Genetically Higher Level of Mannose Has No Impact on Cardiometabolic Risk Factors: Insight from Mendelian Randomization
Abstract
:1. Introduction
2. Methods
2.1. Study Design
2.2. Genetic Instruments for Mannose
2.3. Association of Genetic Instruments with Outcome
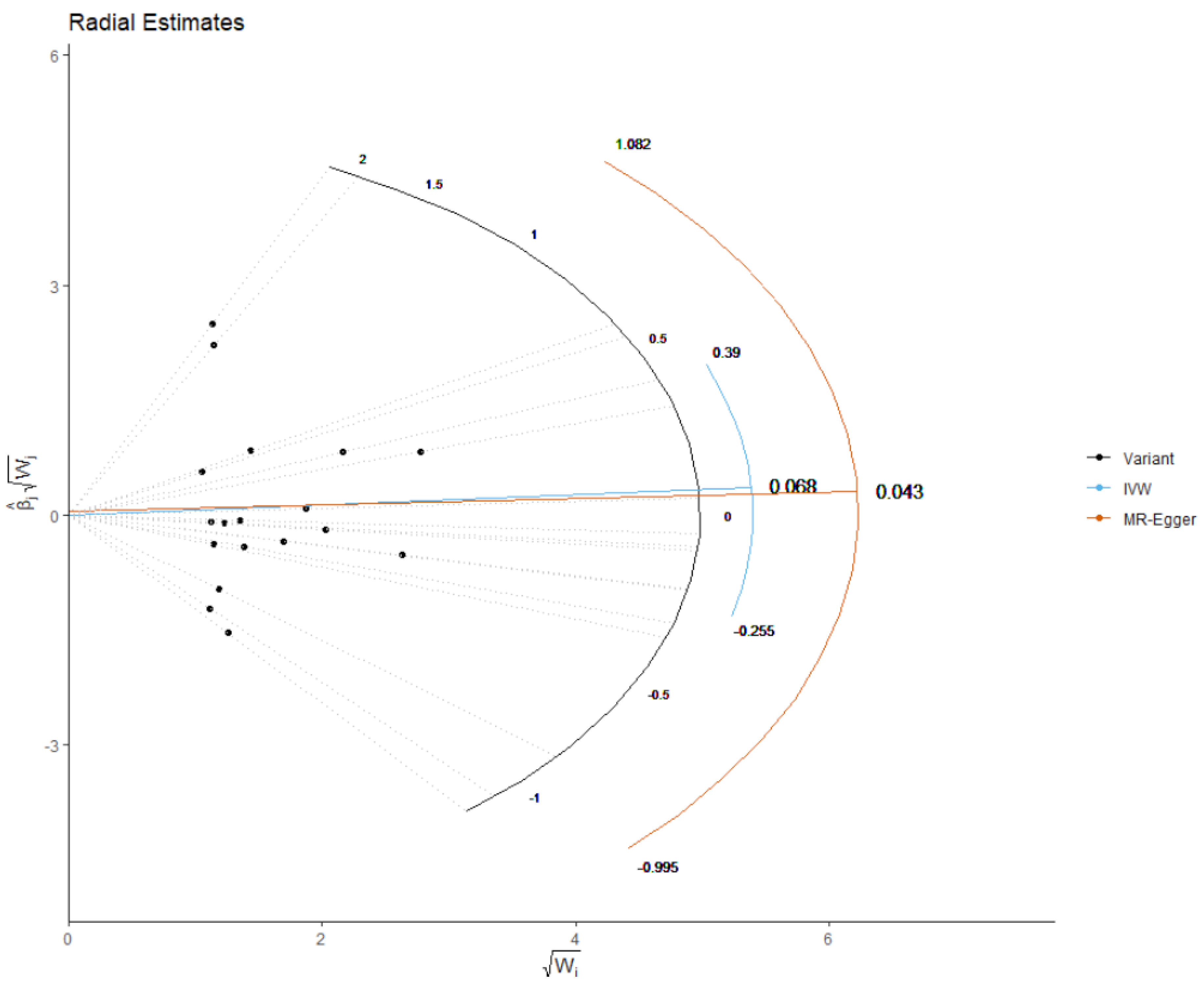
2.4. Mendelian Randomization Analysis
2.5. Ethics
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alton, G.; Hasilik, M.; Niehues, R.; Panneerselvam, K.; Etchison, J.R.; Fana, F.; Freeze, H.H. Direct utilization of mannose for mammalian glycoprotein biosynthesis. Glycobiology 1998, 8, 285–295. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Shi, Y.; Zhang, P.; Miao, M.; Zhang, T.; Jiang, B. D-Mannose: Properties, production, and applications: An overview. Compr. Rev. Food Sci. Food Saf. 2016, 15, 773–785. [Google Scholar] [CrossRef] [Green Version]
- Wood, F.C., Jr.; Cahill, G.F., Jr. Mannose utilization in man. J. Clin. Investig. 1963, 42, 1300–1312. [Google Scholar] [CrossRef] [Green Version]
- Sharma, V.; Freeze, H.H. Mannose efflux from the cells a potential source of mannose in blood. J. Biol. Chem. 2011, 286, 10193–10200. [Google Scholar] [CrossRef] [Green Version]
- Mardinoglu, A.; Stančáková, A.; Lotta, L.A.; Kuusisto, J.; Boren, J.; Blüher, M.; Wareham, N.J.; Ferrannini, E.; Groop, P.H.; Laakso, M. Plasma mannose levels are associated with incident type 2 diabetes and cardiovascular disease. Cell Metab. 2017, 26, 281–283. [Google Scholar] [CrossRef] [Green Version]
- Balkau, B.; Mhamdi, L.; Oppert, J.-M.; Nolan, J.; Golay, A.; Porcellati, F.; Laakso, M.; Ferrannini, E. Physical activity and insulin sensitivity: The RISC study. Diabetes 2008, 57, 2613–2618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hills, S.; Balkau, B.; Coppack, S.; Dekker, J.; Mari, A.; Natali, A.; Walker, M.; Ferrannini, E. The EGIR-RISC STUDY (The European group for the study of insulin resistance: Relationship between insulin sensitivity and cardiovascular disease risk): I. Methodology and objectives. Diabetologia 2004, 47, 566–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Zhang, C.; Kilicarslan, M.; Piening, B.D.; Bjornson, E.; Hallström, B.M.; Groen, A.K.; Ferrannini, E.; Laakso, M.; Snyder, M. Integrated network analysis reveals an association between plasma mannose levels and insulin resistance. Cell Metab. 2016, 24, 172–184. [Google Scholar] [CrossRef] [Green Version]
