Bacterial Diversity of Breast Milk in Healthy Spanish Women: Evolution from Birth to Five Years Postpartum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Breast Milk Samples
2.2. Bacterial DNA Isolation from Milk Samples
2.3. 16S rRNA Amplicon Sequencing
2.4. qPCR Analysis
2.5. Mineral Determination by ICP-MS
2.6. Fatty Acids Analysis by GC–FID
2.7. Statistics
3. Results and Discussion
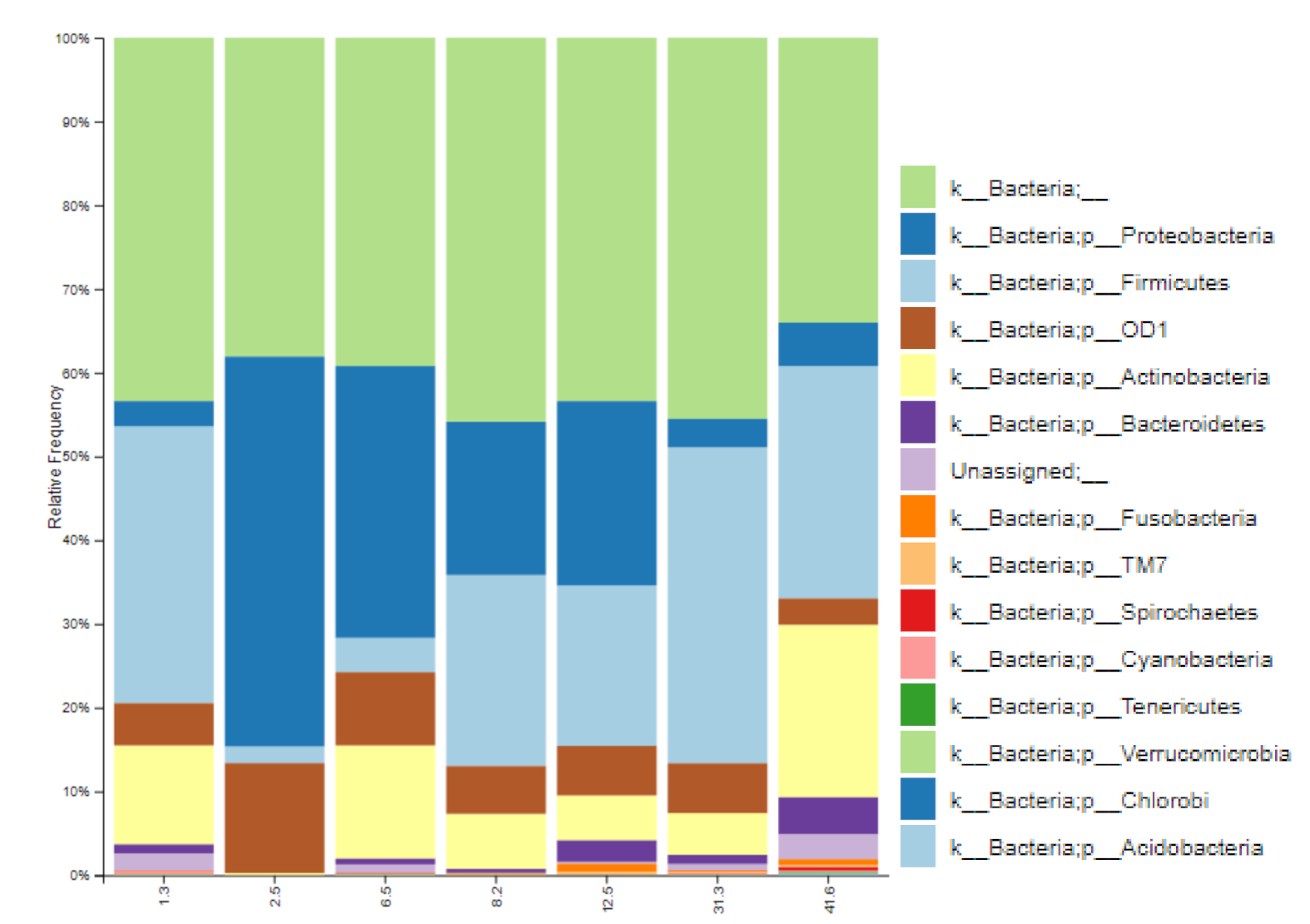
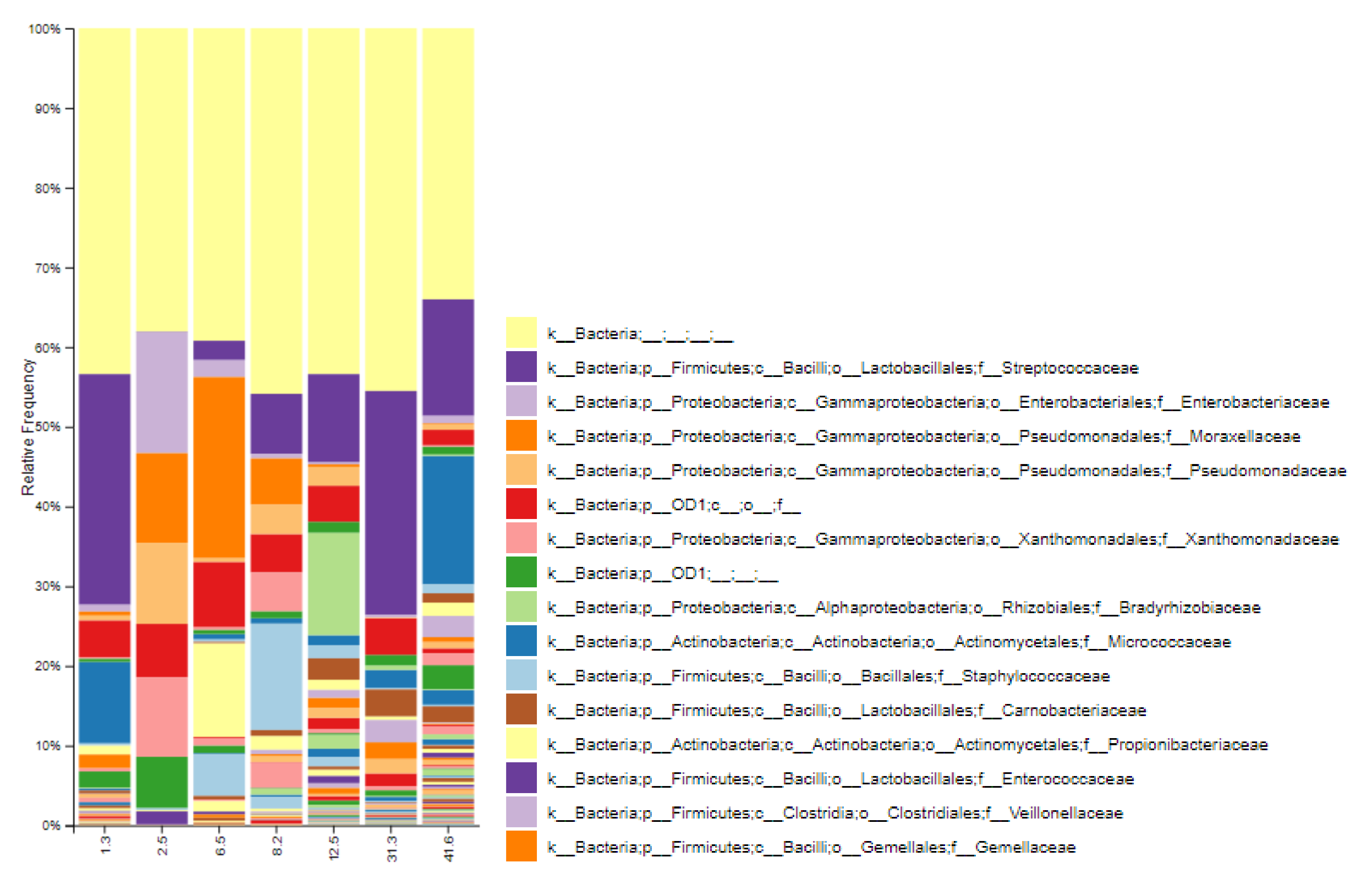
3.1. Bacterial Diversity of Breast Milk in Healthy Spanish Mothers
3.2. Influence of Lactation Time in the Bacterial Diversity of Breast Milk
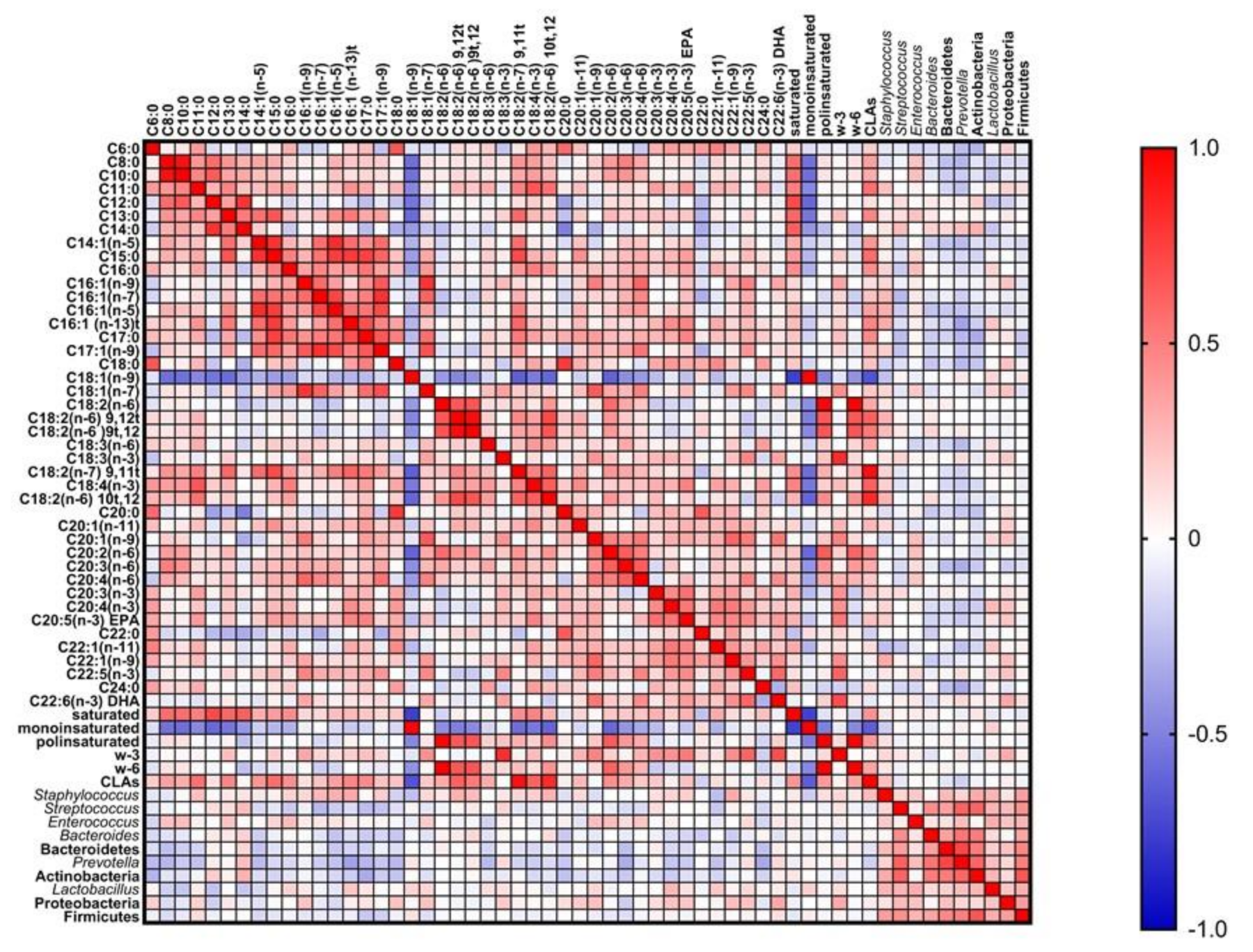
3.3. Influence of Milk Composition in the Bacterial Diversity of Breast Milk: Minerals
3.4. Influence of Milk Composition in the Bacterial Diversity of Breast Milk: Fatty Acids
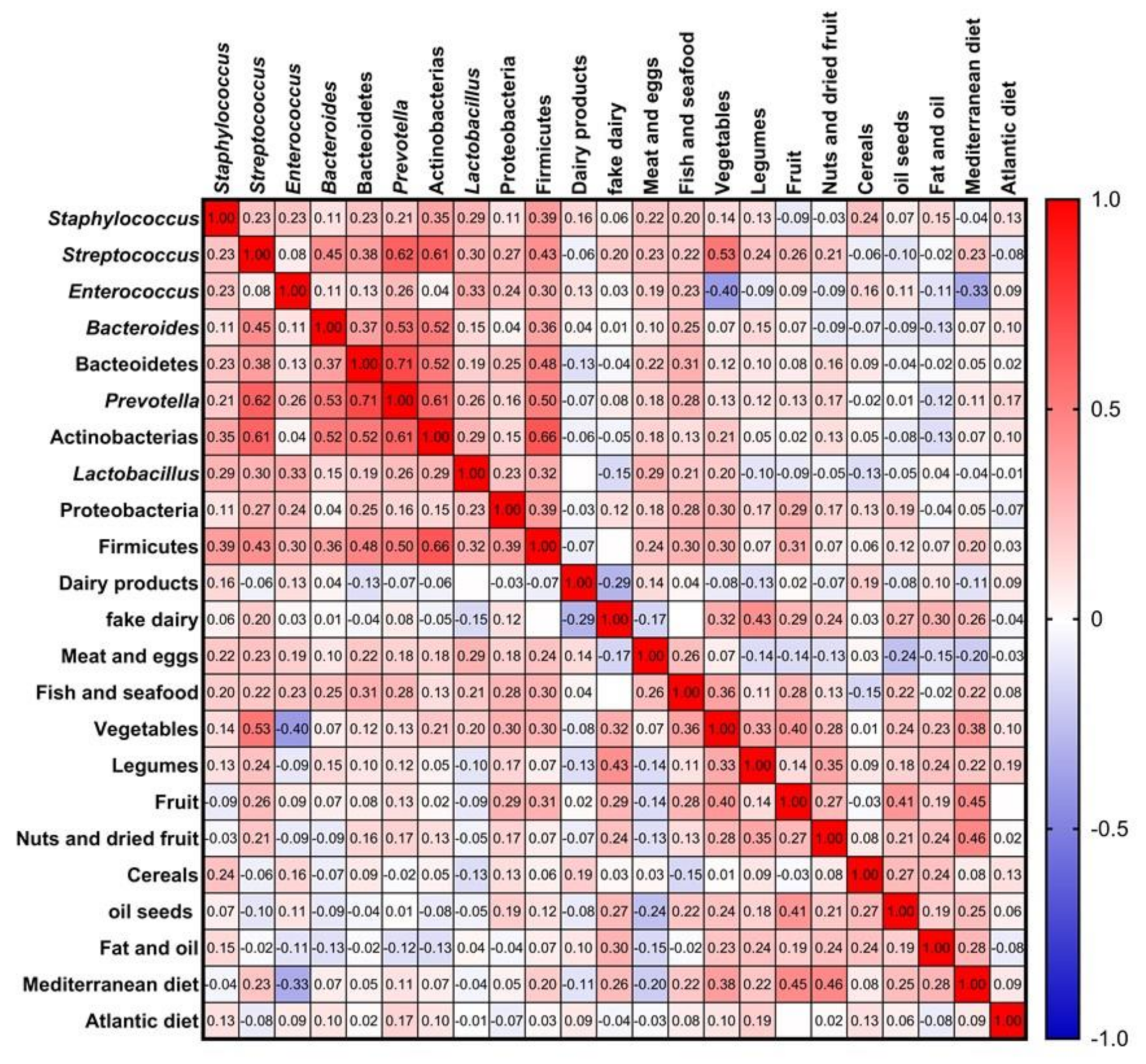
3.5. Influence of Maternal Factors in the Bacterial Diversity of Breast Milk: Diet and Host Factors
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sankar, M.J.; Sinha, B.; Chowdhury, R.; Bhandari, N.; Taneja, S.; Martines, J.; Bahl, R. Optimal breastfeeding practices and infant and child mortality: A systematic review and meta-analysis. Acta Paediatr. 2015, 104, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Curtis, N. Breast milk microbiota: A review of the factors that influence composition. J. Infect. 2020, 81, 17–47. [Google Scholar] [CrossRef] [PubMed]
- Victora, C.G.; Bahl, R.; Barros, A.J.D.; França, G.V.A.; Horton, S.; Krasevec, J.; Murch, S.; Sankar, M.J.; Walker, N.; Rollins, N.C.; et al. Breastfeeding in the 21st century: Epidemiology, mechanisms, and lifelong effect. Lancet 2016, 387, 475–490. [Google Scholar] [CrossRef] [Green Version]
- Hennet, T.; Borsig, L. Breastfed at Tiffany’s. Trends Biochem. Sci. 2016, 41, 508–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barreiro, R.; Regal, P.; López-Racamonde, O.; Cepeda, A.; Fente, C. Evolution of breast milk fatty acids in Spanish mothers after one year of uninterrupted lactation. Prostaglandins Leukot. Essent. Fatty Acids 2020, 159. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, C.; Fente, C.; Barreiro, R.; López-Racamonde, O.; Cepeda, A.; Regal, P. Association between breast milk mineral content and maternal adherence to healthy dietary patterns in Spain: A transversal study. Foods 2020, 9, 659. [Google Scholar] [CrossRef]
- Schwab, C.; Voney, E.; Ramirez Garcia, A.; Vischer, M.; Lacroix, C. Characterization of the Cultivable Microbiota in Fresh and Stored Mature Human Breast Milk. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Damaceno, Q.S.; Souza, J.P.; Nicoli, J.R.; Paula, R.L.; Assis, G.B.; Figueiredo, H.C.; Azevedo, V.; Martins, F.S. Evaluation of Potential Probiotics Isolated from Human Milk and Colostrum. Probiotics Antimicrob. Proteins 2017, 9, 371–379. [Google Scholar] [CrossRef]
- Albesharat, R.; Ehrmann, M.A.; Korakli, M.; Yazaji, S.; Vogel, R.F. Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst. Appl. Microbiol. 2011, 34, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Obermajer, T.; Lipoglavšek, L.; Tompa, G.; Treven, P.; Lorbeg, P.M.; Matijašić, B.B.; Rogelj, I. Colostrum of healthy Slovenian mothers: Microbiota composition and bacteriocin gene prevalence. PLoS ONE 2015, 10, e0123324. [Google Scholar] [CrossRef]
- Urbaniak, C.; Cummins, J.; Brackstone, M.; Macklaim, J.M.; Gloor, G.B.; Baban, C.K.; Scott, L.; O’Hanlon, D.M.; Burton, J.P.; Francis, K.P.; et al. Microbiota of human breast tissue. Appl. Environ. Microbiol. 2014, 80, 3007–3014. [Google Scholar] [CrossRef] [Green Version]
- Bisanz, J.E.; Enos, M.K.; PrayGod, G.; Seney, S.; Macklaim, J.M.; Chilton, S.; Willner, D.; Knight, R.; Fusch, C.; Fusch, G.; et al. Microbiota at multiple body sites during pregnancy in a rural tanzanian population and effects of Moringa-supplemented probiotic yogurt. Appl. Environ. Microbiol. 2015, 81, 4965–4975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, P.; Curtis, N. Factors Influencing the Intestinal Microbiome during the First Year of Life. Pediatr. Infect. Dis. J. 2018, 37, 315–335. [Google Scholar] [CrossRef]
