α-Lipoic Acid Alleviates Hepatic Lipid Deposition by Inhibiting FASN Expression via miR-3548 in Rats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. Serum and Hepatic Biochemical Analyses
2.3. Oil Red-O Staining
2.4. RNA Isolation and mRNA Quantification by Real-Time Polymerase Chain Reaction (PCR)
2.5. Protein Extraction and Western Blot Analysis
2.6. MiRNA Sequencing, Bioinformatics Analyses, and In Vivo Validation
2.7. Cell Culture and Dual-Luciferase Reporter Assay
2.8. Statistical Analysis
3. Results
3.1. α-Lipoic Acid Does Not Affect Rat Growth Performance
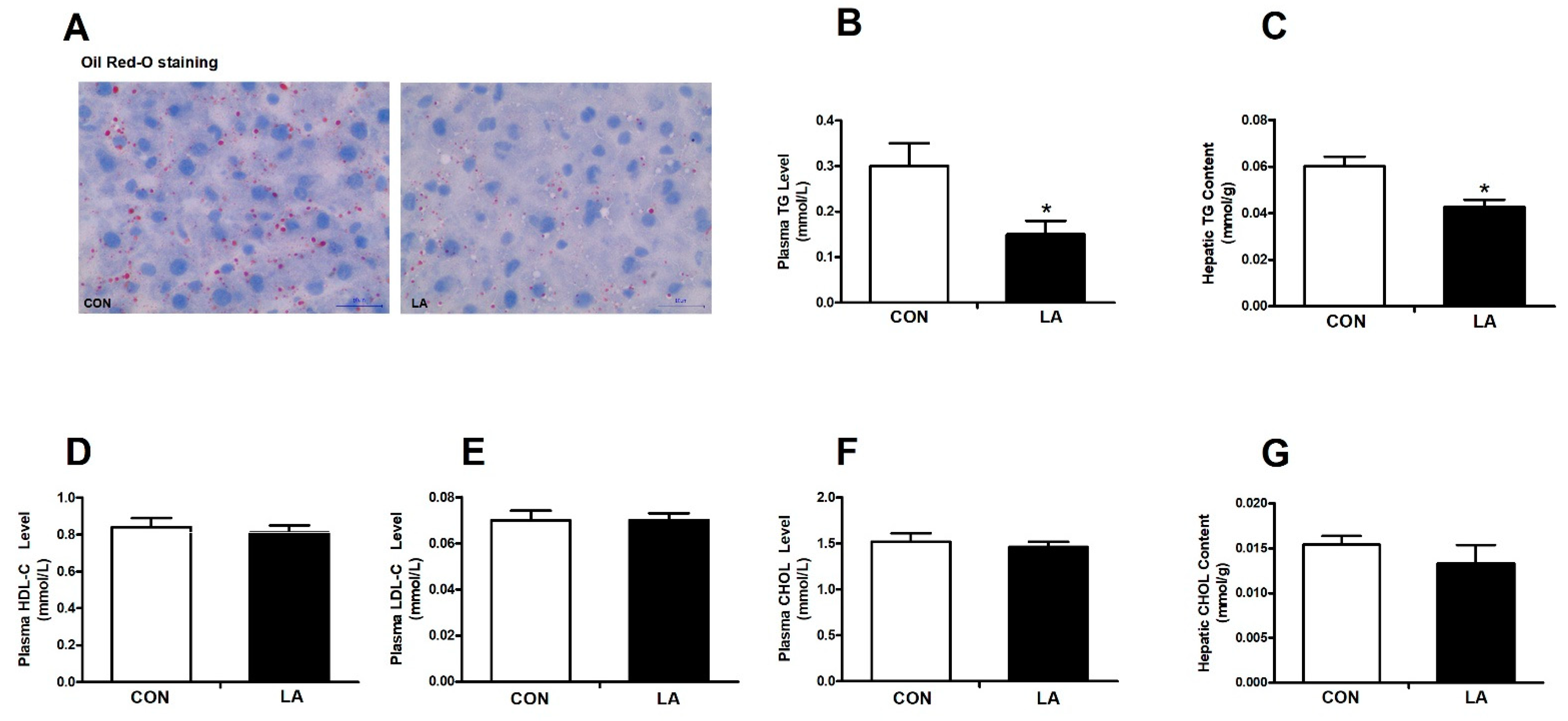
3.2. α-Lipoic Acid Lowers the Plasma Triglyceride Level and Hepatic Triglyceride Content
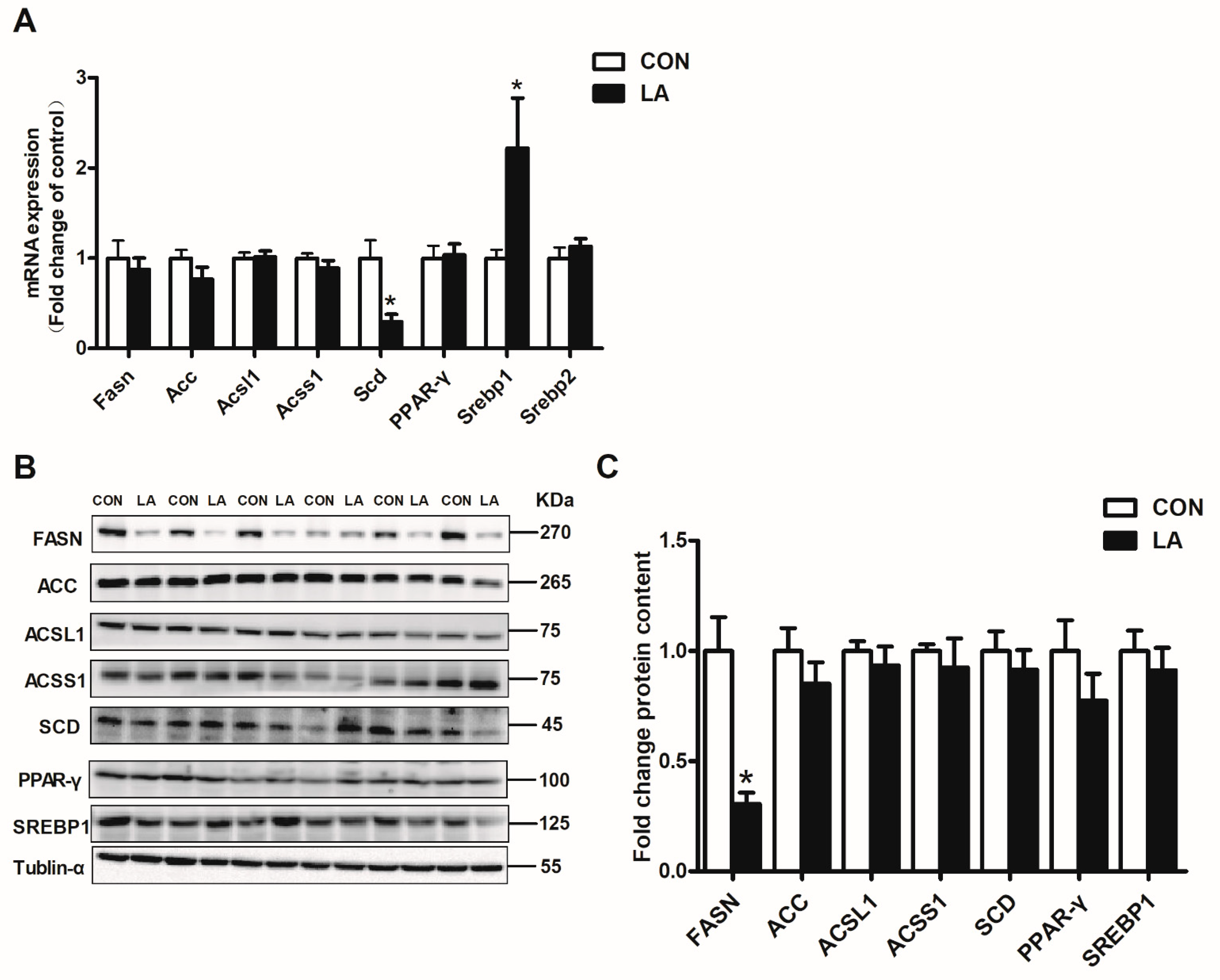
3.3. α-Lipoic Acid Reverses Hepatic Lipid Synthesis-Related Genes and Protein Expression
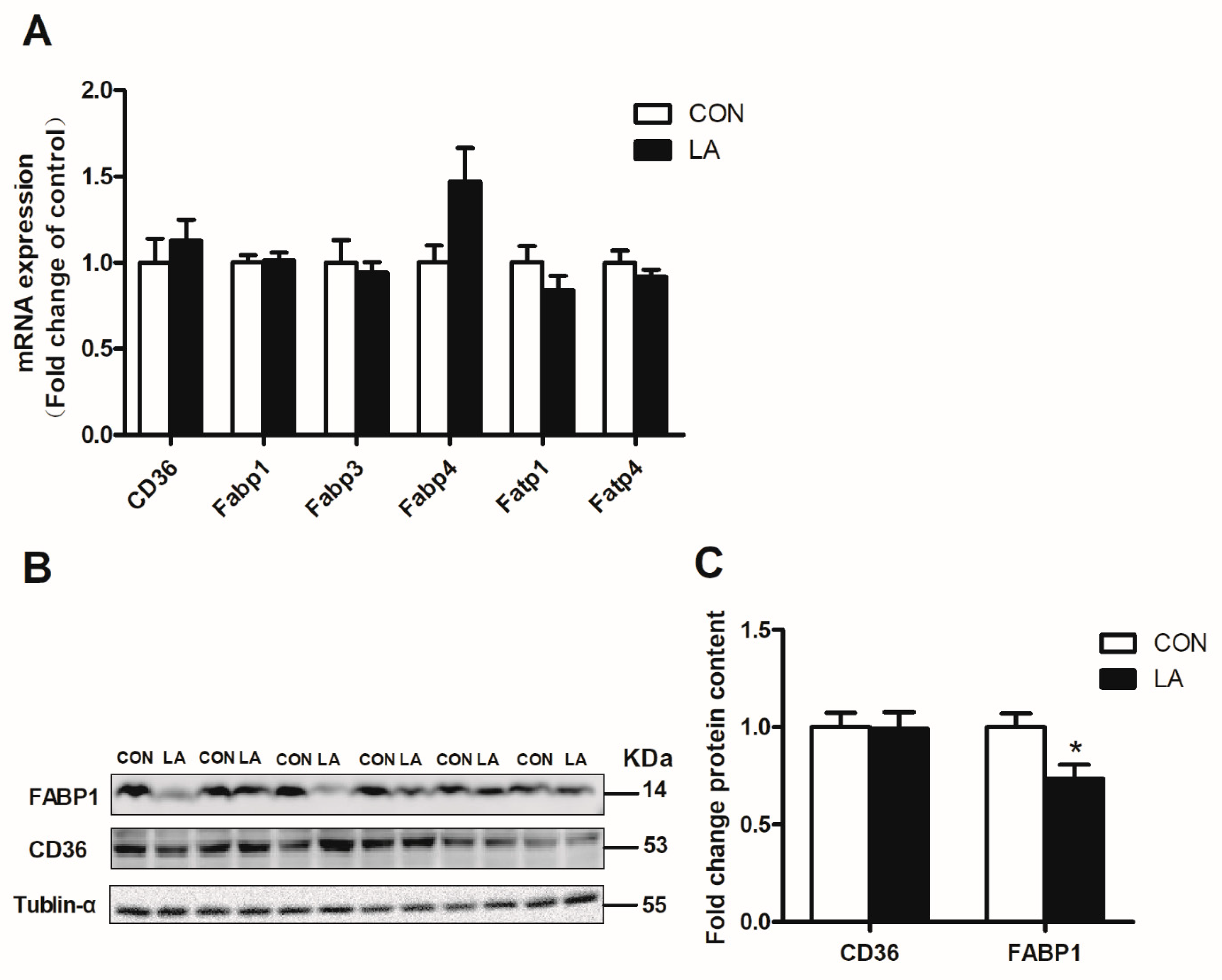
3.4. α-Lipoic Acid Reverses Hepatic Lipolysis and Fatty Acid Transport-Related Gene and Protein Expression
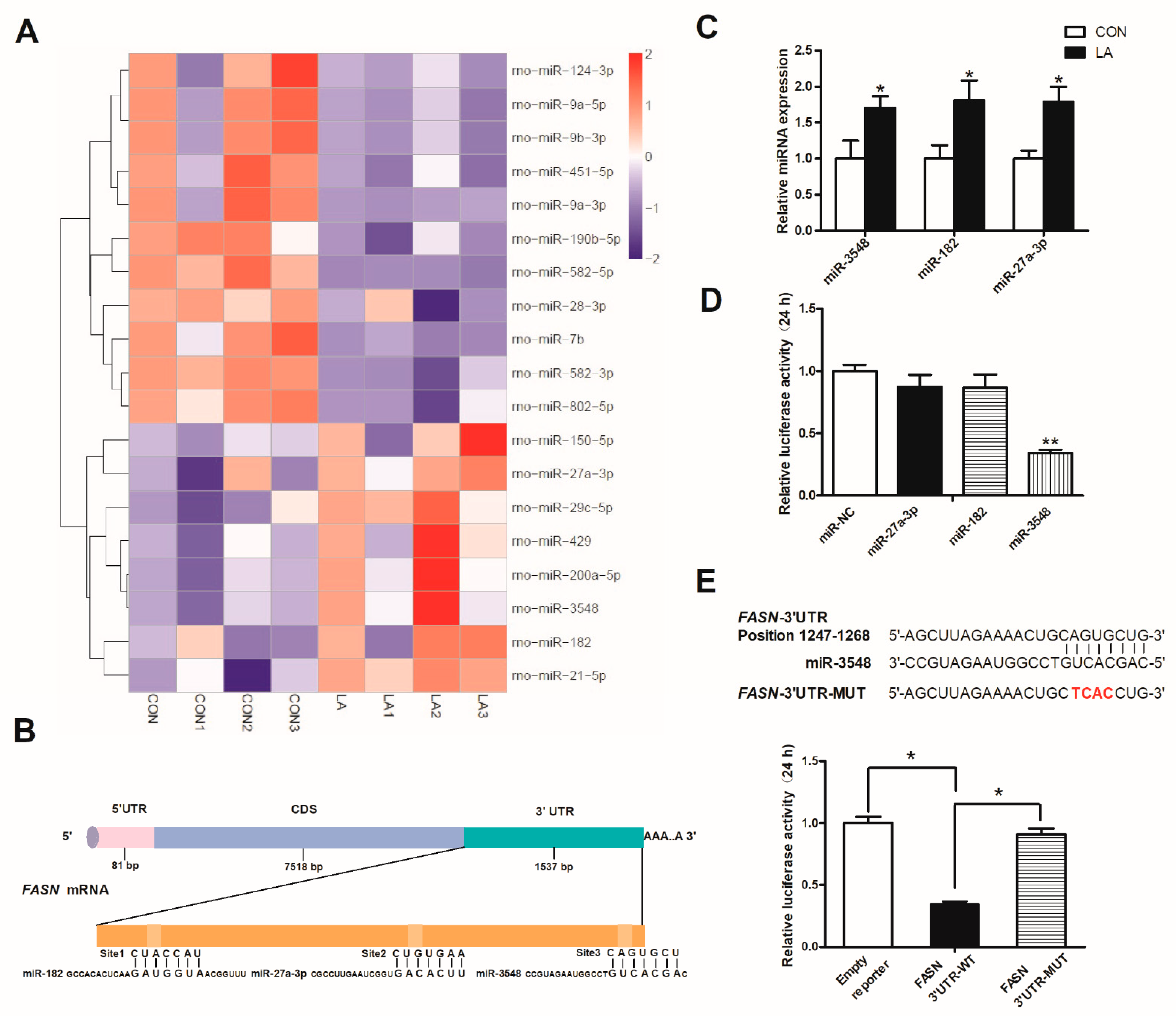
3.5. α-Lipoic Acid Downregulates FASN Expression through miRNA-3548
4. Discussion
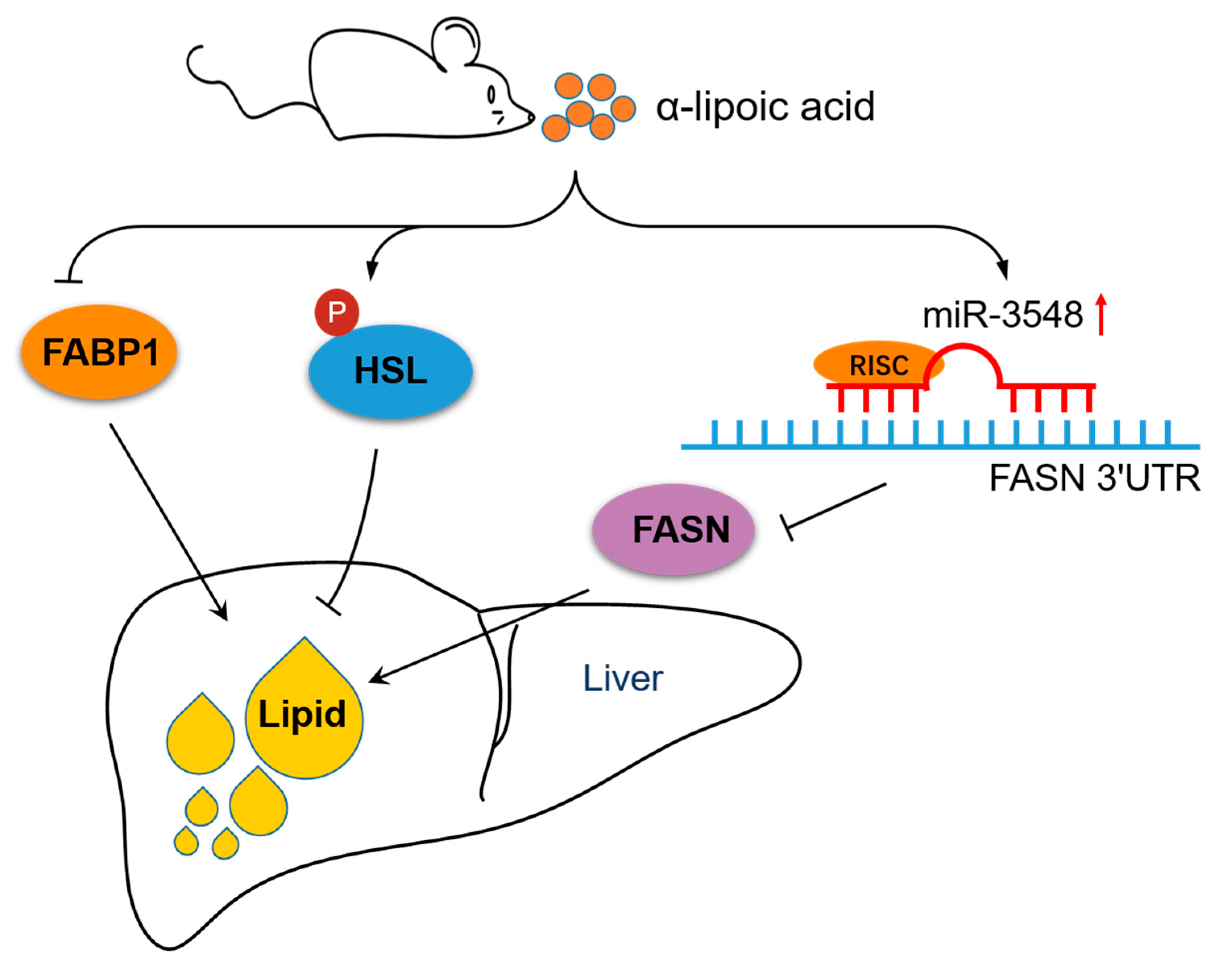
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ibrahim, S.H.; Kohli, R.; Gores, G.J. Mechanisms of lipotoxicity in NAFLD and clinical implications. J. Pediatr. Gastr. Nutr. 2011, 53, 131–140. [Google Scholar] [CrossRef] [Green Version]
- Targher, G.; Bertolini, L.; Padovani, R.; Rodella, S.; Tessari, R.; Zenari, L.; Day, C.; Arcaro, G. Prevalence of nonalcoholic fatty liver disease and its association with cardiovascular disease among type 2 diabetic patients. Diabetes Care 2007, 30, 1212–1218. [Google Scholar] [CrossRef] [Green Version]
- Fan, H.; Ma, X.; Lin, P.; Kang, Q.; Zhao, Z.; Wang, L.; Sun, D.; Cheng, J.; Li, Y. Scutellarin prevents nonalcoholic fatty liver disease (NAFLD) and hyperlipidemia via PI3K/AKT-dependent activation of nuclear factor (Erythroid-derived 2)-like 2 (Nrf2) in rats. Med. Sci. Monit. 2017, 23, 5599–5612. [Google Scholar] [CrossRef] [Green Version]