- Stančáková, A.; Javorský, M.; Kuulasmaa, T.; Haffner, S.M.; Kuusisto, J.; Laakso, M. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6414 Finnish men. Diabetes 2009, 58, 1212–1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sone, H.; Shimano, H.; Ebinuma, H.; Takahashi, A.; Yano, Y.; Iida, K.T.; Suzuki, H.; Toyoshima, H.; Kawakami, Y.; Okuda, Y. Physiological changes in circulating mannose levels in normal, glucose-intolerant, and diabetic subjects. Metabolism 2003, 52, 1019–1027. [Google Scholar] [CrossRef]
- Yoshimura, K.; Hirano, S.; Takata, H.; Funakoshi, S.; Ohmi, S.; Amano, E.; Nishi, Y.; Inoue, M.; Fukuda, Y.; Hayashi, H. Plasma mannose level, a putative indicator of glycogenolysis, and glucose tolerance in Japanese individuals. J. Diabetes Investig. 2017, 8, 489–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amano, E.; Funakoshi, S.; Yoshimura, K.; Hirano, S.; Ohmi, S.; Takata, H.; Terada, Y.; Fujimoto, S. Fasting plasma mannose levels are associated with insulin sensitivity independent of BMI in Japanese individuals with diabetes. Diabetol. Metab. Syndr. 2018, 10, 88. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Smolin, J.; Nayak, J.; Ayala, J.E.; Scott, D.A.; Peterson, S.N.; Freeze, H.H. Mannose Alters Gut Microbiome, Prevents Diet-Induced Obesity, and Improves Host Metabolism. Cell Rep. 2018, 24, 3087–3098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, G.D.; Ebrahim, S. ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003, 32, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, S.Y.; Fauman, E.B.; Petersen, A.K.; Krumsiek, J.; Santos, R.; Huang, J.; Arnold, M.; Erte, I.; Forgetta, V.; Yang, T.P.; et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014, 46, 543–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, T.M.; Lawlor, D.A.; Harbord, R.M.; Sheehan, N.A.; Tobias, J.H.; Timpson, N.J.; Davey Smith, G.; Sterne, J.A. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat. Methods Med. Res. 2012, 21, 223–242. [Google Scholar] [CrossRef] [Green Version]
- Nelson, C.P.; Goel, A.; Butterworth, A.S.; Kanoni, S.; Webb, T.R.; Marouli, E.; Zeng, L.; Ntalla, I.; Lai, F.Y.; Hopewell, J.C.; et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet. 2017, 49, 1385–1391. [Google Scholar] [CrossRef]
- Nikpay, M.; Goel, A.; Won, H.H.; Hall, L.M.; Willenborg, C.; Kanoni, S.; Saleheen, D.; Kyriakou, T.; Nelson, C.P.; Hopewell, J.C.; et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015, 47, 1121–1130. [Google Scholar] [CrossRef] [Green Version]
- Willer, C.J.; Schmidt, E.M.; Sengupta, S.; Peloso, G.M.; Gustafsson, S.; Kanoni, S.; Ganna, A.; Chen, J.; Buchkovich, M.L.; Mora, S.; et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013, 45, 1274–1283. [Google Scholar] [CrossRef] [Green Version]