- Vandenplas, Y.; Carnielli, V.P.; Ksiazyk, J.; Luna, M.S.; Migacheva, N.; Mosselmans, J.M.; Picaud, J.C.; Possner, M.; Singhal, A.; Wabitsch, M. Factors affecting early-life intestinal microbiota development. Nutrition 2020, 78. [Google Scholar] [CrossRef]
- Yuhas, R.; Pramuk, K.; Lien, E.L. Human milk fatty acid composition from nine countries varies most in DHA. Lipids 2006, 41, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Isolauri, E.; Laitinen, K.; Salminen, S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am. J. Clin. Nutr. 2008, 88, 894–899. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, S.H.; Vaidya, Y.H.; Patel, R.J.; Pandit, R.J.; Joshi, C.G.; Kunjadiya, A.P. Culture independent assessment of human milk microbial community in lactational mastitis. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soto, A.; Martín, V.; Jiménez, E.; Mader, I.; Rodríguez, J.M.; Fernández, L. Lactobacilli and bifidobacteria in human breast milk: Influence of antibiotherapy and other host and clinical factors. J. Pediatr. Gastroenterol. Nutr. 2014, 59, 78–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moossavi, S.; Sepehri, S.; Robertson, B.; Bode, L.; Goruk, S.; Field, C.J.; Lix, L.M.; de Souza, R.J.; Becker, A.B.; Mandhane, P.J.; et al. Composition and Variation of the Human Milk Microbiota Are Influenced by Maternal and Early-Life Factors. Cell Host Microbe 2019, 25, 324–335. [Google Scholar] [CrossRef] [Green Version]
- Moossavi, S.; Atakora, F.; Miliku, K.; Sepehri, S.; Robertson, B.; Duan, Q.L.; Becker, A.B.; Mandhane, P.J.; Turvey, S.E.; Moraes, T.J.; et al. Integrated analysis of human milk microbiota with oligosaccharides and fatty acids in the child cohort. Front. Nutr. 2019, 6. [Google Scholar] [CrossRef] [Green Version]
- Kumar, H.; du Toit, E.; Kulkarni, A.; Aakko, J.; Linderborg, K.M.; Zhang, Y.; Nicol, M.P.; Isolauri, E.; Yang, B.; Collado, M.C.; et al. Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Schröder, H.; Fitó, M.; Estruch, R.; Martínez-González, M.A.; Corella, D.; Salas-Salvadó, J.; Lamuela-Raventós, R.; Ros, E.; Salaverría, I.; Fiol, M.; et al. A Short screener is valid for assessing mediterranean diet adherence among older spanish men and women. J. Nutr. 2011, 141, 1140–1145. [Google Scholar] [CrossRef] [Green Version]
- Martínez-González, M.A.; García-Arellano, A.; Toledo, E.; Salas-Salvadó, J.; Buil-Cosiales, P.; Corella, D.; Covas, M.I.; Schröder, H.; Arós, F.; Gómez-Gracia, E.; et al. A 14-item mediterranean diet assessment tool and obesity indexes among high-risk subjects: The PREDIMED trial. PLoS ONE 2012, 7, e43134. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Martín, C.; Garcia-Ortiz, L.; Rodriguez-Sanchez, E.; Martin-Cantera, C.; Soriano-Cano, A.; Arietaleanizbeaskoa, M.S.; Magdalena-Belio, J.F.; Menendez-Suarez, M.; Maderuelo-Fernandez, J.A.; Lugones-Sanchez, C.; et al. The relationship of the atlantic diet with cardiovascular risk factors and markers of arterial stiffness in adults without cardiovascular disease. Nutrients 2019, 11, 742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guallar-Castillón, P.; Oliveira, A.; Lopes, C.; López-García, E.; Rodríguez-Artalejo, F. The Southern European Atlantic Diet is associated with lower concentrations of markers of coronary risk. Atherosclerosis 2013, 226, 502–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Friswell, M.K.; Gika, H.; Stratford, I.J.; Theodoridis, G.; Telfer, B.; Wilson, I.D.; McBain, A.J. Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS ONE 2010, 5, e8584. [Google Scholar] [CrossRef]
- Guo, X.; Xia, X.; Tang, R.; Zhou, J.; Zhao, H.; Wang, K. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett. Appl. Microbiol. 2008, 47, 367–373. [Google Scholar] [CrossRef]
- Stach, J.E.M.; Maldonado, L.A.; Ward, A.C.; Goodfellow, M.; Bull, A.T. New primers for the class Actinobacteria: Application to marine and terrestrial environments. Environ. Microbiol. 2003, 5, 828–841. [Google Scholar] [CrossRef] [Green Version]
- Layton, A.; McKay, L.; Williams, D.; Garrett, V.; Gentry, R.; Sayler, G. Development of Bacteroides 16S rRNA gene taqman-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl. Environ. Microbiol. 2006, 72, 4214–4224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collado, M.C.; Delgado, S.; Maldonado, A.; Rodríguez, J.M. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett. Appl. Microbiol. 2009, 48, 523–528. [Google Scholar] [CrossRef]
