.)
- ∘
- 10701 East Boulevard, Cleveland, OH 44106, USA
- -
- Durham VA Medical Center (Jean Beckham, Ph.D.)
- ∘
- 508 Fulton Street, Durham, NC 27705, USA
- -
- Edith Nourse Rogers Memorial Veterans Hospital (John Wells., Ph.D.)
- ∘
- 200 Springs Road, Bedford, MA 01730, USA
- -
- Edward Hines, Jr. VA Medical Center (Salvador Gutierrez, M.D.)
- ∘
- 5000 South 5th Avenue, Hines, IL 60141, USA
- -
- Veterans Health Care System of the Ozarks (Gretchen Gibson, D.D.S., M.P.H.)
- ∘
- 1100 North College Avenue, Fayetteville, AR 72703, USA
- -
- Fargo VA Health Care System (Kimberly Hammer, Ph.D.)
- ∘
- 2101 N. Elm, Fargo, ND 58102, USA
- -
- VA Health Care Upstate New York (Laurence Kaminsky, Ph.D.)
- ∘
- 113 Holland Avenue, Albany, NY 12208, USA
- -
- New Mexico VA Health Care System (Gerardo Villareal, M.D.)
- ∘
- 1501 San Pedro Drive, S.E. Albuquerque, NM 87108, USA
- -
- VA Boston Healthcare System (Scott Kinlay, M.B.B.S., Ph.D.)
- ∘
- 150 S. Huntington Avenue, Boston, MA 02130, USA
- -
- VA Western New York Healthcare System (Junzhe Xu, M.D.)
- ∘
- 3495 Bailey Avenue, Buffalo, NY 14215-1199, USA
- -
- Ralph H. Johnson VA Medical Center (Mark Hamner, M.D.)
- ∘
- 109 Bee Street, Mental Health Research, Charleston, SC 29401
- -
- Columbia VA Health Care System (Roy Mathew, M.D.)
- ∘
- 6439 Garners Ferry Road, Columbia, SC 29209, USA
- -
- VA North Texas Health Care System (Sujata Bhushan, M.D.)
- ∘
- 4500 S. Lancaster Road, Dallas, TX 75216, USA
- -
- Hampton VA Medical Center (Pran Iruvanti, D.O., Ph.D.)
- ∘
- 100 Emancipation Drive, Hampton, VA 23667, USA
- -
- Richmond VA Medical Center (Michael Godschalk, M.D.)
- ∘
- 1201 Broad Rock Blvd., Richmond, VA 23249, USA
- -
- Iowa City VA Health Care System (Zuhair Ballas, M.D.)
- ∘
- 601 Highway 6 West, Iowa City, IA 52246-2208
- -
- Eastern Oklahoma VA Health Care System (Douglas Ivins, M.D.)
- ∘
- 1011 Honor Heights Drive, Muskogee, OK 74401, USA
- -
- James A. Haley Veterans’ Hospital (Stephen Mastorides, M.D.)
- ∘
- 13000 Bruce B. Downs Blvd, Tampa, FL 33612, USA
- -
- James H. Quillen VA Medical Center (Jonathan Moorman, M.D., Ph.D.)
- ∘
- Corner of Lamont & Veterans Way, Mountain Home, TN 37684, USA
- -
- John D. Dingell VA Medical Center (Saib Gappy, M.D.)
- ∘
- 4646 John R Street, Detroit, MI 48201, USA
- -
- Louisville VA Medical Center (Jon Klein, M.D., Ph.D.)
- ∘
- 800 Zorn Avenue, Louisville, KY 40206, USA
- -
- Manchester VA Medical Center (Nora Ratcliffe, M.D.)
- ∘
- 718 Smyth Road, Manchester, NH 03104, USA
- -
- Miami VA Health Care System (Hermes Florez, M.D., Ph.D.)
- ∘
- 1201 NW 16th Street, 11 GRC, Miami, FL 33125, USA
- -
- Michael E. DeBakey VA Medical Center (Olaoluwa Okusaga, M.D.)
- ∘
- 2002 Holcombe Blvd, Houston, TX 77030, USA
- -
- Minneapolis VA Health Care System (Maureen Murdoch, M.D., M.P.H.)
- ∘
- One Veterans Drive, Minneapolis, MN 55417, USA
- -
- N. FL/S. GA Veterans Health System (Peruvemba Sriram, M.D.)