- Kitade, H.; Chen, G.; Ni, Y.; Ota, T. Nonalcoholic fatty liver disease and insulin resistance: New insights and potential new treatments. Nutrients 2017, 9, 387. [Google Scholar] [CrossRef] [Green Version]
- Ullah, R.; Rauf, N.; Nabi, G.; Ullah, H.; Shen, Y.; Zhou, Y.D.; Fu, J. Role of nutrition in the pathogenesis and prevention of non-alcoholic fatty liver disease: Recent updates. Int. J. Biol. Sci. 2019, 15, 265–276. [Google Scholar] [CrossRef] [Green Version]
- Haslam, D.W.; James, W.P. Obesity. Lancet 2005, 366, 1197–1209. [Google Scholar] [CrossRef]
- Wright, S.M.; Aronne, L.J. Causes of obesity. Abdom. Imaging 2012, 37, 730–732. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.; Anstee, Q.M.; Marietti, M.; Hardy, T.; Henry, L.; Eslam, M.; George, J.; Bugianesi, E. Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Bessone, F.; Razori, M.V.; Roma, M.G. Molecular pathways of nonalcoholic fatty liver disease development and progression. Cell. Mol. Life Sci. 2019, 76, 99–128. [Google Scholar] [CrossRef] [PubMed]
- Pierantonelli, I.; Svegliati-Baroni, G. Nonalcoholic fatty liver disease: Basic pathogenetic mechanisms in the progression from NAFLD to NASH. Transplantation 2019, 103, e1–e13. [Google Scholar] [CrossRef] [PubMed]
- Ipsen, D.H.; Lykkesfeldt, J.; Tveden-Nyborg, P. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell. Mol. Life Sci. 2018, 75, 3313–3327. [Google Scholar] [CrossRef] [Green Version]
- Ameer, F.; Scandiuzzi, L.; Hasnain, S.; Kalbacher, H.; Zaidi, N. De novo lipogenesis in health and disease. Metabolism 2014, 63, 895–902. [Google Scholar] [CrossRef]
- Solinas, G.; Boren, J.; Dulloo, A.G. De novo lipogenesis in metabolic homeostasis: More friend than foe? Mol. Metab. 2015, 4, 367–377. [Google Scholar] [CrossRef]
- Glatz, J.F.; van der Vusse, G.J. Cellular fatty acid-binding proteins: Their function and physiological significance. Prog. Lipid Res. 1996, 35, 243–282. [Google Scholar] [CrossRef]