- Delroisse, J.; Boulvin, A.; Parmentier, I.; Dauphin, R.D.; Vandenbol, M.; Portetelle, D. Quantification of Bifidobacterium spp. and Lactobacillus spp. in rat fecal samples by real-time PCR. Microbiol. Res. 2008, 163, 663–670. [Google Scholar] [CrossRef]
- Bekele, A.Z.; Koike, S.; Kobayashi, Y. Genetic diversity and diet specificity of ruminal Prevotella revealed by 16S rRNA gene-based analysis. FEMS Microbiol. Lett. 2010, 305, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Barreiro, R.; Díaz-Bao, M.; Cepeda, A.; Regal, P.; Fente, C.A. Fatty acid composition of breast milk in Galicia (NW Spain): A cross-country comparison. Prostaglandins Leukot. Essent. Fatty Acids 2018, 135, 102–114. [Google Scholar] [CrossRef]
- Lopez Leyva, L.; Brereton, N.J.B.; Koski, K.G. Emerging frontiers in human milk microbiome research and suggested primers for 16S rRNA gene analysis. Comput. Struct. Biotechnol. J. 2021, 19, 121–133. [Google Scholar] [CrossRef]
- Bukin, Y.S.; Galachyants, Y.P.; Morozov, I.V.; Bukin, S.V.; Zakharenko, A.S.; Zemskaya, T.I. The effect of 16s rRNA region choice on bacterial community metabarcoding results. Sci. Data 2019, 6, 1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sirichoat, A.; Sankuntaw, N.; Engchanil, C.; Buppasiri, P.; Faksri, K.; Namwat, W.; Chantratita, W.; Lulitanond, V. Comparison of different hypervariable regions of 16S rRNA for taxonomic profiling of vaginal microbiota using next-generation sequencing. Arch. Microbiol. 2021, 203, 1159–1166. [Google Scholar] [CrossRef]
- Costantini, L.; Magno, S.; Albanese, D.; Donati, C.; Molinari, R.; Filippone, A.; Masetti, R.; Merendino, N. Characterization of human breast tissue microbiota from core needle biopsies through the analysis of multi hypervariable 16S-rRNA gene regions. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [Green Version]
- Lemay-Nedjelski, L.; Butcher, J.; Ley, S.H.; Asbury, M.R.; Hanley, A.J.; Kiss, A.; Unger, S.; Copeland, J.K.; Wang, P.W.; Zinman, B.; et al. Examining the relationship between maternal body size, gestational glucose tolerance status, mode of delivery and ethnicity on human milk microbiota at three months post-partum. BMC Microbiol. 2020, 20. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.-T.; Deng, W.-F.; Xu, S.-X.; Zhao, J.; Xu, D.; Liu, Y.-H.; Guo, Y.-Y.; Wang, M.-B.; He, F.-S.; Ye, S.-W.; et al. Shotgun metagenomics reveals both taxonomic and tryptophan pathway differences of gut microbiota in major depressive disorder patients. Psychol. Med. 2021, 51, 90–101. [Google Scholar] [CrossRef]
- Lin, P.; Ding, B.; Feng, C.; Yin, S.; Zhang, T.; Qi, X.; Lv, H.; Guo, X.; Dong, K.; Zhu, Y.; et al. Prevotella and Klebsiella proportions in fecal microbial communities are potential characteristic parameters for patients with major depressive disorder. J. Affect. Disord. 2017, 207, 300–304. [Google Scholar] [CrossRef]
- Corona-Cervantes, K.; García-González, I.; Villalobos-Flores, L.E.; Hernández-Quiroz, F.; Piña-Escobedo, A.; Hoyo-Vadillo, C.; Rangel-Calvillo, M.N.; García-Mena, J. Human milk microbiota associated with early colonization of the neonatal gut in Mexican newborns. PeerJ 2020, 8. [Google Scholar] [CrossRef]
- Jian, C.; Luukkonen, P.; Yki-Järvinen, H.; Salonen, A.; Korpela, K. Quantitative PCR provides a simple and accessible method for quantitative microbiota profiling. PLoS ONE 2020, 15, e0227285. [Google Scholar] [CrossRef] [Green Version]
- Aakko, J.; Kumar, H.; Rautava, S.; Wise, A.; Autran, C.; Bode, L.; Isolauri, E.; Salminen, S. Human milk oligosaccharide categories define the microbiota composition in human colostrum. Benef. Microbes 2017, 8, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.; Curley, D.; O’callaghan, T.F.; O’shea, C.; Dempsey, E.M.; O’toole, P.W.; Ross, R.P.; Ryan, C.A.; Stanton, C. The composition of human milk and infant faecal microbiota over the first three months of life: A pilot study. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermansson, H.; Kumar, H.; Collado, M.C.; Salminen, S.; Isolauri, E.; Rautava, S. Breast milk microbiota is shaped by mode of delivery and intrapartum antibiotic exposure. Front. Nutr. 2019, 6. [Google Scholar] [CrossRef] [Green Version]