- ∘
- 1601 SW Archer Road, Gainesville, FL 32608, USA
- -
- Northport VA Medical Center (Shing Shing Yeh, Ph.D., M.D.)
- ∘
- 79 Middleville Road, Northport, NY 11768, USA
- -
- Overton Brooks VA Medical Center (Neeraj Tandon, M.D.)
- ∘
- 510 East Stoner Ave, Shreveport, LA 71101, USA
- -
- Philadelphia VA Medical Center (Darshana Jhala, M.D.)
- ∘
- 3900 Woodland Avenue, Philadelphia, PA 19104, USA
- -
- Phoenix VA Health Care System (Samuel Aguayo, M.D.)
- ∘
- 650 E. Indian School Road, Phoenix, AZ 85012, USA
- -
- Portland VA Medical Center (David Cohen, M.D.)
- ∘
- 3710 SW U.S. Veterans Hospital Road, Portland, OR 97239
- -
- Providence VA Medical Center (Satish Sharma, M.D.)
- ∘
- 830 Chalkstone Avenue, Providence, RI 02908, USA
- -
- Richard Roudebush VA Medical Center (Suthat Liangpunsakul, M.D., M.P.H.)
- ∘
- 1481 West 10th Street, Indianapolis, IN 46202, USA
- -
- Salem VA Medical Center (Kris Ann Oursler, M.D.)
- ∘
- 1970 Roanoke Blvd, Salem, VA 24153, USA
- -
- San Francisco VA Health Care System (Mary Whooley, M.D.)
- ∘
- 4150 Clement Street, San Francisco, CA 94121, USA
- -
- South Texas Veterans Health Care System (Sunil Ahuja, M.D.)
- ∘
- 7400 Merton Minter Boulevard, San Antonio, TX 78229, USA
- -
- Southeast Louisiana Veterans Health Care System (Joseph Constans, Ph.D.)
- ∘
- 2400 Canal Street, New Orleans, LA 70119, USA
- -
- Southern Arizona VA Health Care System (Paul Meyer, M.D., Ph.D.)
- ∘
- 3601 S 6th Avenue, Tucson, AZ 85723, USA
- -
- Sioux Falls VA Health Care System (Jennifer Greco, M.D.)
- ∘
- 2501 W 22nd Street, Sioux Falls, SD 57105, USA
- -
- St. Louis VA Health Care System (Michael Rauchman, M.D.)
- ∘
- 915 North Grand Blvd, St. Louis, MO 63106, USA
- -
- Syracuse VA Medical Center (Richard Servatius, Ph.D.)
- ∘
- 800 Irving Avenue, Syracuse, NY 13210, USA
- -
- VA Eastern Kansas Health Care System (Melinda Gaddy, Ph.D.)
- ∘
- 4101 S 4th Street Trafficway, Leavenworth, KS 66048, USA
- -
- VA Greater Los Angeles Health Care System (Agnes Wallbom, M.D., M.S.)
- ∘
- 11301 Wilshire Blvd, Los Angeles, CA 90073, USA
- -
- VA Long Beach Healthcare System (Timothy Morgan, M.D.)
- ∘
- 5901 East 7th Street Long Beach, CA 90822, USA
- -
- VA Maine Healthcare System (Todd Stapley, D.O.)
- ∘
- 1 VA Center, Augusta, ME 04330, USA
- -
- VA New York Harbor Healthcare System (Scott Sherman, M.D., M.P.H.)
- ∘
- 423 East 23rd Street, New York, NY 10010, USA
- -
- VA Pacific Islands Health Care System (George Ross, M.D.)
- ∘
- 459 Patterson Rd, Honolulu, HI 96819, USA
- -
- VA Palo Alto Health Care System (Philip Tsao, Ph.D.)
- ∘
- 3801 Miranda Avenue, Palo Alto, CA 94304-1290, USA
- -
- VA Pittsburgh Health Care System (Patrick Strollo, Jr., M.D.)
- ∘
- University Drive, Pittsburgh, PA 15240, USA
- -
- VA Puget Sound Health Care System (Edward Boyko, M.D.)
- ∘
- 1660 S. Columbian Way, Seattle, WA 98108-1597, USA
- -
- VA Salt Lake City Health Care System (Laurence Meyer, M.D., Ph.D.)