- Mao, J.; DeMayo, F.J.; Li, H.; Abu-Elheiga, L.; Gu, Z.; Shaikenov, T.E.; Kordari, P.; Chirala, S.S.; Heird, W.C.; Wakil, S.J. Liver-specific deletion of acetyl-CoA carboxylase 1 reduces hepatic triglyceride accumulation without affecting glucose homeostasis. Proc. Natl. Acad. Sci. USA 2006, 103, 8552–8557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, C.; Tran, J.L.; Erion, D.M.; Vera, N.B.; Febbraio, M.; Weiss, E.J. Hepatocyte-specific disruption of CD36 attenuates fatty liver and improves insulin sensitivity in HFD-fed mice. Endocrinology 2016, 157, 570–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koonen, D.P.; Jacobs, R.L.; Febbraio, M.; Young, M.E.; Soltys, C.L.; Ong, H.; Vance, D.E.; Dyck, J.R. Increased hepatic CD36 expression contributes to dyslipidemia associated with diet-induced obesity. Diabetes 2007, 56, 2863–2871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucas, S.; Tavernier, G.; Tiraby, C.; Mairal, A.; Langin, D. Expression of human hormone-sensitive lipase in white adipose tissue of transgenic mice increases lipase activity but does not enhance in vitro lipolysis. J. Lipid Res. 2003, 44, 154–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haemmerle, G.; Zimmermann, R.; Hayn, M.; Theussl, C.; Waeg, G.; Wagner, E. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. J. Biol. Chem. 2002, 277, 4806–4815. [Google Scholar] [CrossRef] [Green Version]
- Newberry, E.P.; Xie, Y.; Kennedy, S.; Han, X.; Buhman, K.K.; Luo, J.; Gross, R.W.; Davidson, N.O. Decreased hepatic triglyceride accumulation and altered fatty acid uptake in mice with deletion of the liver fatty acid-binding protein gene. J. Biol. Chem. 2003, 278, 51664–51672. [Google Scholar] [CrossRef] [Green Version]
- Martin, G.G.; Atshaves, B.P.; Huang, H.; McIntosh, A.L.; Williams, B.J.; Pai, P.J. Hepatic phenotype of liver fatty acid binding protein gene-ablated mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2009, 297, G1053–G1065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moura, F.A.; de Andrade, K.Q.; dos Santos, J.C.; Goulart, M.O. Lipoic acid: Its antioxidant and anti-inflammatory role and clinical applications. Curr. Top. Med. Chem. 2015, 15, 458–483. [Google Scholar] [CrossRef] [PubMed]