- Dupuis, J.; Langenberg, C.; Prokopenko, I.; Saxena, R.; Soranzo, N.; Jackson, A.U.; Wheeler, E.; Glazer, N.L.; Bouatia-Naji, N.; Gloyn, A.L.; et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010, 42, 105–116. [Google Scholar] [CrossRef]
- Locke, A.E.; Kahali, B.; Berndt, S.I.; Justice, A.E.; Pers, T.H.; Day, F.R.; Powell, C.; Vedantam, S.; Buchkovich, M.L.; Yang, J.; et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef] [Green Version]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Davey Smith, G.; Sheehan, N.; Thompson, J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 2017, 36, 1783–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verbanck, M.; Chen, C.Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef]
- Lawlor, D.A.; Harbord, R.M.; Sterne, J.A.; Timpson, N.; Davey Smith, G. Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. Stat. Med. 2008, 27, 1133–1163. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, T.; Yamashita, E.; Mizutani, T.; Nakajima, H.; Yabuuchi, M.; Asano, N.; Miwa, I. Hepatic glycogen breakdown is implicated in the maintenance of plasma mannose concentration. Am. J. Physiol. Endocrinol. Metab. 2005, 288, E534–E540. [Google Scholar] [CrossRef] [PubMed]
- Mori, A.; Sato, T.; Lee, P.; Furuuchi, M.; Tazaki, H.; Katayama, K.; Mizutani, H.; Sako, T.; Arai, T. Clinical significance of plasma mannose concentrations in healthy and diabetic dogs. Vet. Res. Commun. 2009, 33, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Pitkänen, O.M.; Vanhanen, H.; Pitkänen, E. Metabolic syndrome is associated with changes in D-mannose metabolism. Scand. J. Clin. Lab. Investig. 1999, 59, 607–612. [Google Scholar] [CrossRef]
- Chapman, M.J.; Ginsberg, H.N.; Amarenco, P.; Andreotti, F.; Borén, J.; Catapano, A.L.; Descamps, O.S.; Fisher, E.; Kovanen, P.T.; Kuivenhoven, J.A. Triglyceride-rich lipoproteins and high-density lipoprotein cholesterol in patients at high risk of cardiovascular disease: Evidence and guidance for management. Eur. Heart J. 2011, 32, 1345–1361. [Google Scholar] [CrossRef] [Green Version]
- Quispe, R.; Manalac, R.J.; Faridi, K.F.; Blaha, M.J.; Toth, P.P.; Kulkarni, K.R.; Nasir, K.; Virani, S.S.; Banach, M.; Blumenthal, R.S.; et al. Relationship of the triglyceride to high-density lipoprotein cholesterol (TG/HDL-C) ratio to the remainder of the lipid profile: The Very Large Database of Lipids-4 (VLDL-4) study. Atherosclerosis 2015, 242, 243–250. [Google Scholar] [CrossRef]
- Rizzo, M.; Barylski, M.; Rizvi, A.A.; Montalto, G.; Mikhailidis, D.P.; Banach, M. Combined dyslipidemia: Should the focus be LDL cholesterol or atherogenic dyslipidemia? Curr. Pharm. Des. 2013, 19, 3858–3868. [Google Scholar] [CrossRef] [PubMed]