- ∘
- 500 Foothill Drive, Salt Lake City, UT 84148, USA
- -
- VA San Diego Healthcare System (Samir Gupta, M.D., M.S.C.S.)
- ∘
- 3350 La Jolla Village Drive, San Diego, CA 92161, USA
- -
- VA Sierra Nevada Health Care System (Mostaqul Huq, Pharm.D., Ph.D.)
- ∘
- 975 Kirman Avenue, Reno, NV 89502, USA
- -
- VA Southern Nevada Healthcare System (Joseph Fayad, M.D.)
- ∘
- 6900 North Pecos Road, North Las Vegas, NV 89086, USA
- -
- VA Tennessee Valley Healthcare System (Adriana Hung, M.D., M.P.H.)
- ∘
- 1310 24th Avenue, South Nashville, TN 37212, USA
- -
- Washington DC VA Medical Center (Jack Lichy, M.D., Ph.D.)
- ∘
- 50 Irving St, Washington, D. C. 20422, USA
- -
- W.G. (Bill) Hefner VA Medical Center (Robin Hurley, M.D.)
- ∘
- 1601 Brenner Ave, Salisbury, NC 28144, USA
- -
- White River Junction VA Medical Center (Brooks Robey, M.D.)
- ∘
- 163 Veterans Drive, White River Junction, VT 05009, USA
- -
- William S. Middleton Memorial Veterans Hospital (Robert Striker, M.D., Ph.D.)
- ∘
- 2500 Overlook Terrace, Madison, WI 53705, USA
References
- Arnett, D.K.; Blumenthal, R.S.; Albert, M.A.; Buroker, A.B.; Goldberger, Z.D.; Hahn, E.J.; Himmelfarb, C.D.; Khera, A.; Lloyd-Jones, D.; McEvoy, J.W.; et al. 2019 ACC/AHA guideline on the primary prevention of cardiovascular disease: A report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 2019, 74, e177–e232. [Google Scholar] [CrossRef] [PubMed]
- Expert Panel on Detection Evaluation and Treatment of High Blood Cholesterol in Adults. Executive summary of the third report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III). J. Am. Med. Assoc. 2001, 285, 2486–2497. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zeng, F.-F.; Liu, Z.-M.; Zhang, C.-X.; Ling, W.-H.; Chen, Y.-M. Effects of blood triglycerides on cardiovascular and all-cause mortality: A systematic review and meta-analysis of 61 prospective studies. Lipids Health Dis. 2013, 12, 159. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation. Global Status Report on Noncommunicable Diseases 2010: Description of the Global Burden of NCDs, Their Risk Factors and Determinants; World Health Organisation: Geneva, Switzerland, 2011. [Google Scholar]
- Rijkelijkhuizen, J.; Alssema, M.; Nijpels, G.; Stehouwer, C.; Heine, R.; Dekker, J. HbA1c is an independent predictor of non-fatal cardiovascular disease in a Caucasian population without diabetes: A 10-year follow-up of the Hoorn Study. Eur. J. Prev. Cardiol. 2012, 19, 23–31. [Google Scholar]
- Singer, D.E.; Nathan, D.M.; Anderson, K.M.; Wilson, P.W.; Evans, J.C. Association of HbA1c with prevalent cardiovascular disease in the original cohort of the Framingham Heart Study. Diabetes 1992, 41, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.A. Higher “normal” glycated hemoglobin levels were associated with increased risk for diabetes, CVD, stroke, and mortality in adults. Ann. Intern. Med. 2010, 153, JC1-13. [Google Scholar] [CrossRef] [PubMed]
- Lind, M.; Tuomilehto, J.; Uusitupa, M.; Nerman, O.; Eriksson, J.; Ilanne-Parikka, P.; Keinänen-Kiukaanniemi, S.; Peltonen, M.; Pivodic, A.; Lindström, J. The association between HbA1c, fasting glucose, 1-hour glucose and 2-hour glucose during an oral glucose tolerance test and cardiovascular disease in individuals with elevated risk for diabetes. PLoS ONE 2014, 9, e109506. [Google Scholar] [CrossRef]
- Higdon, J.V.; Frei, B. Coffee and health: A review of recent human research. Crit. Rev. Food Sci. Nutr. 2006, 46, 101–123. [Google Scholar] [CrossRef]
- McKay, D.L.; Blumberg, J.B. The role of tea in human health: An update. J. Am. Coll. Nutr. 2002, 21, 1–13. [Google Scholar] [CrossRef]
- Thorp, A.A.; Owen, N.; Neuhaus, M.; Dunstan, D.W. Sedentary behaviors and subsequent health outcomes in adults: A systematic review of longitudinal studies, 1996–2011. Am. J. Prev. Med. 2011, 41, 207–215. [Google Scholar] [CrossRef]
- Siroux, V.; Agier, L.; Slama, R. The exposome concept: A challenge and a potential driver for environmental health research. Eur. Respir. Rev. 2016, 25, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Gaziano, J.M.; Concato, J.; Brophy, M.; Fiore, L.; Pyarajan, S.; Breeling, J.; Whitbourne, S.; Deen, J.; Shannon, C.; Humphries, D.; et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol. 2016, 70, 214–223. [Google Scholar] [CrossRef]