- Biewenga, G.P.; Haenen, G.R.; Bast, A. The pharmacology of the antioxidant lipoic acid. Gen. Pharmacol. 1997, 29, 315–331. [Google Scholar] [CrossRef]
- Gomes, M.B.; Negrato, C.A. Alpha-lipoic acid as a pleiotropic compound with potential therapeutic use in diabetes and other chronic diseases. Diabetol. Metab. Syndr. 2014, 6, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.H.; Wang, D.W.; Xu, S.F.; Zhang, S.; Fan, Y.G.; Yang, Y.Y.; Guo, S.Q.; Wang, S.; Guo, T.; Wang, Z.Y.; et al. Alpha-Lipoic acid improves abnormal behavior by mitigation of oxidative stress, inflammation, ferroptosis, and tauopathy in P301S Tau transgenic mice. Redox Biol. 2018, 14, 535–548. [Google Scholar] [CrossRef]
- Lai, Y.S.; Shih, C.Y.; Huang, Y.F.; Chou, T.C. Antiplatelet activity of alpha-lipoic acid. J. Agric. Food Chem. 2010, 58, 8596–8603. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Cai, X.; Xu, Y.; Zhu, C.; Wang, L.; Wang, S.; Zhu, X.; Gao, P.; Zhang, Y.; Jiang, Q.; et al. Alpha-Lipoic acids promote the protein synthesis of C2C12 myotubes by the TLR2/PI3K signaling pathway. J. Agric. Food Chem. 2016, 64, 1720–1729. [Google Scholar] [CrossRef] [PubMed]
- Park, K.G.; Min, A.K.; Koh, E.H.; Kim, H.S.; Kim, M.O.; Park, H.S.; Kim, Y.D.; Yoon, T.S.; Jang, B.K.; Hwang, J.S.; et al. Alpha-lipoic acid decreases hepatic lipogenesis through adenosine monophosphate-activated protein kinase (AMPK)-dependent and AMPK-independent pathways. Hepatology 2008, 48, 1477–1486. [Google Scholar] [CrossRef] [Green Version]
- Valdecantos, M.P.; Perez-Matute, P.; Gonzalez-Muniesa, P.; Prieto-Hontoria, P.L.; Moreno-Aliaga, M.J.; Martinez, J.A. Lipoic acid administration prevents nonalcoholic steatosis linked to long-term high-fat feeding by modulating mitochondrial function. J. Nutr. Biochem. 2012, 23, 1676–1684. [Google Scholar] [CrossRef]
