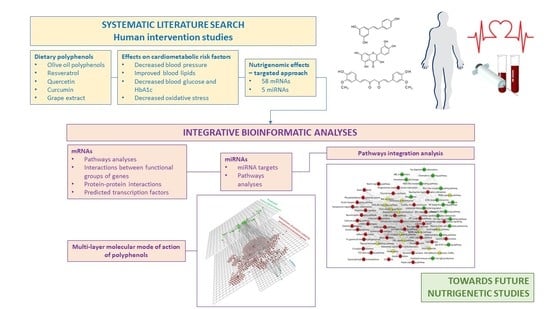
Systematic Bioinformatic Analyses of Nutrigenomic Modifications by Polyphenols Associated with Cardiometabolic Health in Humans—Evidence from Targeted Nutrigenomic Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strategy for Literature search and Data Extraction
2.2. Bioinformatic Analyses
3. Results
3.1. Studies, Bioactives and Differentially Expressed Genes
3.2. mRNAs—Bioinformatic Analyses
3.2.1. Pathways Analyses
3.2.2. Interactions between Functional Groups of Genes
3.2.3. Protein-Protein Interactions
3.2.4. Transcription Factors
3.3. miRNAs—Bioinformatic Analyses
3.3.1. miRNA Targets
3.3.2. Pathways Analyses of miRNA Targets
3.4. Integration Analyses
3.4.1. Integration of mRNAs and miRNA Targets
3.4.2. Integration of mRNA and miRNA-Target Pathways
3.4.3. mRNAs, miRNAs and Transcription Factors Integration Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Del Rio, D.; Rodriguez-Mateos, A.; Spencer, J.P.; Tognolini, M.; Borges, G.; Crozier, A. Dietary (poly)phenolics in human health: Structures, bioavailability, and evidence of protective effects against chronic diseases. Antioxid. Redox Signal. 2013, 18, 1818–1892. [Google Scholar] [CrossRef] [Green Version]
- Tresserra-Rimbau, A.; Lamuela-Raventos, R.M.; Moreno, J.J. Polyphenols, food and pharma. Current knowledge and directions for future research. Biochem. Pharmacol. 2018, 156, 186–195. [Google Scholar] [CrossRef]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of Phenylpropanoid Pathway and the Role of Polyphenols in Plants under Abiotic Stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cory, H.; Passarelli, S.; Szeto, J.; Tamez, M.; Mattei, J. The Role of Polyphenols in Human Health and Food Systems: A Mini-Review. Front. Nutr. 2018, 5, 87. [Google Scholar] [CrossRef] [Green Version]
- Neveu, V.; Perez-Jimenez, J.; Vos, F.; Crespy, V.; du Chaffaut, L.; Mennen, L.; Knox, C.; Eisner, R.; Cruz, J.; Wishart, D.; et al. Phenol-Explorer: An online comprehensive database on polyphenol contents in foods. Database 2010, 2010, bap024. [Google Scholar] [CrossRef] [PubMed]
- Scalbert, A.; Williamson, G. Dietary intake and bioavailability of polyphenols. J. Nutr. 2000, 130, 2073S–2085S. [Google Scholar] [CrossRef]
- Miranda, A.M.; Steluti, J.; Fisberg, R.M.; Marchioni, D.M. Dietary intake and food contributors of polyphenols in adults and elderly adults of Sao Paulo: A population-based study. Br. J. Nutr. 2016, 115, 1061–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosso, G.; Stepaniak, U.; Topor-Madry, R.; Szafraniec, K.; Pajak, A. Estimated dietary intake and major food sources of polyphenols in the Polish arm of the HAPIEE study. Nutrition 2014, 30, 1398–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hertog, M.G.; Feskens, E.J.; Hollman, P.C.; Katan, M.B.; Kromhout, D. Dietary antioxidant flavonoids and risk of coronary heart disease: The Zutphen Elderly Study. Lancet 1993, 342, 1007–1011. [Google Scholar] [CrossRef]
- Wedick, N.M.; Pan, A.; Cassidy, A.; Rimm, E.B.; Sampson, L.; Rosner, B.; Willett, W.; Hu, F.B.; Sun, Q.; van Dam, R.M. Dietary flavonoid intakes and risk of type 2 diabetes in US men and women. Am. J. Clin. Nutr. 2012, 95, 925–933. [Google Scholar] [CrossRef] [Green Version]
- Loffredo, L.; Baratta, F.; Ludovica, P.; Battaglia, S.; Carnevale, R.; Nocella, C.; Novo, M.; Pannitteri, G.; Ceci, F.; Angelico, F.; et al. Effects of dark chocolate on endothelial function in patients with non-alcoholic steatohepatitis. Nutr. Metab. Cardiovasc. Dis. 2017, 28, 143–149. [Google Scholar] [CrossRef]
- Grassi, D.; Desideri, G.; Necozione, S.; Lippi, C.; Casale, R.; Properzi, G.; Blumberg, J.B.; Ferri, C. Blood pressure is reduced and insulin sensitivity increased in glucose-intolerant, hypertensive subjects after 15 days of consuming high-polyphenol dark chocolate. J. Nutr. 2008, 138, 1671–1676. [Google Scholar] [CrossRef]