- Urbaniak, C.; Angelini, M.; Gloor, G.B.; Reid, G. Human milk microbiota profiles in relation to birthing method. Gestation and Infant Gender.Microbiome 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- Cabrera-Rubio, R.; Mira-Pascual, L.; Mira, A.; Collado, M.C. Impact of mode of delivery on the milk microbiota composition of healthy women. J. Dev. Orig. Health Dis. 2016, 7, 54–60. [Google Scholar] [CrossRef] [Green Version]
- Davé, V.; Street, K.; Francis, S.; Bradman, A.; Riley, L.; Eskenazi, B.; Holland, N. Bacterial microbiome of breast milk and child saliva from low-income Mexican-American women and children. Pediatr. Res. 2016, 79, 846–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, M.R.; Avershina, E.; Storrø, O.; Johnsen, R.; Rudi, K.; Øien, T. Breastfeeding-associated microbiota in human milk following supplementation with Lactobacillus rhamnosus GG, Lactobacillus acidophilus La-5, and Bifidobacterium animalis ssp. lactis Bb-12. J. Dairy Sci. 2018, 101, 889–899. [Google Scholar] [CrossRef]
- Sakwinska, O.; Moine, D.; Delley, M.; Combremont, S.; Rezzonico, E.; Descombes, P.; Vinyes-Pares, G.; Zhang, Y.; Wang, P.; Thakkar, S.K. Microbiota in breast milk of Chinese lactating mothers. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [Green Version]
- Boix-Amorós, A.; Collado, M.C.; Mira, A. Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Ballard, O.; Morrow, A.L. Human milk composition: Nutrients and bioactive factors. Pediatr. Clin. North Am. 2013, 60, 49–74. [Google Scholar] [CrossRef] [Green Version]
- Khodayarpardo, P.; Mira-Pascual, L.; Collado, M.C.; Martínez-Costa, C. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J. Perinatol. 2014, 34, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Solís, G.; de los Reyes-Gavilan, C.G.; Fernández, N.; Margolles, A.; Gueimonde, M. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe 2010, 16, 307–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drago, L.; Toscano, M.; De Grandi, R.; Grossi, E.; Padovani, E.M.; Peroni, D.G. Microbiota network and mathematic microbe mutualism in colostrum and mature milk collected in two different geographic areas: Italy versus Burundi. ISME J. 2017, 11, 875–884. [Google Scholar] [CrossRef]
- Cabrera-Rubio, R.; Collado, M.C.; Laitinen, K.; Salminen, S.; Isolauri, E.; Mira, A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am. J. Clin. Nutr. 2012, 96, 544–551. [Google Scholar] [CrossRef] [Green Version]
- Al-Shehri, S.S.; Sweeney, E.L.; Cowley, D.M.; Liley, H.G.; Ranasinghe, P.D.; Charles, B.G.; Shaw, P.N.; Vagenas, D.; Duley, J.A.; Knox, C.L. Deep sequencing of the 16S ribosomal RNA of the neonatal oral microbiome: A comparison of breast-fed and formula-fed infants. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [Green Version]
- Holgerson, P.L.; Esberg, A.; Sjödin, A.; West, C.E.; Johansson, I. A longitudinal study of the development of the saliva microbiome in infants 2 days to 5 years compared to the microbiome in adolescents. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Nierhaus, K.H. Mg2+, K+, and the ribosome. J. Bacteriol. 2014, 196, 3817–3819. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.D.; Galera-Laporta, L.; Bialecka-Fornal, M.; Moon, E.C.; Shen, Z.; Briggs, S.P.; Garcia-Ojalvo, J.; Süel, G.M. Magnesium Flux Modulates Ribosomes to Increase Bacterial Survival. Cell 2019, 177, 352–360. [Google Scholar] [CrossRef] [Green Version]
- Cunrath, O.; Bumann, D. Host resistance factor SLC11A1 restricts Salmonella growth through magnesium deprivation. Science 2019, 366, 995–999. [Google Scholar] [CrossRef]
- Cheng, X.; Redanz, S.; Treerat, P.; Qin, H.; Choi, D.; Zhou, X.; Xu, X.; Merritt, J.; Kreth, J. Magnesium-Dependent Promotion of H2O2 Production Increases Ecological Competitiveness of Oral Commensal Streptococci. J. Dent. Res. 2020, 99, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Webster, T.J.; Tran, P. Selenium nanoparticles inhibit Staphylococcus aureus growth. Int. J. Nanomed. 2011, 6, 1553–1558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malbe, M.; Attila, M.; Atroshi, F. Possible involvement of selenium in Staphylococcus aureus inhibition in cow’s whey. J. Anim. Physiol. Anim. Nutr. 2006, 90, 159–164. [Google Scholar] [CrossRef]