- Yang, Y.; Li, W.; Liu, Y.; Sun, Y.; Li, Y.; Yao, Q.; Li, J.; Zhang, Q.; Gao, Y.; Gao, L.; et al. Alpha-lipoic acid improves high-fat diet-induced hepatic steatosis by modulating the transcription factors SREBP-1, FoxO1 and Nrf2 via the SIRT1/LKB1/AMPK pathway. J. Nutr. Biochem. 2014, 25, 1207–1217. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Galilea, M.; Perez-Matute, P.; Prieto-Hontoria, P.L.; Sainz, N.; Lopez-Yoldi, M.; Houssier, M.; Martínez, J.A.; Langin, D.; Moreno-Aliaga, M.J. Alpha-lipoic acid reduces fatty acid esterification and lipogenesis in adipocytes from overweight/obese subjects. Obesity 2014, 22, 2210–2215. [Google Scholar] [CrossRef] [PubMed]
- Teachey, M.K.; Taylor, Z.C.; Maier, T.; Saengsirisuwan, V.; Sloniger, J.A.; Jacob, S.; Klatt, M.J.; Ptock, A.; Kraemer, K.; Hasselwander, O.; et al. Interactions of conjugated linoleic acid and lipoic acid on insulin action in the obese Zucker rat. Metabolism 2003, 52, 1167–1174. [Google Scholar] [CrossRef] [Green Version]
- Guo, S.; Yang, C.; Jiang, S.; Ni, Y.; Zhao, R.; Ma, W. Repeated Restraint Stress Enhances Hepatic TFR2 Expression and Induces Hepatic Iron Accumulation in Rats. Biol. Trace Elem. Res. 2020, 196, 590–596. [Google Scholar] [CrossRef]
- Mehlem, A.; Hagberg, C.E.; Muhl, L.; Eriksson, U.; Falkevall, A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat. Protoc. 2013, 8, 1149–1154. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Guo, S.; Li, H.; Ni, Y.; Ma, W.; Zhao, R. Identification and functional verification of microRNA-16 family targetingintestinal divalent metal transporter 1 (DMT1) In Vitro and In Vivo. Front. Physiol. 2019, 10, 819. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Fang, X.; Liu, M.; Ni, Y.; Ma, W.; Zhao, R. MiR-20b down-regulates intestinal ferroportin expression In Vitro and In Vivo. Cells 2019, 8, 1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Q.W.; Jones, C.S.; Kalchayanand, N.; Zhu, M.J.; Du, M. Effect of dietary alpha-lipoic acid on growth, body composition, muscle pH, and AMP-activated protein kinase phosphorylation in mice. J. Anim. Sci. 2005, 83, 2611–2617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.J.; Bird, K.E.; McMillen, T.S.; LeBoeuf, R.C.; Hagen, T.M.; Frei, B. Dietary alpha-lipoic acid supplementation inhibits atherosclerotic lesion development in apolipoprotein E-deficient and apolipoprotein E/low-density lipoprotein receptor-deficient mice. Circulation 2008, 117, 421–428. [Google Scholar] [CrossRef] [Green Version]
- Seo, E.Y.; Ha, A.W.; Kim, W.K. Alpha-Lipoic acid reduced weight gain and improved the lipid profile in rats fed with high fat diet. Nutr. Res. Pract. 2012, 6, 195–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arshad, M.S.; Anjum, F.M.; Khan, M.I.; Shahid, M. Wheat germ oil and alpha-lipoic acid predominantly improve the lipid profile of broiler meat. J. Agric. Food Chem. 2013, 61, 11158–11165. [Google Scholar] [CrossRef] [PubMed]
- Carbonelli, M.G.; Di Renzo, L.; Bigioni, M.; Di Daniele, N.; De Lorenzo, A.; Fusco, M.A. Alpha-lipoic acid supplementation: A tool for obesity therapy? Curr. Pharm. Des. 2010, 16, 840–846. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Park, D.W.; Choi, S.H.; Kim, J.J.; Cho, H.S. A preliminary investigation of alpha-lipoic acid treatment of antipsychotic drug-induced weight gain in patients with schizophrenia. J. Clin. Psychopharmacol. 2008, 28, 138–146. [Google Scholar] [CrossRef]
- Li, N.; Yan, W.; Hu, X.; Huang, Y.; Wang, F.; Zhang, W.; Wang, Q.; Wang, X.; Sun, W. Effects of oral alpha-lipoic acid administration on body weight in overweight or obese subjects: A crossover randomized, double-blind, placebo-controlled trial. Clin. Endocrinol. 2017, 86, 680–687. [Google Scholar] [CrossRef]
- Jayakumar, A.; Chirala, S.S.; Chinault, A.C.; Baldini, A.; Abu-Elheiga, L.; Wakil, S.J. Isolation and chromosomal mapping of genomic clones encoding the human fatty acid synthase gene. Genomics 1994, 23, 420–424. [Google Scholar] [CrossRef]