- Park, E.; Edirisinghe, I.; Choy, Y.Y.; Waterhouse, A.; Burton-Freeman, B. Effects of grape seed extract beverage on blood pressure and metabolic indices in individuals with pre-hypertension: A randomised, double-blinded, two-arm, parallel, placebo-controlled trial. Br. J. Nutr. 2016, 115, 226–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hokayem, M.; Blond, E.; Vidal, H.; Lambert, K.; Meugnier, E.; Feillet-Coudray, C.; Coudray, C.; Pesenti, S.; Luyton, C.; Lambert-Porcheron, S.; et al. Grape polyphenols prevent fructose-induced oxidative stress and insulin resistance in first-degree relatives of type 2 diabetic patients. Diabetes Care 2013, 36, 1454–1461. [Google Scholar] [CrossRef] [Green Version]
- Pinto, X.; Fanlo-Maresma, M.; Corbella, E.; Corbella, X.; Mitjavila, M.T.; Moreno, J.J.; Casas, R.; Estruch, R.; Corella, D.; Bullo, M.; et al. A Mediterranean Diet Rich in Extra-Virgin Olive Oil Is Associated with a Reduced Prevalence of Nonalcoholic Fatty Liver Disease in Older Individuals at High Cardiovascular Risk. J. Nutr. 2019, 149, 1920–1929. [Google Scholar] [CrossRef] [PubMed]
- Storniolo, C.E.; Casillas, R.; Bullo, M.; Castaner, O.; Ros, E.; Saez, G.T.; Toledo, E.; Estruch, R.; Ruiz-Gutierrez, V.; Fito, M.; et al. A Mediterranean diet supplemented with extra virgin olive oil or nuts improves endothelial markers involved in blood pressure control in hypertensive women. Eur. J. Nutr. 2017, 56, 89–97. [Google Scholar] [CrossRef]
- Menezes, R.; Rodriguez-Mateos, A.; Kaltsatou, A.; Gonzalez-Sarrias, A.; Greyling, A.; Giannaki, C.; Andres-Lacueva, C.; Milenkovic, D.; Gibney, E.R.; Dumont, J.; et al. Impact of Flavonols on Cardiometabolic Biomarkers: A Meta-Analysis of Randomized Controlled Human Trials to Explore the Role of Inter-Individual Variability. Nutrients 2017, 9, 117. [Google Scholar] [CrossRef]
- Morand, C.; De Roos, B.; Garcia-Conesa, M.T.; Gibney, E.R.; Landberg, R.; Manach, C.; Milenkovic, D.; Rodriguez-Mateos, A.; Van de Wiele, T.; Tomas-Barberan, F. Why interindividual variation in response to consumption of plant food bioactives matters for future personalised nutrition. Proc. Nutr. Soc. 2020, 79, 225–235. [Google Scholar] [CrossRef]
- Gibney, E.R.; Milenkovic, D.; Combet, E.; Ruskovska, T.; Greyling, A.; Gonzalez-Sarrias, A.; de Roos, B.; Tomas-Barberan, F.; Morand, C.; Rodriguez-Mateos, A. Factors influencing the cardiometabolic response to (poly)phenols and phytosterols: A review of the COST Action POSITIVe activities. Eur. J. Nutr. 2019, 58, 37–47. [Google Scholar] [CrossRef] [Green Version]
- Milenkovic, D.; Morand, C.; Cassidy, A.; Konic-Ristic, A.; Tomas-Barberan, F.; Ordovas, J.M.; Kroon, P.; De Caterina, R.; Rodriguez-Mateos, A. Interindividual Variability in Biomarkers of Cardiometabolic Health after Consumption of Major Plant-Food Bioactive Compounds and the Determinants Involved. Adv. Nutr. 2017, 8, 558–570. [Google Scholar] [CrossRef] [PubMed]
- Caradonna, F.; Consiglio, O.; Luparello, C.; Gentile, C. Science and Healthy Meals in the World: Nutritional Epigenomics and Nutrigenetics of the Mediterranean Diet. Nutrients 2020, 12, 1748. [Google Scholar] [CrossRef]
- Caradonna, F.; Cruciata, I.; Luparello, C. Nutrigenetics, nutrigenomics and phenotypic outcomes of dietary low-dose alcohol consumption in the suppression and induction of cancer development: Evidence from in vitro studies. Crit. Rev. Food Sci. Nutr. 2020, 8, 1–32. [Google Scholar] [CrossRef]
- Ruskovska, T.; Maksimova, V.; Milenkovic, D. Polyphenols in human nutrition: From the in vitro antioxidant capacity to the beneficial effects on cardiometabolic health and related inter-individual variability—An overview and perspective. Br. J. Nutr. 2020, 123, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Krga, I.; Tamaian, R.; Mercier, S.; Boby, C.; Monfoulet, L.E.; Glibetic, M.; Morand, C.; Milenkovic, D. Anthocyanins and their gut metabolites attenuate monocyte adhesion and transendothelial migration through nutrigenomic mechanisms regulating endothelial cell permeability. Free Radic. Biol. Med. 2018, 124, 364–379. [Google Scholar] [CrossRef] [PubMed]