- Nagasaka, H.; Yorifuji, T.; Bandsma, R.H.; Takatani, T.; Asano, H.; Mochizuki, H.; Takuwa, M.; Tsukahara, H.; Inui, A.; Tsunoda, T. Sustained high plasma mannose less sensitive to fluctuating blood glucose in glycogen storage disease type Ia children. J. Inherit. Metab. Dis. Off. J. Soc. Study Inborn Errors Metab. 2013, 36, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Reiner, Ž. Hypertriglyceridaemia and risk of coronary artery disease. Nat. Rev. Cardiol. 2017, 14, 401. [Google Scholar] [CrossRef] [PubMed]
- Ferrannini, E.; Bokarewa, M.; Brembeck, P.; Baboota, R.; Hedjazifar, S.; Andersson, K.; Baldi, S.; Campi, B.; Muscelli, E.; Saba, A. Mannose is an insulin-regulated metabolite reflecting whole-body insulin sensitivity in man. Metabolism 2020, 102, 153974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| SNP | GX | GX SE | EA | OA | EAF |
|---|---|---|---|---|---|
| rs1275504 | 0.0168 | 0.0025 | A | T | 0.2976 |
| rs11676911 | −0.0429 | 0.0084 | A | G | 0.0216 |
| rs4980268 | 0.019 | 0.0039 | A | G | 0.9184 |
| rs12474099 | −0.0202 | 0.0043 | A | G | 0.0649 |
| rs7747345 | 0.0122 | 0.0026 | A | T | 0.8153 |
| rs13116473 | −0.0448 | 0.0096 | A | G | 0.9853 |
| rs10736 | −0.0118 | 0.0026 | T | C | 0.2011 |
| rs12414366 | 0.0214 | 0.0047 | C | G | 0.7696 |
| rs16932803 | −0.0121 | 0.0026 | T | G | 0.1673 |
| rs1181723 | 0.0116 | 0.0026 | A | G | 0.2086 |
| rs1841535 | 0.0252 | 0.0056 | T | G | 0.1482 |
| rs991670 | −0.0111 | 0.0025 | T | C | 0.6624 |
| rs2272172 | −0.0648 | 0.0145 | T | C | 0.0226 |
| rs2270451 | 0.0152 | 0.0034 | A | G | 0.8941 |
| rs7007251 | −0.015 | 0.0034 | A | G | 0.1057 |
| rs17061914 | −0.0146 | 0.0033 | A | G | 0.8581 |
| rs10982244 | −0.0177 | 0.004 | A | G | 0.6098 |
| rs2723713 | −0.0356 | 0.008 | T | C | 0.0254 |
| Exposures | MR | Heterogeneity | Pleiotropy | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Method | Beta | SE | p-Value | Method | Q | p-Value | Intercept | SE | p-Value | |
| CAD | MR Egger | −0.609 | 0.346 | 0.097 | MR Egger | 15.683 | 0.475 | 0.0122 | 0.006 | 0.084 |
| WM | −0.189 | 0.199 | 0.342 | |||||||
| IVW | −0.031 | 0.153 | 0.835 | IVW | 19.060 | 0.324 | ||||
| RAPS | −0.095 | 0.153 | 0.532 | |||||||
| TC | MR Egger | −0.569 | 0.409 | 0.182 | MR Egger | 42.170 | 0.0003 | 0.006 | 0.007 | 0.360 |
| WM | −0.010 | 0.155 | 0.946 | |||||||
| IVW | −0.212 | 0.151 | 0.161 | IVW | 44.503 | 0.0002 | ||||
| RAPS | −0.242 | 0.157 | 0.122 | |||||||
| LDL | MR Egger | −0.709 | 0.336 | 0.051 | MR Egger | 27.361 | 0.037 | 0.010 | 0.005 | 0.077 |
| WM | −0.044 | 0.140 | 0.753 | |||||||
| IVW | −0.119 | 0.134 | 0.375 | IVW | 33.450 | 0.009 | ||||
| RAPS | −0.140 | 0.131 | 0.283 | |||||||