- Ducheix, S.; Peres, C.; Hardfeldt, J.; Frau, C.; Mocciaro, G.; Piccinin, E.; Lobaccaro, J.M.; De Santis, S.; Chieppa, M.; Bertrand-Michel, J.; et al. Deletion of stearoyl-coA desaturase-1 from the intestinal epithelium promotes inflammation and tumorigenesis, reversed by dietary oleate. Gastroenterology 2018, 155, 1524–1538. [Google Scholar] [CrossRef] [PubMed]
- Zhen, M.; Xu, J.; Zhang, Z.; Zou, X.; Wang, X.; Wang, X. Htd2 deficiency-associated suppression of α-lipoic acid production provokes mitochondrial dysfunction and insulin resistance in adipocytes. Redox Biol. 2021, 41, 101948. [Google Scholar]
- Chen, A.; Tang, Y.; Davis, V.; Hsu, F.F.; Kennedy, S.M.; Song, H.; Turk, J.; Brunt, E.M.; Newberry, E.P.; Davidson, N.O. L-FABP modulates murine stellate cell activation and diet induced nonalcoholic fatty liver disease. Hepatology 2013, 57, 2202–2212. [Google Scholar] [CrossRef] [Green Version]
- Newberry, E.P.; Kennedy, S.M.; Xie, Y.; Sternard, B.T.; Luo, J.; Davidson, N.O. Diet-induced obesity and hepatic steatosis in L-FABP-/- mice is abrogated with SF, but not PUFA, feeding and attenuated after cholesterol supplementation. Am. J. Physiol. Gastrointest. Liver Phys. 2008, 294, G307–G314. [Google Scholar] [CrossRef] [PubMed]
- Duncan, R.E.; Ahmadian, M.; Jaworski, K.; Sarkadi-Nagy, E.; Sul, H.S. Regulation of lipolysis in adipocytes. Annu. Rev. Nutr. 2007, 27, 79–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, T.S.; Jessen, N.; Jorgensen, J.O.; Moller, N.; Lund, S. Dissecting adipose tissue lipolysis: Molecular regulation and implications for metabolic disease. J. Mol. Endocrinol. 2014, 52, R199–R222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabbrini, E.; Sullivan, S.; Klein, S. Obesity and nonalcoholic fatty liver disease: Biochemical, metabolic, and clinical implications. Hepatology 2010, 51, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Song, L.R.; Li, D.; Weng, J.C.; Li, C.B.; Wang, L.; Wu, Z.; Zhang, J.T. MicroRNA-195 functions as a tumor suppressor by directly targeting fatty acid synthase in malignant meningioma. World Neurosurg. 