- Ruskovska, T.; Massaro, M.; Carluccio, M.A.; Arola-Arnal, A.; Muguerza, B.; Vanden Berghe, W.; Declerck, K.; Bravo, F.I.; Calabriso, N.; Combet, E.; et al. Systematic bioinformatic analysis of nutrigenomic data of flavanols in cell models of cardiometabolic disease. Food Funct. 2020, 11, 5040–5064. [Google Scholar] [CrossRef] [PubMed]
- Mitjavila, M.T.; Moreno, J.J. The effects of polyphenols on oxidative stress and the arachidonic acid cascade. Implications for the prevention/treatment of high prevalence diseases. Biochem. Pharmacol. 2012, 84, 1113–1122. [Google Scholar] [CrossRef] [PubMed]
- Tarragon, E.; Moreno, J.J. Polyphenols and taste 2 receptors. Physiological, pathophysiological and pharmacological implications. Biochem. Pharmacol. 2020, 178, 114086. [Google Scholar] [CrossRef]
- Most, J.; Timmers, S.; Warnke, I.; Jocken, J.W.; van Boekschoten, M.; de Groot, P.; Bendik, I.; Schrauwen, P.; Goossens, G.H.; Blaak, E.E. Combined epigallocatechin-3-gallate and resveratrol supplementation for 12 wk increases mitochondrial capacity and fat oxidation, but not insulin sensitivity, in obese humans: A randomized controlled trial. Am. J. Clin. Nutr. 2016, 104, 215–227. [Google Scholar] [CrossRef] [Green Version]
- Most, J.; Warnke, I.; Boekschoten, M.V.; Jocken, J.W.E.; de Groot, P.; Friedel, A.; Bendik, I.; Goossens, G.H.; Blaak, E.E. The effects of polyphenol supplementation on adipose tissue morphology and gene expression in overweight and obese humans. Adipocyte 2018, 7, 190–196. [Google Scholar] [CrossRef]
- Gaj, S.; Eijssen, L.; Mensink, R.P.; Evelo, C.T. Validating nutrient-related gene expression changes from microarrays using RT(2) PCR-arrays. Genes Nutr. 2008, 3, 153–157. [Google Scholar] [CrossRef] [Green Version]
- Morey, J.S.; Ryan, J.C.; Van Dolah, F.M. Microarray validation: Factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol. Proced. Online 2006, 8, 175–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. Ann. Intern. Med. 2009, 151, 264–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stockel, D.; Kehl, T.; Trampert, P.; Schneider, L.; Backes, C.; Ludwig, N.; Gerasch, A.; Kaufmann, M.; Gessler, M.; Graf, N.; et al. Multi-omics enrichment analysis using the GeneTrail2 web service. Bioinformatics 2016, 32, 1502–1508. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Cho, J.W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8–13. [Google Scholar] [CrossRef] [Green Version]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Koopmans, F.; van Nierop, P.; Andres-Alonso, M.; Byrnes, A.; Cijsouw, T.; Coba, M.P.; Cornelisse, L.N.; Farrell, R.J.; Goldschmidt, H.L.; Howrigan, D.P.; et al. SynGO: An Evidence-Based, Expert-Curated Knowledge Base for the Synapse. Neuron 2019, 103, 217–234.e214. [Google Scholar] [CrossRef] [Green Version]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.; Xia, J. OmicsNet: A web-based tool for creation and visual analysis of biological networks in 3D space. Nucleic Acids Res. 2018, 46, W514–W522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, G.; Xia, J. Using OmicsNet for Network Integration and 3D Visualization. Curr. Protoc. Bioinform. 2019, 65, e69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Castaner, O.; Covas, M.I.; Khymenets, O.; Nyyssonen, K.; Konstantinidou, V.; Zunft, H.F.; de la Torre, R.; Munoz-Aguayo, D.; Vila, J.; Fito, M. Protection of LDL from oxidation by olive oil polyphenols is associated with a downregulation of CD40-ligand expression and its downstream products in vivo in humans. Am. J. Clin. Nutr. 2012, 95, 1238–1244. [Google Scholar] [CrossRef] [Green Version]
- Farras, M.; Valls, R.M.; Fernandez-Castillejo, S.; Giralt, M.; Sola, R.; Subirana, I.; Motilva, M.J.; Konstantinidou, V.; Covas, M.I.; Fito, M. Olive oil polyphenols enhance the expression of cholesterol efflux related genes in vivo in humans. A randomized controlled trial. J. Nutr. Biochem. 2013, 24, 1334–1339. [Google Scholar] [CrossRef]