- Rotterdam, E.; Katan, M.B.; Knuiman, J.T. Importance of time interval between repeated measurements of total or high-density lipoprotein cholesterol when estimating an individual's baseline concentrations. Clin. Chem. 1987, 33, 1913–1915. [Google Scholar] [CrossRef]
- Schreiber, M.; Malesios, C.; Psarakis, S. Exploratory factor analysis for the Hirsch index, 17 h-type variants, and some traditional bibliometric indicators. J. Informetr. 2012, 6, 347–358. [Google Scholar] [CrossRef]
- Kang, H.S.; Ahn, I.S.; Kim, J.H.; Kim, D.K. Neuropsychiatric symptoms in Korean patients with Alzheimer’s disease: Exploratory factor analysis and confirmatory factor analysis of the neuropsychiatric inventory. Dement. Geriatr. Cogn. Disord. 2010, 29, 82–87. [Google Scholar] [CrossRef]
- Panaretos, D.; Tzavelas, G.; Vamvakari, M.; Panagiotakos, D. Factor analysis as a tool for pattern recognition in biomedical research; a review with application in R software. J. Data Sci. 2017, 16, 615–630. [Google Scholar]
- Yong, A.G.; Pearce, S. A beginner’s guide to factor analysis: Focusing on exploratory factor analysis. Tutor. Quant. Methods Psychol. 2013, 9, 79–94. [Google Scholar] [CrossRef]
- Hendrickson, A.E.; White, P.O. Promax: A quick method for rotation to oblique simple structure. Br. J. Stat. Psychol. 1964, 12, 65–70. [Google Scholar] [CrossRef]
- SAS Institute Inc. SAS/STAT® 15.1 User’s Guide; SAS Institute Inc.: Cary, NC, USA, 2018. [Google Scholar]
- Fuller, W. Measurement Error Models; JohnWiley: New York, NY, USA, 1987. [Google Scholar]
- Horn, J.L. A rationale and test for the number of factors in factor analysis. Psychometrika 1965, 30, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-O.; Ahtola, O.; Spector, P.E.; Mueller, C.W. Introduction to Factor Analysis: What It Is and How to Do It; Sage: Thousand Oaks, CA, USA, 1978. [Google Scholar]
- Villalpando, S.; Zamudio, Y.L.; Shamah-Levy, T.; Mundo-Rosas, V.; Manzano, A.C.; Lamadrid-Figueroa, H. Substitution of whole cows’ milk with defatted milk for 4 months reduced serum total cholesterol, HDL-cholesterol and total apoB in a sample of Mexican school-age children (6–16 years of age). Br. J. Nutr. 2015, 114, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, K.A.; Childs, M.T.; Stimson, C.; Kushi, L.H.; McGovern, P.G.; Potter, J.D.; Yamanaka, W.K. Effect of consumption of whole milk and skim milk on blood lipid profiles in healthy men. Am. J. Clin. Nutr. 1994, 59, 612–618. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Manson, J.E.; Buring, J.E.; Lee, I.-M.; Sesso, H.D. Dietary intake of dairy products, calcium, and vitamin D and the risk of hypertension in middle-aged and older women. Hypertension 2008, 51, 1073–1079. [Google Scholar] [CrossRef] [PubMed]
- Appel, L.J.; Moore, T.J.; Obarzanek, E.; Vollmer, W.M.; Svetkey, L.P.; Sacks, F.M.; Bray, G.A.; Vogt, T.M.; Cutler, J.A.; Windhauser, M.M. A clinical trial of the effects of dietary patterns on blood pressure. N. Engl. J. Med. 1997, 336, 1117–1124. [Google Scholar] [CrossRef] [PubMed]
- German, J.B.; Gibson, R.A.; Krauss, R.M.; Nestel, P.; Lamarche, B.; Van Staveren, W.A.; Steijns, J.M.; De Groot, L.C.; Lock, A.L.; Destaillats, F. A reappraisal of the impact of dairy foods and milk fat on cardiovascular disease risk. Eur. J. Nutr. 2009, 48, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Lichtenstein, A.H.; Appel, L.J.; Brands, M.; Carnethon, M.; Daniels, S.; Franch, H.A.; Franklin, B.; Kris-Etherton, P.; Harris, W.S.; Howard, B. Summary of American Heart Association diet and lifestyle recommendations revision 2006. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 2186–2191. [Google Scholar] [CrossRef]
- U.S. Department of Health and Human Services; U.S. Department of Agriculture. 2015–2020 Dietary Guidelines for Americans, 8th ed.; U.S. Department of Health and Human Services: Rockville, MD, USA, 2015.