- Liu, K.; Ding, T.; Fang, L.; Cui, L.; Li, J.; Meng, X.; Zhu, G.; Qian, C.; Wang, H.; Li, J. Organic Selenium Ameliorates Staphylococcus aureus-Induced Mastitis in Rats by Inhibiting the Activation of NF-κB and MAPK Signaling Pathways. Front. Vet. Sci. 2020, 7. [Google Scholar] [CrossRef]
- EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). Scientific Opinion on the substantiation of health claims related to selenium and protection of DNA, proteins and lipids from oxidative damage (ID 277, 283, 286, 1289, 1290, 1291, 1293, 1751), function of the immune system (ID 278), thyroid function (ID 279, 282, 286, 1289, 1290, 1291, 1293), function of the heart and blood vessels (ID 280), prostate function (ID 284), cognitive function (ID 285) and spermatogenesis (ID 396) pursuant to Article 13(1) of Regulation (EC) No 1924/2006. EFSA J. 2009, 7, 1220. [Google Scholar] [CrossRef]
- Allemann, M.N.; Shulse, C.N.; Allen, E.E. Linkage of marine bacterial polyunsaturated fatty acid and long-chain hydrocarbon biosynthesis. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, T.; Hirose, K.; Yusuf, Y.; Kawamoto, J.; Kurihara, T. Bioconversion From Docosahexaenoic Acid to Eicosapentaenoic Acid in the Marine Bacterium Shewanella livingstonensis Ac10. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Chin, S.F.; Storkson, J.M.; Liu, W.; Albright, K.J.; Pariza, M.W. Conjugated linoleic acid (9,11- and 10,12-octadecadienoic acid) is produced in conventional but not germ-free rats fed linoleic acid. J. Nutr. 1994, 124, 694–701. [Google Scholar] [CrossRef]
- Kumar, P.; Lee, J.-H.; Beyenal, H.; Lee, J. Fatty Acids as Antibiofilm and Antivirulence Agents. Trends Microbiol. 2020, 28, 753–768. [Google Scholar] [CrossRef]
- Salsinha, A.S.; Pimentel, L.L.; Fontes, A.L.; Gomes, A.M.; Rodríguez-Alcalá, L.M. Microbial production of conjugated linoleic acid and conjugated linolenic acid relies on a multienzymatic system. Microbiol. Mol. Biol. Rev. 2018, 82. [Google Scholar] [CrossRef] [Green Version]
- Rox, K.; Jansen, R.; Loof, T.G.; Gillen, C.M.; Bernecker, S.; Walker, M.J.; Chhatwal, G.S.; Müller, R. Linoleic and palmitoleic acid block streptokinase-mediated plasminogen activation and reduce severity of invasive group A streptococcal infection. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Ferraris, C.; Elli, M.; Tagliabue, A. Gut microbiota for health: How can diet maintain a healthy gut microbiota? Nutrients 2020, 12, 3596. [Google Scholar] [CrossRef] [PubMed]
- Redondo-Useros, N.; Nova, E.; González-Zancada, N.; Díaz, L.E.; Gómez-Martínez, S.; Marcos, A. Microbiota and lifestyle: A special focus on diet. Nutrients 2020, 12, 1776. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Lian, Y.; Zhao, C.; Du, H.; Han, Y.; Gao, W.; Xiao, H.; Zheng, J. Dietary Fibers from Fruits and Vegetables and Their Health Benefits via Modulation of Gut Microbiota. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1514–1532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, D.L.; Gill, S.K.; Brown, K.; Tasnim, N.; Ghosh, S.; Innis, S.; Jacobson, K. Maternal exposure to fish oil primes offspring to harbor intestinal pathobionts associated with altered immune cell balance. Gut Microbes 2015, 6, 24–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Z.; Hao, W.; Kwek, E.; Lei, L.; Liu, J.; Zhu, H.; Ma, K.Y.; Zhao, Y.; Ho, H.M.; He, W.-S.; et al. Fish Oil Is More Potent than Flaxseed Oil in Modulating Gut Microbiota and Reducing Trimethylamine- N-oxide-Exacerbated Atherogenesis. J. Agric. Food Chem. 2019, 67, 13635–13647. [Google Scholar] [CrossRef] [PubMed]