- Martin-Pelaez, S.; Castaner, O.; Konstantinidou, V.; Subirana, I.; Munoz-Aguayo, D.; Blanchart, G.; Gaixas, S.; de la Torre, R.; Farre, M.; Saez, G.T.; et al. Effect of olive oil phenolic compounds on the expression of blood pressure-related genes in healthy individuals. Eur. J. Nutr. 2017, 56, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Seyyedebrahimi, S.; Khodabandehloo, H.; Nasli Esfahani, E.; Meshkani, R. The effects of resveratrol on markers of oxidative stress in patients with type 2 diabetes: A randomized, double-blind, placebo-controlled clinical trial. Acta Diabetol. 2018, 55, 341–353. [Google Scholar] [CrossRef]
- Khorshidi, M.; Moini, A.; Alipoor, E.; Rezvan, N.; Gorgani-Firuzjaee, S.; Yaseri, M.; Hosseinzadeh-Attar, M.J. The effects of quercetin supplementation on metabolic and hormonal parameters as well as plasma concentration and gene expression of resistin in overweight or obese women with polycystic ovary syndrome. Phytother. Res. 2018, 32, 2282–2289. [Google Scholar] [CrossRef]
- Barber-Chamoux, N.; Milenkovic, D.; Verny, M.A.; Habauzit, V.; Pereira, B.; Lambert, C.; Richard, D.; Boby, C.; Mazur, A.; Lusson, J.R.; et al. Substantial Variability Across Individuals in the Vascular and Nutrigenomic Response to an Acute Intake of Curcumin: A Randomized Controlled Trial. Mol. Nutr. Food Res. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Tome-Carneiro, J.; Larrosa, M.; Yanez-Gascon, M.J.; Davalos, A.; Gil-Zamorano, J.; Gonzalvez, M.; Garcia-Almagro, F.J.; Ruiz Ros, J.A.; Tomas-Barberan, F.A.; Espin, J.C.; et al. One-year supplementation with a grape extract containing resveratrol modulates inflammatory-related microRNAs and cytokines expression in peripheral blood mononuclear cells of type 2 diabetes and hypertensive patients with coronary artery disease. Pharmacol. Res. 2013, 72, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Tome-Carneiro, J.; Gonzalvez, M.; Larrosa, M.; Yanez-Gascon, M.J.; Garcia-Almagro, F.J.; Ruiz-Ros, J.A.; Tomas-Barberan, F.A.; Garcia-Conesa, M.T.; Espin, J.C. Grape resveratrol increases serum adiponectin and downregulates inflammatory genes in peripheral blood mononuclear cells: A triple-blind, placebo-controlled, one-year clinical trial in patients with stable coronary artery disease. Cardiovasc. Drugs Ther. 2013, 27, 37–48. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Candales, A.; Hernandez Burgos, P.M.; Hernandez-Suarez, D.F.; Harris, D. Linking Chronic Inflammation with Cardiovascular Disease: From Normal Aging to the Metabolic Syndrome. J. Nat. Sci. 2017, 3, e341. [Google Scholar]
- Hotamisligil, G.S. Inflammation, metaflammation and immunometabolic disorders. Nature 2017, 542, 177–185. [Google Scholar] [CrossRef]
- Ormazabal, V.; Nair, S.; Elfeky, O.; Aguayo, C.; Salomon, C.; Zuniga, F.A. Association between insulin resistance and the development of cardiovascular disease. Cardiovasc. Diabetol. 2018, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Caputo, T.; Gilardi, F.; Desvergne, B. From chronic overnutrition to metaflammation and insulin resistance: Adipose tissue and liver contributions. FEBS Lett. 2017, 591, 3061–3088. [Google Scholar] [CrossRef] [Green Version]
- Longo, M.; Zatterale, F.; Naderi, J.; Parrillo, L.; Formisano, P.; Raciti, G.A.; Beguinot, F.; Miele, C. Adipose Tissue Dysfunction as Determinant of Obesity-Associated Metabolic Complications. Int. J. Mol. Sci. 2019, 20, 2358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruskovska, T.; Bernlohr, D.A. Oxidative stress and protein carbonylation in adipose tissue—Implications for insulin resistance and diabetes mellitus. J. Proteom. 2013, 92, 323–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nandipati, K.C.; Subramanian, S.; Agrawal, D.K. Protein kinases: Mechanisms and downstream targets in inflammation-mediated obesity and insulin resistance. Mol. Cell. Biochem. 2017, 426, 27–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, H.; Ullah, H.; Castilho, P.; Gomila, A.S.; D’Onofrio, G.; Filosa, R.; Wang, F.; Nabavi, S.M.; Daglia, M.; Silva, A.S.; et al. Targeting NF-kappaB signaling pathway in cancer by dietary polyphenols. Crit. Rev. Food Sci. Nutr. 2020, 60, 2790–2800. [Google Scholar] [CrossRef]