- Djoussé, L.; Arnett, D.K.; Coon, H.; Province, M.A.; Moore, L.L.; Ellison, R.C. Fruit and vegetable consumption and LDL cholesterol: The National Heart, Lung, and Blood Institute Family Heart Study. Am. J. Clin. Nutr. 2004, 79, 213–217. [Google Scholar] [CrossRef]
- Singh, R.B.; Rastogi, S.S.; Niaz, M.A.; Ghosh, S.; Singh, R.; Gupta, S. Effect of fat-modified and fruit-and vegetable-enriched diets on blood lipids in the Indian Diet Heart Study. Am. J. Cardiol. 1992, 70, 869–874. [Google Scholar] [CrossRef]
- Singh, R.B.; Ghosh, S.; Singh, R. Effects on serum lipids of adding fruits and vegetables to prudent diet in the Indian Experiment of Infarct Survival (IEIS). Cardiology 1992, 80, 283–293. [Google Scholar] [CrossRef]
- DASH Research Group. Effects on blood lipids of a blood pressure–lowering diet: The Dietary Approaches to Stop Hypertension (DASH) Trial. Am. J. Clin. Nutr. 2001, 74, 80–89. [Google Scholar] [CrossRef]
- Ballesteros, M.N.; Cabrera, R.M.; Saucedo, M.S.; Yepiz-Plascencia, G.M.; Ortega, M.I.; Valencia, M.E. Dietary fiber and lifestyle influence serum lipids in free living adult men. J. Am. Coll. Nutr. 2001, 20, 649–655. [Google Scholar] [CrossRef]
- Stone, N.J. Lowering low-density cholesterol with diet: The important role of functional foods as adjuncts. Coron. Artery Dis. 2001, 12, 547–552. [Google Scholar] [CrossRef]
- Jensen, E.N.; Buch-Andersen, T.; Ravn-Haren, G.; Dragsted, L.O. Mini-review: The effects of apples on plasma cholesterol levels and cardiovascular risk—A review of the evidence. J. Hortic. Sci. Biotechnol. 2009, 84, 34–41. [Google Scholar] [CrossRef]
- Vidal, R.; Hernandez-Vallejo, S.; Pauquai, T.; Texier, O.; Rousset, M.; Chambaz, J.; Demignot, S.; Lacorte, J.-M. Apple procyanidins decrease cholesterol esterification and lipoprotein secretion in Caco-2/TC7 enterocytes. J. Lipid Res. 2005, 46, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Lam, C.K.; Zhang, Z.; Yu, H.; Tsang, S.Y.; Huang, Y.; Chen, Z.Y. Apple polyphenols inhibit plasma CETP activity and reduce the ratio of non-HDL to HDL cholesterol. Mol. Nutr. Food Res. 2008, 52, 950–958. [Google Scholar] [CrossRef] [PubMed]
- Ogino, Y.; Osada, K.; Nakamura, S.; Ohta, Y.; Kanda, T.; Sugano, M. Absorption of dietary cholesterol oxidation products and their downstream metabolic effects are reduced by dietary apple polyphenols. Lipids 2007, 42, 151. [Google Scholar] [CrossRef]
- Serra, A.T.; Rocha, J.; Sepodes, B.; Matias, A.A.; Feliciano, R.P.; de Carvalho, A.; Bronze, M.R.; Duarte, C.M.; Figueira, M. Evaluation of cardiovascular protective effect of different apple varieties–correlation of response with composition. Food Chem. 2012, 135, 2378–2386. [Google Scholar] [CrossRef]
- Gonzalez, M.; Rivas, C.; Caride, B.; Lamas, M.A.; Taboada, M.C. Effects of orange and apple pectin on cholesterol concentration in serum, liver and faeces. J. Physiol. Biochem. 1998, 54, 99–104. [Google Scholar]