- Balfegò, M.; Canivell, S.; Hanzu, F.A.; Sala-Vila, A.; Martínez-Medina, M.; Murillo, S.; Mur, T.; Ruano, E.G.; Linares, F.; Porras, N.; et al. Effects of sardine-enriched diet on metabolic control, inflammation and gut microbiota in drug-naïve patients with type 2 diabetes: A pilot randomized trial. Lipids Health Dis. 2016, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, H.; Liu, J.; Jackson, M.I.; Zhao, F.; Yan, L.; Combs, G.F., Jr. Fatty liver accompanies an increase in lactobacillus species in the hind gut of C57BL/6 mice fed a high-fat diet. J. Nutr. 2013, 143, 627–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Lin, X.; Zhao, F.; Shi, X.; Li, H.; Li, Y.; Zhu, W.; Xu, X.; Lu, C.; Zhou, G. Meat, dairy and plant proteins alter bacterial composition of rat gut bacteria. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.B.; Madan, J.; Coker, M.; Hoen, A.; Baker, E.R.; Karagas, M.R.; Mueller, N.T. Does birth mode modify associations of maternal pre-pregnancy BMI and gestational weight gain with the infant gut microbiome? Int. J. Obes. 2020, 44, 23–32. [Google Scholar] [CrossRef] [PubMed]


| Target | Sequence (5′-3′) | Reference | |
|---|---|---|---|
| Proteobacteria | F | CATGACGTTACCCGCAGAAGAAG | [29] |
| R | CTCTACGAGACTCAAGCTTGC | ||
| Firmicutes | F | ATGTGGTTTAATTCGAAGCA | [30] |
| R | AGCTGACGACAACCATGCAC | ||
| Actinobacteria | F | GCGKCCTATCAGCTTGTT | [31] |
| R | CCGCCTACGAGCYCTTTACGC | ||
| Bacteroidetes | F | CATGTGGTTTAATTCGATGAT | [30] |
| R | AGCTGACGACAACCATGCAG | ||
| Bacteroides | F | GAGAGGAAGGTCCCCCAC | [32] |
| R | CGCTACTTGGCTGGTTCAG | ||
| Enterococcus | F | CCTTATTGTTAGTTGCCATCATT | [33] |
| R | ACTCGTTGTACTTCCCATTGT | ||
| Staphylococcus | F | GGCCGTGTTGAACGTGGTCAAATCA | [33] |
| R | TIACCATTTCAGTACCTTCTGGTAA | ||
| Streptococcus | F | GAAGAATTGCTTGAATTGGTTGAA | [33] |
| R | GGACGGTAGTTGTTGAAGAATGG | ||
| Lactobacillus | F | GAGGCAGCAGTAGGGAATCTTC | [34] |
| R | GGCCAGTTACTACCTCTATCCTTCTTC | ||
| Prevotella | F | GGTTCTGAGAGGAAGGTCCCC | [35] |
| R | TCCTGCACGCTACTTGGCTG | ||
| Group | Reference Strain | Broth Media | Temperature | Time |
|---|---|---|---|---|
| Proteobacteria | Salmonella Typhimurium CECT 4594 | Nutrient broth | 37 °C | 24 h |
| Firmicutes | S. aureus CECT 59 | Nutrient broth | 37 °C | 24 h |
| Actinobacteria | Corynebacterium tuberculostearicum CECT 763 | Tryptic soy broth | 37 °C | 48 h |
| Bacteroidetes | Bacteroides vulgatus LMG 17767 | Columbia blood agar | 37 °C | 48 h |
| Bacteroides | Bacteroides vulgatus LMG 17767 | Columbia blood agar | 37 °C | 48 h |
| Enterococcus | Enterococcus faecalis LMG 20863 | Tryptic soy broth | 37 °C | 24 h |
| Staphylococcus | S. aureus CECT 59 | Nutrient broth | 37 °C | 24 h |
| Streptococcus | S. agalactiae CECT 183 | Nutrient broth | 37 °C | 24 h |
| Lactobacillus | Lactobacillus delbrueckii CECT 4005 T | Man Rogosa Sharpe broth | 37 °C | 24 h |
| Prevotella | Prevotella copri DSM 18205 | Schaedler broth | 37 °C | 48 h |
| Variable 1 | Mean | SD | Median | Min | Max |
|---|---|---|---|---|---|
| Gestational age at birth (weeks) | 39.76 | 1.33 | 40.00 | 36.00 | 42.29 |
| Maternal age (years) | 35.46 | 4.02 | 35.00 | 26.00 | 46.00 |
| Maternal height (m) | 1.64 | 0.06 | 1.64 | 1.50 | 1.77 |
| Weight (kg) | 66.15 | 11.61 | 65.00 | 44.00 | 96.00 |
| Maternal BMI (kg/m2) | 24.48 | 3.85 | 24.39 | 17.85 | 35.03 |
| Lactating time (months) | 4.21 | 11.17 | 3.20 | 0.50 | 59.00 |
| Pregnancy weight gain (kg) | 13.25 | 3.63 | 13.00 | 5.00 | 23.00 |
| Newborn weight (kg) | 3.26 | 0.42 | 3.28 | 2.29 | 4.33 |
| MD adherence (score: 0–1) | 0.51 | 0.22 | 0.57 | 0.14 | 0.86 |
| SEAD adherence (score: 0–1) | 0.38 | 0.17 | 0.44 | 0.00 | 0.78 |
| Newborn sex (%) | |||||
| • Female | 54.74 | ||||
| • Male | 45.26 | ||||
| Delivery mode (%) | |||||
| • Vaginal | 86.21 | ||||
| • C-section | 13.79 | ||||
| Lactation group (%) | |||||
| • <6 months | 70.71 | ||||
| • ≥6 months | 29.29 | ||||
| Tandem breastfeeding (%) | 6.93 |
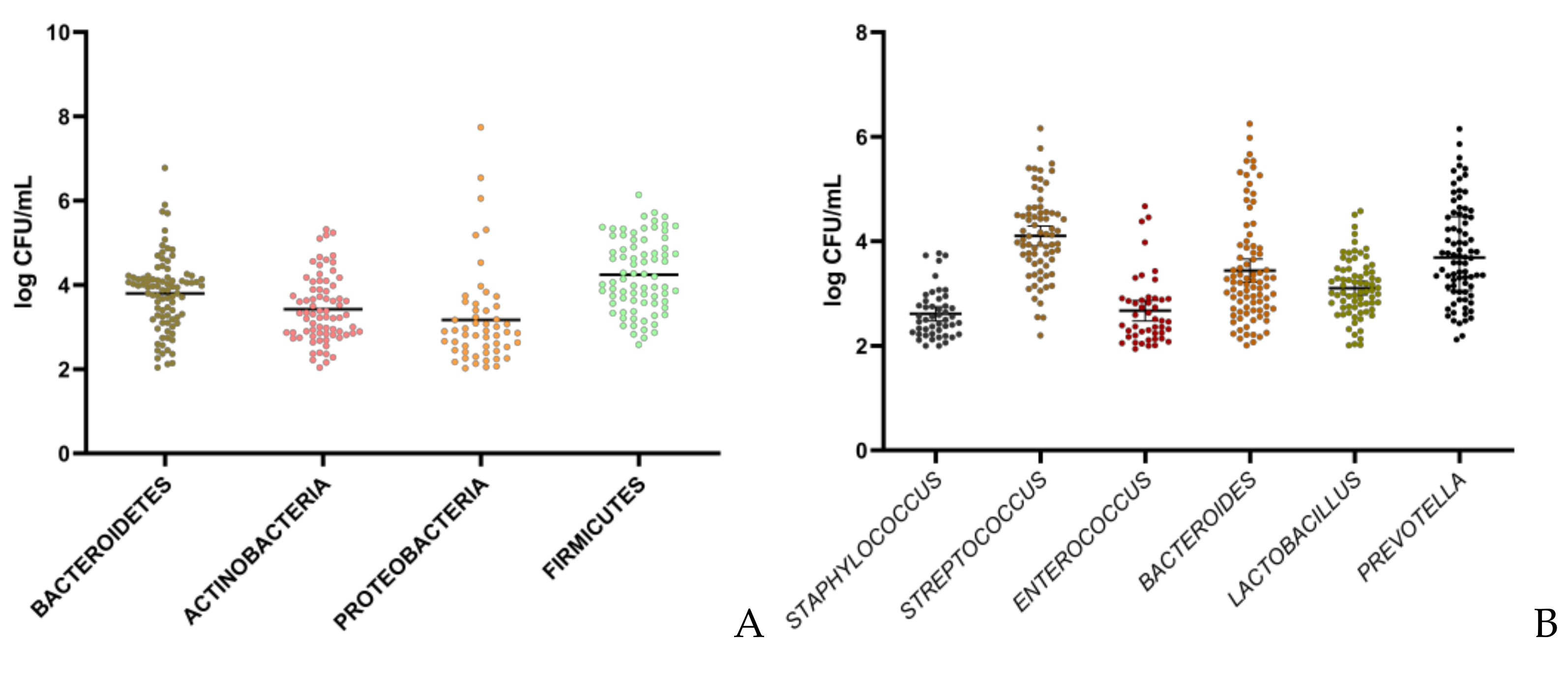
| Microbiota | All Samples 1 (0.5–59 Months) N = 99 | Conventional Lactation n = 70 | Prolonged 2 Lactation n = 29 | Spearman Correlation | ||
|---|---|---|---|---|---|---|
| Genera | Log CFU/mL Mean ± SD | % Prevalence (n/N) | Log CFU/mL Mean ± SD | Log CFU/mL Mean ± SD | r | p |
| Staphylococcus | 2.61 ± 0.46 d | 47.47 (47/99) | 2.57 ± 0.43 | 2.71 ± 0.56 | 0.175 | 0.239 |
| Streptococcus | 4.10 ± 0.80 a | 72.73 (72/99) | 3.96 ± 0.79 | 4.42 ± 0.78 * | 0.267 | 0.026 |
| Enterococcus | 2.67 ± 0.66 d | 46.46 (46/99) | 2.74 ± 0.76 | 2.60 ± 0.43 | −0.151 | 0.322 |