- Milenkovic, D.; Vanden Berghe, W.; Boby, C.; Leroux, C.; Declerck, K.; Szarc vel Szic, K.; Heyninck, K.; Laukens, K.; Bizet, M.; Defrance, M.; et al. Dietary flavanols modulate the transcription of genes associated with cardiovascular pathology without changes in their DNA methylation state. PLoS ONE 2014, 9, e95527. [Google Scholar] [CrossRef] [Green Version]
- Milenkovic, D.; Berghe, W.V.; Morand, C.; Claude, S.; van de Sandt, A.; Gorressen, S.; Monfoulet, L.E.; Chirumamilla, C.S.; Declerck, K.; Szic, K.S.V.; et al. A systems biology network analysis of nutri(epi)genomic changes in endothelial cells exposed to epicatechin metabolites. Sci. Rep. 2018, 8, 15487. [Google Scholar] [CrossRef]
- Claude, S.; Boby, C.; Rodriguez-Mateos, A.; Spencer, J.P.; Gerard, N.; Morand, C.; Milenkovic, D. Flavanol metabolites reduce monocyte adhesion to endothelial cells through modulation of expression of genes via p38-MAPK and p65-Nf-kB pathways. Mol. Nutr. Food Res. 2014, 58, 1016–1027. [Google Scholar] [CrossRef]
- Moreno-Ulloa, A.; Mendez-Luna, D.; Beltran-Partida, E.; Castillo, C.; Guevara, G.; Ramirez-Sanchez, I.; Correa-Basurto, J.; Ceballos, G.; Villarreal, F. The effects of (-)-epicatechin on endothelial cells involve the G protein-coupled estrogen receptor (GPER). Pharmacol. Res. 2015, 100, 309–320. [Google Scholar] [CrossRef] [Green Version]
- Yeung, F.; Hoberg, J.E.; Ramsey, C.S.; Keller, M.D.; Jones, D.R.; Frye, R.A.; Mayo, M.W. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004, 23, 2369–2380. [Google Scholar] [CrossRef] [Green Version]
- Yoshizaki, T.; Milne, J.C.; Imamura, T.; Schenk, S.; Sonoda, N.; Babendure, J.L.; Lu, J.C.; Smith, J.J.; Jirousek, M.R.; Olefsky, J.M. SIRT1 exerts anti-inflammatory effects and improves insulin sensitivity in adipocytes. Mol. Cell. Biol. 2009, 29, 1363–1374. [Google Scholar] [CrossRef] [Green Version]
- Yoshizaki, T.; Schenk, S.; Imamura, T.; Babendure, J.L.; Sonoda, N.; Bae, E.J.; Oh, D.Y.; Lu, M.; Milne, J.C.; Westphal, C.; et al. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am. J. Physiol. Endocrinol. Metab. 2010, 298, E419–E428. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.; Tang, X.; Chen, H.Z. Sirtuins and Insulin Resistance. Front. Endocrinol. 2018, 9, 748. [Google Scholar] [CrossRef] [Green Version]
- Olmos, Y.; Sanchez-Gomez, F.J.; Wild, B.; Garcia-Quintans, N.; Cabezudo, S.; Lamas, S.; Monsalve, M. SirT1 regulation of antioxidant genes is dependent on the formation of a FoxO3a/PGC-1alpha complex. Antioxid. Redox Signal. 2013, 19, 1507–1521. [Google Scholar] [CrossRef]
- Ren, Z.; He, H.; Zuo, Z.; Xu, Z.; Wei, Z.; Deng, J. The role of different SIRT1-mediated signaling pathways in toxic injury. Cell. Mol. Biol. Lett. 2019, 24, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, S.; Matter, C.M. Protective roles of SIRT1 in atherosclerosis. Cell. Cycle 2011, 10, 640–647. [Google Scholar] [CrossRef] [Green Version]
- Kane, A.E.; Sinclair, D.A. Sirtuins and NAD(+) in the Development and Treatment of Metabolic and Cardiovascular Diseases. Circ. Res. 2018, 123, 868–885. [Google Scholar] [CrossRef] [PubMed]
- Mazloom, H.; Alizadeh, S.; Esfahani, E.N.; Razi, F.; Meshkani, R. Decreased expression of microRNA-21 is associated with increased cytokine production in peripheral blood mononuclear cells (PBMCs) of obese type 2 diabetic and non-diabetic subjects. Mol. Cell. Biochem. 2016, 419, 11–17. [Google Scholar] [CrossRef]
- Hijmans, J.G.; Diehl, K.J.; Bammert, T.D.; Kavlich, P.J.; Lincenberg, G.M.; Greiner, J.J.; Stauffer, B.L.; DeSouza, C.A. Association between hypertension and circulating vascular-related microRNAs. J. Hum. Hypertens. 2018, 32, 440–447. [Google Scholar] [CrossRef]
- Ling, H.Y.; Hu, B.; Hu, X.B.; Zhong, J.; Feng, S.D.; Qin, L.; Liu, G.; Wen, G.B.; Liao, D.F. MiRNA-21 reverses high glucose and high insulin induced insulin resistance in 3T3-L1 adipocytes through targeting phosphatase and tensin homologue. Exp. Clin. Endocrinol. Diabetes 2012, 120, 553–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, A.; Yan, H.; Liang, J.; Du, C.; Zhao, X.; Sun, L.; Chen, Y. MicroRNA-21 regulates hepatic glucose metabolism by targeting FOXO1. Gene 2017, 627, 194–201. [Google Scholar] [CrossRef]