- Wang, X.; Ouyang, Y.; Liu, J.; Zhu, M.; Zhao, G.; Bao, W.; Hu, F.B. Fruit and vegetable consumption and mortality from all causes, cardiovascular disease, and cancer: Systematic review and dose-response meta-analysis of prospective cohort studies. Br. Med. J. 2014, 349, g4490. [Google Scholar] [CrossRef] [PubMed]
- Lesack, K.; Naugler, C. An open-source software program for performing Bonferroni and related corrections for multiple comparisons. J. Pathol. Inform. 2011, 2, 52. [Google Scholar]
- Freeland-Graves, J.H.; Nitzke, S. Position of the Academy of Nutrition and Dietetics: Total diet approach to healthy eating. J. Acad. Nutr. Diet. 2013, 113, 307–317. [Google Scholar] [CrossRef]
- Tucker, K.L. Dietary patterns, approaches, and multicultural perspective. Appl. Physiol. Nutr. Metab. 2010, 35, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Schulze, M.B.; Hoffmann, K. Methodological approaches to study dietary patterns in relation to risk of coronary heart disease and stroke. Br. J. Nutr. 2006, 95, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Ocké, M.C. Evaluation of methodologies for assessing the overall diet: Dietary quality scores and dietary pattern analysis. Proc. Nutr. Soc. 2013, 72, 191–199. [Google Scholar] [CrossRef] [PubMed]

| Value | |
|---|---|
| DEMOGRAPHICS | |
| Age (years) | 62.40 ± 13.41 |
| Gender (% males) | 86 |
| Caucasian (%) | 85 |
| Current smoking (number of cigarettes smoked/day) | 0.26 ± 0.77 |
| SUPPLEMENT USE | |
| Omega-3 supplement use (%) | 23 |
| Vitamin D supplement use (%) | 36 |
| Multivitamin supplement use (%) | 54 |
| DIETARY INTAKE | |
| Dairy | |
| Whole milk (serves/day) | 0.17 ± 0.55 |
| Skim milk (serves/day) | 0.51 ± 0.87 |
| Meat | |
| Red meat in main dish (serves/day) | 0.20 ± 0.28 |
| Red meat in mixed dish (serves/day) | 0.19 ± 0.26 |
| Hamburgers (serves/day) | 0.16 ± 0.24 |
| Processed meat (serves/day) | 0.16 ± 0.28 |
| Hot dogs (serves/day) | 0.08 ± 0.19 |
| Bacon (serves/day) | 0.15 ± 0.29 |
| Sweets and other foods | |
| French fries (serves/day) | 0.12 ± 0.23 |
| Potato chips (serves/day) | 0.18 ± 0.32 |
| Cake (serves/day) | 0.06 ± 0.15 |
| Home-made pie (serves/day) | 0.05 ± 0.13 |
| Ready-made pie (serves/day) | 0.05 ± 0.14 |
| Alcoholic beverages | |
| Liquor (serves/day) | 0.15 ± 0.52 |
| Beer (serves/day) | 0.31 ± 0.84 |
| Wine (serves/day) | 0.17 ± 0.49 |
| Fruit | |
| Peaches (serves/day) | 0.13 ± 0.32 |
| Oranges (serves/day) | 0.19 ± 0.37 |
| Apples (serves/day) | 0.27 ± 0.43 |
| Vegetables | |
| Peas (serves/day) | 0.14 ± 0.25 |
| Spinach (serves/day) | 0.15 ± 0.32 |
| Yams (serves/day) | 0.09 ± 0.23 |
| Squash (serves/day) | 0.07 ± 0.21 |
| Cooked carrot (serves/day) | 0.13 ± 0.25 |
| Corn (serves/day) | 0.15 ± 0.25 |
| String beans (serves/day) | 0.17 ± 0.26 |
| Beans (serves/day) | 0.18 ± 0.32 |
| Cabbage (serves/day) | 0.14 ± 0.28 |
| Broccoli (serves/day) | 0.19 ± 0.31 |
| PHYSICAL ACTIVITY | |
| Vigorous physical activity during leisure time (hours/day) | 0.85 ± 1.68 |