2020, 136, e355–e364. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Tang, Y.; Tang, E.; Lu, W. MicroRNA-103 represses hepatic de novo lipogenesis and alleviates NAFLD via targeting FASN and SCD1. Biochem. Biophys. Res. Commun. 2020, 524, 716–722. [Google Scholar] [CrossRef]
- Yang, Z.; Zhong, L.; Xian, R.; Yuan, B. MicroRNA-223 regulates inflammation and brain injury via feedback to NLRP3 inflammasome after intracerebral hemorrhage. Mol. Immunol. 2015, 65, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Yan, K.; Sun, B.; Gao, S.; Yang, X.; Ni, Y.; Ma, W.; Zhao, R. Long-Term high-fat diet decreases hepatic iron storage associated with suppressing TFR2 and ZIP14 expression in rats. J. Agric. Food Chem. 2018, 66, 11612–11621. [Google Scholar] [CrossRef] [PubMed]

| Parameters | CON | LA | p-Value |
|---|---|---|---|
| Initial body weight (g) | 179.07 ± 3.37 | 175.33 ± 2.01 | 0.43 |
| Final body weight (g) | 416.51 ± 12.68 | 401.86 ± 9.51 | 0.49 |
| ADG (g/d) | 6.99 ± 0.32 | 6.66 ± 0.24 | 0.43 |
| ADFI (g/d) | 24.84 ± 0.86 | 23.55 ± 0.63 | 0.16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Yan, K.; Fang, X.; Ni, Y.; Ma, W.; Zhao, R. α-Lipoic Acid Alleviates Hepatic Lipid Deposition by Inhibiting FASN Expression via miR-3548 in Rats. Nutrients 2021, 13, 2331. https://doi.org/10.3390/nu13072331
Guo S, Yan K, Fang X, Ni Y, Ma W, Zhao R. α-Lipoic Acid Alleviates Hepatic Lipid Deposition by Inhibiting FASN Expression via miR-3548 in Rats. Nutrients. 2021; 13(7):2331. https://doi.org/10.3390/nu13072331
Chicago/Turabian StyleGuo, Shihui, Kai Yan, Xi Fang, Yingdong Ni, Wenqiang Ma, and Ruqian Zhao. 2021. "α-Lipoic Acid Alleviates Hepatic Lipid Deposition by Inhibiting FASN Expression via miR-3548 in Rats" Nutrients 13, no. 7: 2331. https://doi.org/10.3390/nu13072331