- Wang, S.; Liang, C.; Ai, H.; Yang, M.; Yi, J.; Liu, L.; Song, Z.; Bao, Y.; Li, Y.; Sun, L.; et al. Hepatic miR-181b-5p Contributes to Glycogen Synthesis Through Targeting EGR1. Dig. Dis. Sci. 2019, 64, 1548–1559. [Google Scholar] [CrossRef]
- Hori, D.; Dunkerly-Eyring, B.; Nomura, Y.; Biswas, D.; Steppan, J.; Henao-Mejia, J.; Adachi, H.; Santhanam, L.; Berkowitz, D.E.; Steenbergen, C.; et al. miR-181b regulates vascular stiffness age dependently in part by regulating TGF-beta signaling. PLoS ONE 2017, 12, e0174108. [Google Scholar] [CrossRef] [Green Version]
- Witkowski, M.; Witkowski, M.; Saffarzadeh, M.; Friebel, J.; Tabaraie, T.; Ta Bao, L.; Chakraborty, A.; Dorner, A.; Stratmann, B.; Tschoepe, D.; et al. Vascular miR-181b controls tissue factor-dependent thrombogenicity and inflammation in type 2 diabetes. Cardiovasc. Diabetol. 2020, 19, 20. [Google Scholar] [CrossRef]
- Oses, M.; Margareto Sanchez, J.; Portillo, M.P.; Aguilera, C.M.; Labayen, I. Circulating miRNAs as Biomarkers of Obesity and Obesity-Associated Comorbidities in Children and Adolescents: A Systematic Review. Nutrients 2019, 11, 2890. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Xu, H.; Pan, X.; Wu, W.; Wang, H.; Yan, L.; Zhang, M.; Liu, X.; Xia, S.; Shao, Q. miR-34a and miR-125b are upregulated in peripheral blood mononuclear cells from patients with type 2 diabetes mellitus. Exp. Ther. Med. 2017, 14, 5589–5596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Xu, Y.; Zhu, Y.; Sun, H.; Juguilon, C.; Li, F.; Fan, D.; Yin, L.; Zhang, Y. Macrophage miR-34a Is a Key Regulator of Cholesterol Efflux and Atherosclerosis. Mol. Ther. 2020, 28, 202–216. [Google Scholar] [CrossRef]
- Pan, Y.; Hui, X.; Hoo, R.L.C.; Ye, D.; Chan, C.Y.C.; Feng, T.; Wang, Y.; Lam, K.S.L.; Xu, A. Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. J. Clin. Investig. 2019, 129, 834–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scoditti, E.; Carpi, S.; Massaro, M.; Pellegrino, M.; Polini, B.; Carluccio, M.A.; Wabitsch, M.; Verri, T.; Nieri, P.; De Caterina, R. Hydroxytyrosol Modulates Adipocyte Gene and miRNA Expression Under Inflammatory Condition. Nutrients 2019, 11, 2493. [Google Scholar] [CrossRef] [Green Version]
- Oldenburg, J.; de Rooij, J. Mechanical control of the endothelial barrier. Cell Tissue Res. 2014, 355, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Orekhov, A.N.; Bobryshev, Y.V. Endothelial Barrier and Its Abnormalities in Cardiovascular Disease. Front. Physiol. 2015, 6, 365. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.R.; Ohlmeyer, M. Protein phosphatase 2A as a therapeutic target in inflammation and neurodegeneration. Pharmacol. Ther. 2019, 201, 181–201. [Google Scholar] [CrossRef]
- Afzal, M.; Redha, A.; AlHasan, R. Anthocyanins Potentially Contribute to Defense against Alzheimer’s Disease. Molecules 2019, 24, 4255. [Google Scholar] [CrossRef] [Green Version]
- Vauzour, D. Dietary polyphenols as modulators of brain functions: Biological actions and molecular mechanisms underpinning their beneficial effects. Oxid. Med. Cell. Longev. 2012, 2012, 914273. [Google Scholar] [CrossRef] [PubMed] [Green Version]










| Intervention | Participants | Study Design and Outcomes Related to Cardiometabolic Health | Gene Expression; Significantly Modulated Genes | Ref. | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plant Food/Extract/Bioactive | Dose | Duration of Intervention | Gender | Age (Years) | Number of Volunteers | Health Status | Study Design | Significantly Altered Biomarkers | RNA Type Studied | Method | Cells Analyzed for Gene Expression | Official Gene Symbol | Official Gene Name (from GeneCards) | |
| Olive oil polyphenols | 25 mL olive oil/day with high polyphenol content (366 mg/kg) vs. 25 mL olive oil/day with low polyphenol content (2.7 mg/kg) | 3 weeks | M | 20–60 | 18 | Healthy | Randomized, crossover, controlled study | Decreased diastolic blood pressure, BMI, total cholesterol, LDL-c, oxLDL, MCP1 | mRNA | RT-qPCR | PBMCs | CD40LG | CD40 Ligand | [51] |