| Moderate physical activity during leisure time (hours/day) | 1.20 ± 1.92 |
| Vigorous physical activity at home (hours/day) | 0.85 ± 1.51 |
| Moderate physical activity at home (hours/day) | 1.14 ± 1.75 |
| Vigorous physical activity at work (hours/day) | 0.9 ± 1.87 |
| Moderate physical activity at work (hours/day) | 1.77 ± 2.53 |
| Light physical activity during leisure time (hours/day) | 2.37 ± 2.65 |
| Light physical activity at home (hours/day) | 2.93 ± 2.71 |
| Light physical activity at work (hours/day) | 2.96 ± 3.19 |
| Mean ± SD | |
|---|---|
| Total cholesterol | |
| Mean of measurements (mg/dL) | 177.40 ± 33.96 |
| Maximum measurement (mg/dL) | 187.28 ± 36.98 |
| Low-density lipoprotein cholesterol | |
| Mean of measurements (mg/dL) | 104.57 ± 29.36 |
| Maximum measurement (mg/dL) | 113.15 ± 31.90 |
| Triglycerides | |
| Mean of measurements (mg/dL) | 123.48 ± 63.96 |
| Maximum measurement (mg/dL) | 145.81 ± 80.31 |
| High-density lipoprotein cholesterol | |
| Mean of measurements (mg/dL) | 49.79 ± 13.94 |
| Maximum measurement (mg/dL) | 53.02 ± 15.10 |
| Systolic blood pressure | |
| Mean of measurements (mmHg) | 130.27 ± 12.43 |
| Maximum measurement (mmHg) | 145.71 ± 17.93 |
| Diastolic blood pressure | |
| Mean of measurements (mmHg) | 76.72 ± 7.67 |
| Maximum measurement (mmHg) | 85.74 ± 10.24 |
| HbA1c | |
| Mean of measurements (DCCT %) | 5.64 ± 0.58 |
| Maximum measurement (DCCT %) | 5.74 ± 0.66 |
| Glucose | |
| Mean of measurements (mg/dL) | 101.92 ± 17.70 |
| Maximum measurement (mg/dL) | 113.45 ± 26.93 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivey, K.L.; Nguyen, X.-M.T.; Posner, D.; Rogers, G.B.; Tobias, D.K.; Song, R.; Ho, Y.-L.; Li, R.; Wilson, P.W.F.; Cho, K.; et al. The Structure of Relationships between the Human Exposome and Cardiometabolic Health: The Million Veteran Program. Nutrients 2021, 13, 1364. https://doi.org/10.3390/nu13041364
Ivey KL, Nguyen X-MT, Posner D, Rogers GB, Tobias DK, Song R, Ho Y-L, Li R, Wilson PWF, Cho K, et al. The Structure of Relationships between the Human Exposome and Cardiometabolic Health: The Million Veteran Program. Nutrients. 2021; 13(4):1364. https://doi.org/10.3390/nu13041364
Chicago/Turabian StyleIvey, Kerry L., Xuan-Mai T. Nguyen, Daniel Posner, Geraint B. Rogers, Deirdre K. Tobias, Rebecca Song, Yuk-Lam Ho, Ruifeng Li, Peter W. F. Wilson, Kelly Cho, and et al. 2021. "The Structure of Relationships between the Human Exposome and Cardiometabolic Health: The Million Veteran Program" Nutrients 13, no. 4: 1364. https://doi.org/10.3390/nu13041364
APA StyleIvey, K. L., Nguyen, X.-M. T., Posner, D., Rogers, G. B., Tobias, D. K., Song, R., Ho, Y.-L., Li, R., Wilson, P. W. F., Cho, K., Gaziano, J. M., Hu, F. B., Willett, W. C., & Djoussé, L. (2021). The Structure of Relationships between the Human Exposome and Cardiometabolic Health: The Million Veteran Program. Nutrients, 13(4), 1364. https://doi.org/10.3390/nu13041364