| Lactobacillus | 3.10 ± 0.56 c | 73.73 (73/99) | 3.13 ± 0.59 | 3.02 ± 0.52 | −0.089 | 0.461 |
| Bacteroides | 3.43 ± 1.02 c | 85.86 (85/99) | 3.26 ± 0.98 | 4.02 ± 1.19 ** | 0.275 | 0.011 |
| Prevotella | 3.78 ± 0.93 b | 80.80 (80/99) | 3.38 ± 0.76 | 4.54 ± 0.85 **** | 0.451 | 3 × 10−5 |
| Phyla | ||||||
| Firmicutes | 4.24 ± 0.87 a | 77.77 (77/99) | 4.21 ± 0.86 | 4.26 ± 0.92 | 0.105 | 0.372 |
| Proteobacteria | 3.17 ± 1.15 c | 53.54 (53/99) | 3.26 ± 1.29 | 2.97 ± 0.74 | −0.005 | 0.971 |
| Actinobacteria | 3.42 ± 0.78 c | 74.75 (74/99) | 3.23 ± 0.64 | 3.86 ± 0.88 *** | 0.334 | 0.004 |
| Bacteroidetes | 3.80 ± 0.88 b | 89.90 (89/99) | 3.55 ± 0.80 | 4.23 ± 0.94 *** | 0.403 | 1.7 × 10−4 |
| Conventional Lactation (n = 43) | Prolonged Lactation (n = 26) | Spearman Correlation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mineral | Mean | SD | Median | Min | Max | Mean | SD | Median | Min | Max | r | p |
| Na (mg/L) | 134.6 | 65.5 | 117.1 | 44.75 | 303.9 | 161.2 | 82.15 | 130.2 | 72.57 | 389.2 | 0.000 | 0.999 |
| K (mg/L) | 461 | 66.43 | 453.8 | 345.9 | 599.7 | 454.6 | 57.36 | 439.8 | 343 | 622.3 | −0.324 | 0.009 |
| Ca (mg/L) | 275.9 | 63.52 | 263.3 | 136.4 | 463.3 | 283.9 | 56.64 | 265.3 | 192.6 | 383.5 | −0.496 | <0.001 |
| P (mg/L) | 120.4 | 26.9 | 117.2 | 73.0 | 176.5 | 137.4 | 33.66 | 123.6 | 79.82 | 219.6 | −0.113 | 0.371 |
| Mg (mg/L) | 33.03 | 4.96 | 33.68 | 21.7 | 42.98 | 32.91 | 7.03 | 31.06 | 19.86 | 48.23 | 0.151 | 0.230 |
| Fe (mg/L) | 0.20 | 0.09 | 0.20 | 0.07 | 0.45 | 0.18 | 0.15 | 0.19 | 0.06 | 0.75 | −0.200 | 0.113 |
| Se (µg/L) | 12.01 | 7.07 | 9.94 | 4.37 | 41.35 | 12.07 | 5.15 | 10.51 | 6.74 | 23.95 | −0.085 | 0.508 |
| Conventional Lactation n = 70 | Prolonged Lactation n = 29 | Spearman Correlation | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fatty Acid 1 | Mean ± SD | Median | Range | Mean ± SD | Median | Range | r | p | ||
| C6:0 * | 0.496 ± 0.285 | 0.410 | 0.210 | 1.48 | 0.358 ± 0.293 | 0.260 | 0.093 | 1.481 | −0.270 | 0.0140 |
| C8:0 ** | 0.304 ± 0.071 | 0.308 | 0.156 | 0.48 | 0.262 ± 0.060 | 0.265 | 0.139 | 0.434 | −0.311 | 0.0017 |
| C10:0 * | 1.773 ± 0.415 | 1.787 | 0.854 | 2.83 | 1.605 ± 0.326 | 1.571 | 0.978 | 2.328 | −0.199 | 0.0481 |
| C11:0 | 0.046 ± 0.018 | 0.043 | 0.000 | 0.11 | 0.042 ± 0.023 | 0.036 | 0.017 | 0.107 | −0.056 | 0.6015 |
| C12:0 | 9.524 ± 3.161 | 8.993 | 3.852 | 22.25 | 10.020 ± 1.995 | 9.941 | 6.185 | 13.810 | 0.181 | 0.0726 |
| C13:0 | 0.040 ± 0.013 | 0.040 | 0.012 | 0.06 | 0.041 ± 0.015 | 0.039 | 0.019 | 0.074 | −0.002 | 0.9826 |
| C14:0 **** | 6.552 ± 2.125 | 6.343 | 3.075 | 12.18 | 8.731 ± 2.399 | 8.362 | 4.565 | 15.060 | 0.460 | 0.0000 |
| C14:1 (n-5) | 0.214 ± 0.096 | 0.198 | 0.042 | 0.47 | 0.189 ± 0.106 | 0.167 | 0.042 | 0.424 | −0.116 | 0.2527 |
| C15:0 | 0.248 ± 0.083 | 0.235 | 0.072 | 0.45 | 0.218 ± 0.083 | 0.186 | 0.096 | 0.410 | −0.220 | 0.0290 |
| C16:0 ** | 19.480 ± 2.653 | 19.640 | 13.640 | 26.30 | 17.840 ± 3.102 | 17.140 | 13.540 | 25.170 | −0.318 | 0.0013 |
| C16:1 (n-9) | 0.537 ± 0.114 | 0.516 | 0.318 | 0.84 | 0.549 ± 0.145 | 0.543 | 0.304 | 0.831 | −0.014 | 0.8920 |
| C16:1 (n-7) | 2.227 ± 0.739 | 2.086 | 0.835 | 4.25 | 2.132 ± 0.725 | 1.991 | 1.153 | 4.090 | −0.073 | 0.4715 |
| C16:1 (n-5) | 0.065 ± 0.024 | 0.059 | 0.020 | 0.15 | 0.058 ± 0.023 | 0.052 | 0.026 | 0.111 | −0.186 | 0.0651 |
| C16:1 (n-13)t † | 0.078 ± 0.040 | 0.073 | 0.015 | 0.17 | 0.060 ± 0.028 | 0.052 | 0.015 | 0.124 | −0.333 | 0.0024 |
| C17:0 ** | 0.289 ± 0.060 | 0.279 | 0.158 | 0.45 | 0.254 ± 0.066 | 0.249 | 0.129 | 0.453 | −0.277 | 0.0054 |
| C17:1 (n-9) | 0.202 ± 0.061 | 0.198 | 0.098 | 0.45 | 0.189 ± 0.053 | 0.188 | 0.102 | 0.324 | −0.087 | 0.3896 |
| C18:0 *** | 6.664 ± 1.610 | 6.209 | 4.246 | 11.32 | 5.438 ± 1.348 | 5.111 | 3.410 | 9.053 | −0.362 | 0.0002 |
| C18:1 (n-9) | 28.490 ± 5.917 | 27.700 | 13.110 | 39.28 | 30.310 ± 8.723 | 28.140 | 14.520 | 44.380 | 0.035 | 0.7311 |
| C18:1 (n-7) | 0.709 ± 0.126 | 0.701 | 0.379 | 0.99 | 0.685 ± 0.128 | 0.695 | 0.408 | 0.873 | −0.087 | 0.3892 |
| C18:2 (n-6) | 16.110 ± 4.001 | 15.350 | 9.749 | 27.31 | 15.310 ± 4.052 | 14.880 | 8.730 | 23.860 | −0.074 | 0.4680 |
| C18:2 (n-6)9,12t | 0.157 ± 0.044 | 0.148 | 0.063 | 0.29 | 0.154 ± 0.050 | 0.149 | 0.082 | 0.270 | −0.033 | 0.7478 |