| IL23A | Interleukin 23 Subunit Alpha | |||||||||||||
| IL7R | Interleukin 7 Receptor | |||||||||||||
| CXCR1 | C-X-C Motif Chemokine Receptor 1 | |||||||||||||
| ADRB2 | Adrenoceptor Beta 2 | |||||||||||||
| OLR1 | Oxidized Low Density Lipoprotein Receptor 1 | |||||||||||||
| Olive oil polyphenols | 30 mL olive oil with high polyphenol content—HPC (961 mg/kg) vs. 30 mL olive oil with moderate polyphenol content—MPC (289 mg/kg) | 5 h acute study | F, M | 20–75 | 13 | Prehypertension or stage 1 hypertension without antihypertensive treatment | Randomized, double-blind, crossover, controlled study | Decreased glucose and oxidized LDL after both interventions. Multiple regression analyses showed that with HPC intervention changes in gene expression were related to a decrease in oxidized low-density lipoproteins and with an increase in oxygen radical absorbance capacity and olive oil polyphenols. These associations were not found after MPC ingestion. | mRNA | RT-qPCR | White blood cells | ABCA1 | ATP Binding Cassette Subfamily A Member 1 | [52] |
| SCARB1 | Scavenger Receptor Class B Member 1 | |||||||||||||
| MED1 | Mediator Complex Subunit 1 | |||||||||||||
| PPARA | Peroxisome Proliferator Activated Receptor Alpha | |||||||||||||
| PPARG | Peroxisome Proliferator Activated Receptor Gamma | |||||||||||||
| PPARD | Peroxisome Proliferator Activated Receptor Delta | |||||||||||||
| CD36 | CD36 Molecule | |||||||||||||
| PTGS1 | Prostaglandin-Endoperoxide Synthase 1 | |||||||||||||
| Olive oil polyphenols | 25 mL olive oil/day with high polyphenol content (366 mg/kg) vs. 25 mL olive oil/day with low polyphenol content (2.7 mg/kg) | 3 weeks | M | 20–60 | 18 | Healthy | Randomized, double-blind, crossover, controlled study | Decreased diastolic blood pressure, total cholesterol, LDL-c and oxLDL | mRNA | RT-qPCR | PBMCs | CXCR2 | C-X-C Motif Chemokine Receptor 2 | [53] |
| Resveratrol | 800 mg/day | 2 months | F, M | 30–70 | 46 | Type 2 diabetes | Randomized, double-blind, placebo-controlled, parallel study | Increased plasma total thiol and total antioxidant capacity. Decreased plasma protein carbonyl, systolic and diastolic blood pressure, body weight, BMI, and intracellular superoxide anion in PBMCs. | mRNA | RT-qPCR | PBMCs | NFE2L2 | Nuclear Factor, Erythroid 2 Like 2 | [54] |
| SOD2 | Superoxide Dismutase 2 | |||||||||||||
| Quercetin | 1000 mg/day | 12 weeks | F | 20–40 | 78 | Overweight or obese with polycystic ovary syndrome | Randomized, double-blind, placebo-controlled, parallel study | Decreased plasma resistin | mRNA | RT-qPCR | PBMCs | RETN | Resistin | [55] |
| Curcumin | 5 g | 2 h acute study | F, M | 50–64 | 5, 5 | Healthy smokers, postmenopausal | Randomized, double-blind, placebo-controlled, crossover study | Increased FMD, decreased pulse pressure | mRNA | RT-qPCR | PBMCs | CXCR6 | C-X-C Motif Chemokine Receptor 6 | [56] |
| CXCR3 | C-X-C Motif Chemokine Receptor 3 | |||||||||||||
| CXCL9 | C-X-C Motif Chemokine Ligand 9 | |||||||||||||
| CXCL17 | C-X-C Motif Chemokine Ligand 17 | |||||||||||||
| CXCL16 | C-X-C Motif Chemokine Ligand 16 | |||||||||||||
| CXCL10 | C-X-C Motif Chemokine Ligand 10 | |||||||||||||
| CX3CR1 | C-X3-C Motif Chemokine Receptor 1 | |||||||||||||
| CCR7 | C-C Motif Chemokine Receptor 7 | |||||||||||||
| CCR1 | C-C Motif Chemokine Receptor 1 | |||||||||||||
| CCL3 | C-C Motif Chemokine Ligand 3 | |||||||||||||
| RAC1 | Rac Family Small GTPase 1 | |||||||||||||
| PLA2G7 | Phospholipase A2 Group VII | |||||||||||||
| PECAM1 | Platelet And Endothelial Cell Adhesion Molecule 1 | |||||||||||||
| PCDH12 | Protocadherin 12 | |||||||||||||
| ITGB3 | Integrin Subunit Beta 3 | |||||||||||||
| ITGB2 | Integrin Subunit Beta 2 | |||||||||||||
| ITGA5 | Integrin Subunit Alpha 5 | |||||||||||||
| ICAM3 | Intercellular Adhesion Molecule 3 | |||||||||||||
| ICAM2 | Intercellular Adhesion Molecule 2 | |||||||||||||
| GJA3 | Gap Junction Protein Alpha 3 | |||||||||||||
| CD40 | CD40 Molecule | |||||||||||||
