Association between Polygenetic Risk Scores of Low Immunity and Interactions between These Scores and Moderate Fat Intake in a Large Cohort
Abstract
:1. Introduction
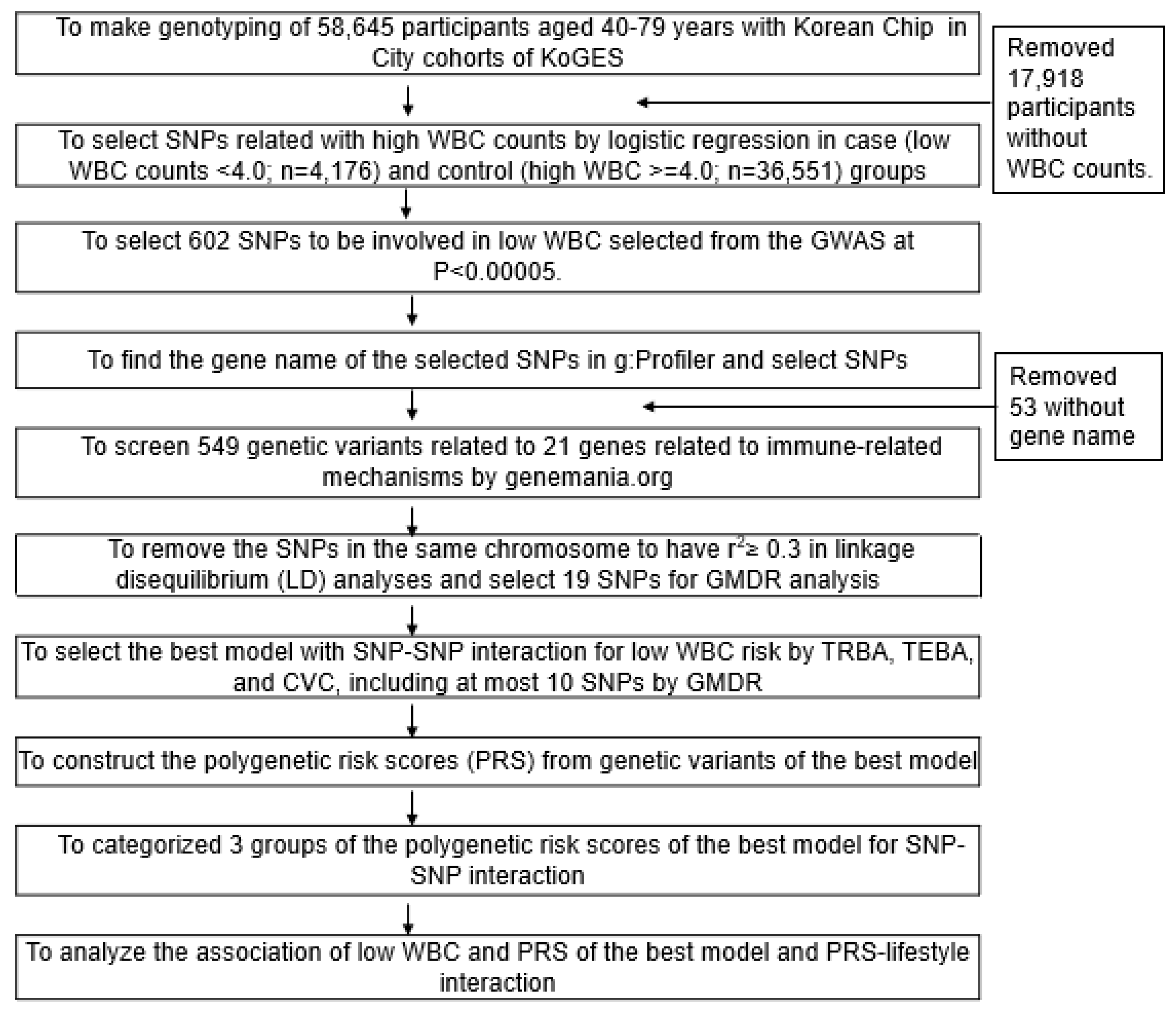
2. Methods
2.1. Participants
2.2. Anthropometric and Biochemical Measurements
2.3. Definition of Immunity and MetS
2.4. Dietary Pattern Analysis from Semi-Quantitative Food Frequency Questionnaire (SQFFQ)
2.5. Dietary Inflammatory Index (DII)
2.6. Quality Control of Genotyping
2.7. Genetic Variants for Low-WBC Count Risk