| C18:2 (n-6)9t,12 | 0.129 ± 0.035 | 0.124 | 0.049 | 0.24 | 0.131 ± 0.041 | 0.130 | 0.068 | 0.222 | 0.010 | 0.9245 |
| C18:3 (n-6) ††† | 0.128 ± 0.065 | 0.138 | 0.019 | 0.32 | 0.084 ± 0.046 | 0.086 | 0.013 | 0.200 | −0.202 | 0.0492 |
| C18:3 (n-3) | 0.741 ± 0.485 | 0.606 | 0.250 | 3.59 | 0.895 ± 0.708 | 0.777 | 0.314 | 4.124 | 0.096 | 0.3462 |
| C18:2 (n-7)9,11t | 0.578 ± 0.158 | 0.551 | 0.211 | 0.94 | 0.519 ± 0.191 | 0.464 | 0.263 | 0.888 | −0.181 | 0.0725 |
| C18:4 (n-3) | 0.131 ± 0.042 | 0.123 | 0.052 | 0.25 | 0.117 ± 0.051 | 0.106 | 0.055 | 0.236 | −0.157 | 0.1382 |
| C18:2 (n-6)10t,12 | 0.344 ± 0.113 | 0.319 | 0.095 | 0.66 | 0.317 ± 0.119 | 0.279 | 0.162 | 0.568 | −0.050 | 0.6255 |
| C20:0 *** | 0.159 ± 0.034 | 0.156 | 0.091 | 0.25 | 0.133 ± 0.027 | 0.131 | 0.081 | 0.188 | −0.280 | 0.0050 |
| C20:1 (n-11) | 0.073 ± 0.029 | 0.071 | 0.000 | 0.19 | 0.079 ± 0.042 | 0.062 | 0.030 | 0.212 | 0.002 | 0.9858 |
| C20:1 (n-9) * | 0.473 ± 0.110 | 0.469 | 0.218 | 0.79 | 0.417 ± 0.124 | 0.389 | 0.210 | 0.794 | −0.339 | 0.0006 |
| C20:2 (n-6) | 0.337 ± 0.090 | 0.313 | 0.140 | 0.57 | 0.303 ± 0.081 | 0.294 | 0.202 | 0.552 | −0.250 | 0.0126 |
| C20:3 (n-6) **** | 0.529 ± 0.148 | 0.509 | 0.204 | 0.95 | 0.393 ± 0.139 | 0.360 | 0.236 | 0.743 | −0.447 | 0.0000 |
| C20:4 (n-6) | 0.593 ± 0.143 | 0.574 | 0.217 | 0.98 | 0.560 ± 0.198 | 0.495 | 0.241 | 0.997 | −0.181 | 0.0722 |
| C20:3 (n-3) | 0.069 ± 0.038 | 0.058 | 0.017 | 0.21 | 0.063 ± 0.045 | 0.054 | 0.018 | 0.211 | 0.020 | 0.8543 |
| C20:4 (n-3) †††† | 0.108 ± 0.047 | 0.101 | 0.022 | 0.26 | 0.078 ± 0.065 | 0.064 | 0.019 | 0.317 | −0.346 | 0.0006 |
| C20:5 (n-3) | 0.139 ± 0.088 | 0.117 | 0.042 | 0.52 | 0.120 ± 0.087 | 0.083 | 0.039 | 0.345 | −0.107 | 0.2965 |
| C22:0 | 0.068 ± 0.022 | 0.063 | 0.034 | 0.15 | 0.063 ± 0.016 | 0.063 | 0.037 | 0.109 | −0.049 | 0.6270 |
| C22:1 (n-11) †††† | 0.085 ± 0.055 | 0.074 | 0.017 | 0.34 | 0.044 ± 0.025 | 0.041 | 0.000 | 0.126 | −0.343 | 0.0006 |
| C22:1 (n-9) | 0.093 ± 0.030 | 0.095 | 0.000 | 0.17 | 0.087 ± 0.027 | 0.080 | 0.040 | 0.150 | −0.186 | 0.0983 |
| C22:5 (n-3) | 0.131 ± 0.048 | 0.119 | 0.043 | 0.27 | 0.141 ± 0.066 | 0.124 | 0.075 | 0.341 | 0.042 | 0.6805 |
| C24:0 †† | 0.063 ± 0.058 | 0.037 | 0.013 | 0.27 | 0.034 ± 0.021 | 0.027 | 0.013 | 0.126 | −0.199 | 0.0492 |
| C22:6 (n-3) | 0.414 ± 0.311 | 0.348 | 0.049 | 1.61 | 0.462 ± 0.287 | 0.374 | 0.130 | 1.226 | 0.004 | 0.9682 |
| Total SFA | 45.530 ± 5.919 | 44.630 | 35.300 | 63.56 | 44.860 ± 5.814 | 43.610 | 35.230 | 57.910 | −0.026 | 0.8007 |
| Total MUFA | 33.850 ± 6.111 | 33.200 | 16.590 | 44.54 | 35.470 ± 8.186 | 33.350 | 19.340 | 48.680 | 0.035 | 0.7294 |
| Total PUFA | 20.560 ± 4.210 | 19.680 | 13.400 | 31.89 | 19.610 ± 5.062 | 19.520 | 11.180 | 33.120 | −0.099 | 0.3306 |
| Total PUFA n-3 | 1.661 ± 0.725 | 1.469 | 0.758 | 5.19 | 1.837 ± 1.003 | 1.568 | 0.858 | 5.359 | 0.055 | 0.5893 |
| Total PUFA n-6 | 17.970 ± 4.145 | 17.240 | 11.520 | 29.27 | 16.950 ± 4.413 | 16.720 | 9.703 | 26.830 | −0.093 | 0.3585 |
| CLAs | 0.922 ± 0.233 | 0.869 | 0.321 | 1.38 | 0.836 ± 0.296 | 0.722 | 0.455 | 1.422 | −0.154 | 0.1273 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanjulián, L.; Lamas, A.; Barreiro, R.; Cepeda, A.; Fente, C.A.; Regal, P. Bacterial Diversity of Breast Milk in Healthy Spanish Women: Evolution from Birth to Five Years Postpartum. Nutrients 2021, 13, 2414. https://doi.org/10.3390/nu13072414
Sanjulián L, Lamas A, Barreiro R, Cepeda A, Fente CA, Regal P. Bacterial Diversity of Breast Milk in Healthy Spanish Women: Evolution from Birth to Five Years Postpartum. Nutrients. 2021; 13(7):2414. https://doi.org/10.3390/nu13072414
Chicago/Turabian StyleSanjulián, Laura, Alexandre Lamas, Rocío Barreiro, Alberto Cepeda, Cristina A. Fente, and Patricia Regal. 2021. "Bacterial Diversity of Breast Milk in Healthy Spanish Women: Evolution from Birth to Five Years Postpartum" Nutrients 13, no. 7: 2414. https://doi.org/10.3390/nu13072414
APA StyleSanjulián, L., Lamas, A., Barreiro, R., Cepeda, A., Fente, C. A., & Regal, P. (2021). Bacterial Diversity of Breast Milk in Healthy Spanish Women: Evolution from Birth to Five Years Postpartum. Nutrients, 13(7), 2414. https://doi.org/10.3390/nu13072414