| ABCG1 | ATP Binding Cassette Subfamily G Member 1 | |||||||||||||
| ABCC2 | ATP Binding Cassette Subfamily C Member 2 | |||||||||||||
| ABCB4 | ATP Binding Cassette Subfamily B Member 4 | |||||||||||||
| ABCA4 | ATP Binding Cassette Subfamily A Member 4 | |||||||||||||
| ABCA2 | ATP Binding Cassette Subfamily A Member 2 | |||||||||||||
| ADIPOR1 | Adiponectin Receptor 1 | |||||||||||||
| ADIPOR2 | Adiponectin Receptor 2 | |||||||||||||
| FASN | Fatty Acid Synthase | |||||||||||||
| LIPA | Lipase A, Lysosomal Acid Type | |||||||||||||
| IL6 | Interleukin 6 | |||||||||||||
| STAT1 | Signal Transducer And Activator Of Transcription 1 | |||||||||||||
| CCL20 | C-C Motif Chemokine Ligand 20 | |||||||||||||
| CCL22 | C-C Motif Chemokine Ligand 22 | |||||||||||||
| CCR5 | C-C Motif Chemokine Receptor 5 (Gene/Pseudogene) | |||||||||||||
| CXCL6 | C-X-C Motif Chemokine Ligand 6 | |||||||||||||
| ABCA2 | ATP Binding Cassette Subfamily A Member 2 | |||||||||||||
| IL1R2 | Interleukin 1 Receptor Type 2 | |||||||||||||
| Grape extract (GE) or grape extract plus resveratrol (GE-Res) | 1 capsule/day of GE, GE-Res or placebo in the morning for the first 6 months, and 2 capsules/day for the following 6 months. The phenolic content of the GE and the GE-Res was very similar (151 ± 17 mg and 139 ± 18 mg phenolics per capsule, respectively) but GE-Res also contained 8.1 ± 0.5 mg of resveratrol per capsule. | 1 year | M | Adults, up to 80 years old | 18 | Type 2 diabetes, hypertension, and coronary artery disease | Randomized, triple-blind, placebo-controlled, dose-response, 1-year follow-up study with three parallel arms designated as placebo (maltodextrin), GE (conventional grape extract) and GE-Res (grape extract containing resveratrol) | The following data is extracted from the previous paper [58]: 1. GE-Res vs. Placebo-increased adiponectin, decreased PAI1, total cholesterol, glucose and HbA1c | mRNA, miRNA | RT-qPCR | PBMCs | IL1B | Interleukin 1 Beta | [57] |
| TNF | Tumor Necrosis Factor | |||||||||||||
| NFKBIA | NFKB Inhibitor Alpha | |||||||||||||
| hsa-miR-21-5p | ||||||||||||||
| hsa-miR-181b-5p | ||||||||||||||
| hsa-miR-663a | ||||||||||||||
| hsa-miR-30c-2-3p | ||||||||||||||
| hsa-miR-34a-5p | ||||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruskovska, T.; Budić-Leto, I.; Corral-Jara, K.F.; Ajdžanović, V.; Arola-Arnal, A.; Bravo, F.I.; Deligiannidou, G.-E.; Havlik, J.; Janeva, M.; Kistanova, E.; et al. Systematic Bioinformatic Analyses of Nutrigenomic Modifications by Polyphenols Associated with Cardiometabolic Health in Humans—Evidence from Targeted Nutrigenomic Studies. Nutrients 2021, 13, 2326. https://doi.org/10.3390/nu13072326
Ruskovska T, Budić-Leto I, Corral-Jara KF, Ajdžanović V, Arola-Arnal A, Bravo FI, Deligiannidou G-E, Havlik J, Janeva M, Kistanova E, et al. Systematic Bioinformatic Analyses of Nutrigenomic Modifications by Polyphenols Associated with Cardiometabolic Health in Humans—Evidence from Targeted Nutrigenomic Studies. Nutrients. 2021; 13(7):2326. https://doi.org/10.3390/nu13072326
Chicago/Turabian StyleRuskovska, Tatjana, Irena Budić-Leto, Karla Fabiola Corral-Jara, Vladimir Ajdžanović, Anna Arola-Arnal, Francisca Isabel Bravo, Georgia-Eirini Deligiannidou, Jaroslav Havlik, Milkica Janeva, Elena Kistanova, and et al. 2021. "Systematic Bioinformatic Analyses of Nutrigenomic Modifications by Polyphenols Associated with Cardiometabolic Health in Humans—Evidence from Targeted Nutrigenomic Studies" Nutrients 13, no. 7: 2326. https://doi.org/10.3390/nu13072326
APA StyleRuskovska, T., Budić-Leto, I., Corral-Jara, K. F., Ajdžanović, V., Arola-Arnal, A., Bravo, F. I., Deligiannidou, G.-E., Havlik, J., Janeva, M., Kistanova, E., Kontogiorgis, C., Krga, I., Massaro, M., Miler, M., Milosevic, V., Morand, C., Scoditti, E., Suárez, M., Vauzour, D., & Milenkovic, D. (2021). Systematic Bioinformatic Analyses of Nutrigenomic Modifications by Polyphenols Associated with Cardiometabolic Health in Humans—Evidence from Targeted Nutrigenomic Studies. Nutrients, 13(7), 2326. https://doi.org/10.3390/nu13072326