| HDL | MR Egger | 0.308 | 0.235 | 0.207 | MR Egger | 13.746 | 0.617 | −0.005 | 0.004 | 0.191 |
| WM | 0.068 | 0.127 | 0.589 | |||||||
| IVW | 0.011 | 0.088 | 0.896 | IVW | 15.605 | 0.551 | ||||
| RAPS | 0.007 | 0.097 | 0.939 | |||||||
| TG | MR Egger | −0.522 | 0.729 | 0.484 | MR Egger | 159.485 | <0.001 | 0.001 | 0.012 | 0.900 |
| WM | −0.13 | 0.137 | 0.337 | |||||||
| IVW | −0.436 | 0.263 | 0.097 | IVW | 159.645 | <0.001 | ||||
| RAPS | −0.143 | 0.160 | 0.370 | |||||||
| WC | MR Egger | −0.094 | 0.257 | 0.717 | MR Egger | 25.005 | 0.094 | 0.004 | 0.004 | 0.353 |
| WM | 0.150 | 0.116 | 0.198 | |||||||
| IVW | 0.133 | 0.095 | 0.162 | IVW | 26.345 | 0.092 | ||||
| RAPS | 0.110 | 0.099 | 0.267 | |||||||
| BMI | MR Egger | −0.318 | 0.199 | 0.128 | MR Egger | 20.020 | 0.273 | 0.007 | 0.003 | 0.037 |
| WM | 0.086 | 0.101 | 0.391 | |||||||
| IVW | 0.098 | 0.083 | 0.234 | IVW | 26.004 | 0.099 | ||||
| RAPS | 0.089 | 0.080 | 0.269 | |||||||
| FBG | MR Egger | −0.177 | 0.236 | 0.464 | MR Egger | 29.217 | 0.032 | 0.005 | 0.004 | 0.210 |
| WM | −0.018 | 0.103 | 0.855 | |||||||
| IVW | 0.106 | 0.094 | 0.259 | IVW | 32.132 | 0.021 | ||||
| RAPS | 0.046 | 0.095 | 0.627 | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazidi, M.; Dehghan, A.; Banach, M.; on behalf of the Lipid and Blood Pressure Meta-Analysis Collaboration (LBPMC) Group and the International Lipid Expert Panel (ILEP). Genetically Higher Level of Mannose Has No Impact on Cardiometabolic Risk Factors: Insight from Mendelian Randomization. Nutrients 2021, 13, 2563. https://doi.org/10.3390/nu13082563
Mazidi M, Dehghan A, Banach M, on behalf of the Lipid and Blood Pressure Meta-Analysis Collaboration (LBPMC) Group and the International Lipid Expert Panel (ILEP). Genetically Higher Level of Mannose Has No Impact on Cardiometabolic Risk Factors: Insight from Mendelian Randomization. Nutrients. 2021; 13(8):2563. https://doi.org/10.3390/nu13082563
Chicago/Turabian StyleMazidi, Mohsen, Abbas Dehghan, Maciej Banach, and on behalf of the Lipid and Blood Pressure Meta-Analysis Collaboration (LBPMC) Group and the International Lipid Expert Panel (ILEP). 2021. "Genetically Higher Level of Mannose Has No Impact on Cardiometabolic Risk Factors: Insight from Mendelian Randomization" Nutrients 13, no. 8: 2563. https://doi.org/10.3390/nu13082563
APA StyleMazidi, M., Dehghan, A., Banach, M., & on behalf of the Lipid and Blood Pressure Meta-Analysis Collaboration (LBPMC) Group and the International Lipid Expert Panel (ILEP). (2021). Genetically Higher Level of Mannose Has No Impact on Cardiometabolic Risk Factors: Insight from Mendelian Randomization. Nutrients, 13(8), 2563. https://doi.org/10.3390/nu13082563