2.8. The Best Model for Gene–Gene Interactions of Genetic Variants by Generalized Multifactor Dimensionality Reduction (GMDR)
2.9. Statistical Analyses
3. Results
3.1. General Characteristics of Participants in the WBC Count Groups
3.2. Lifestyles and Nutrient Intakes
3.3. Genetic Variants Associated with Low-WBC Count Risk and Gene–Gene Interactions between Genetic Variants by GMDR
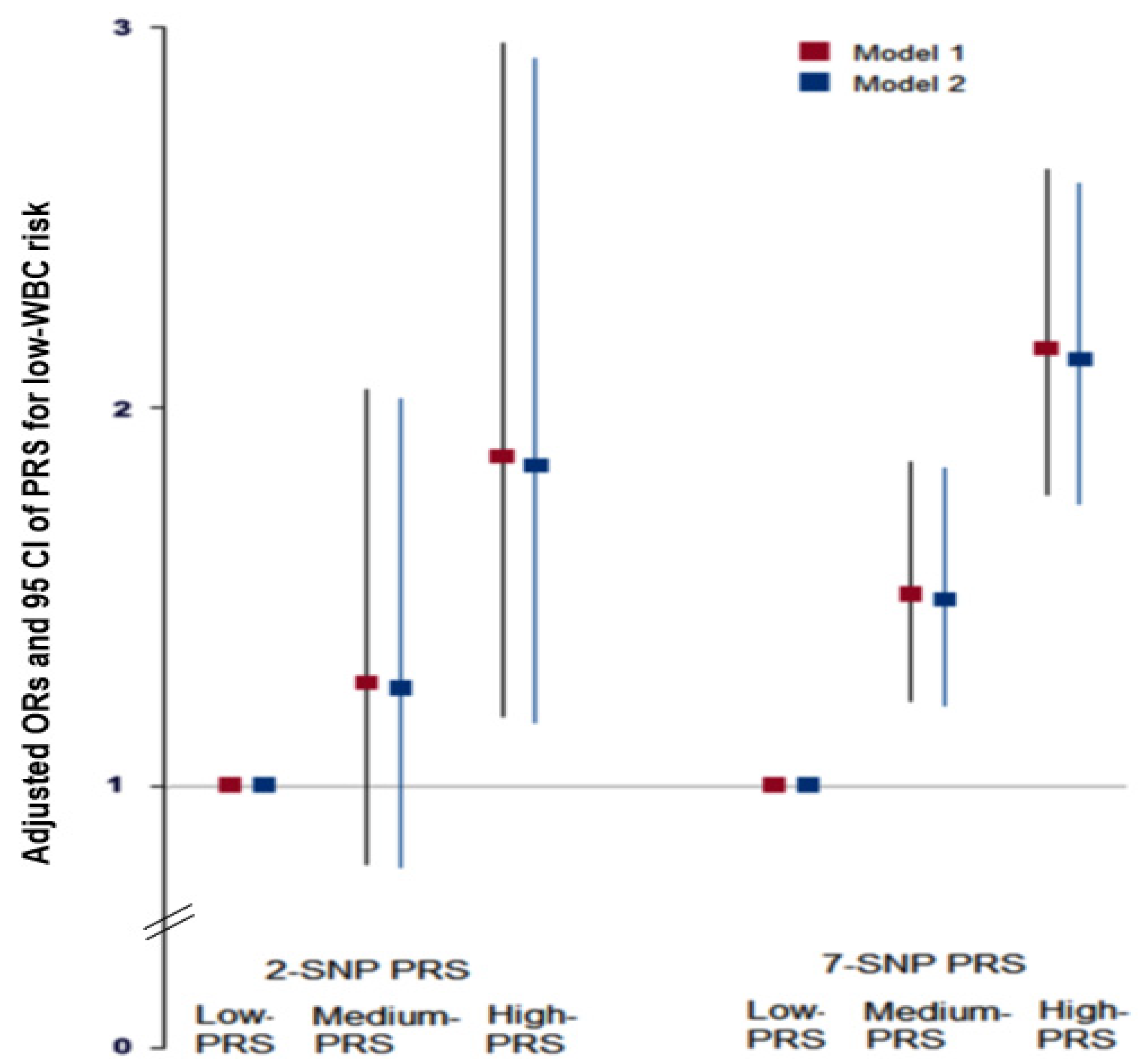
3.4. Associations between PRS Derived from the 7-SNP Model and MetS and Its Components
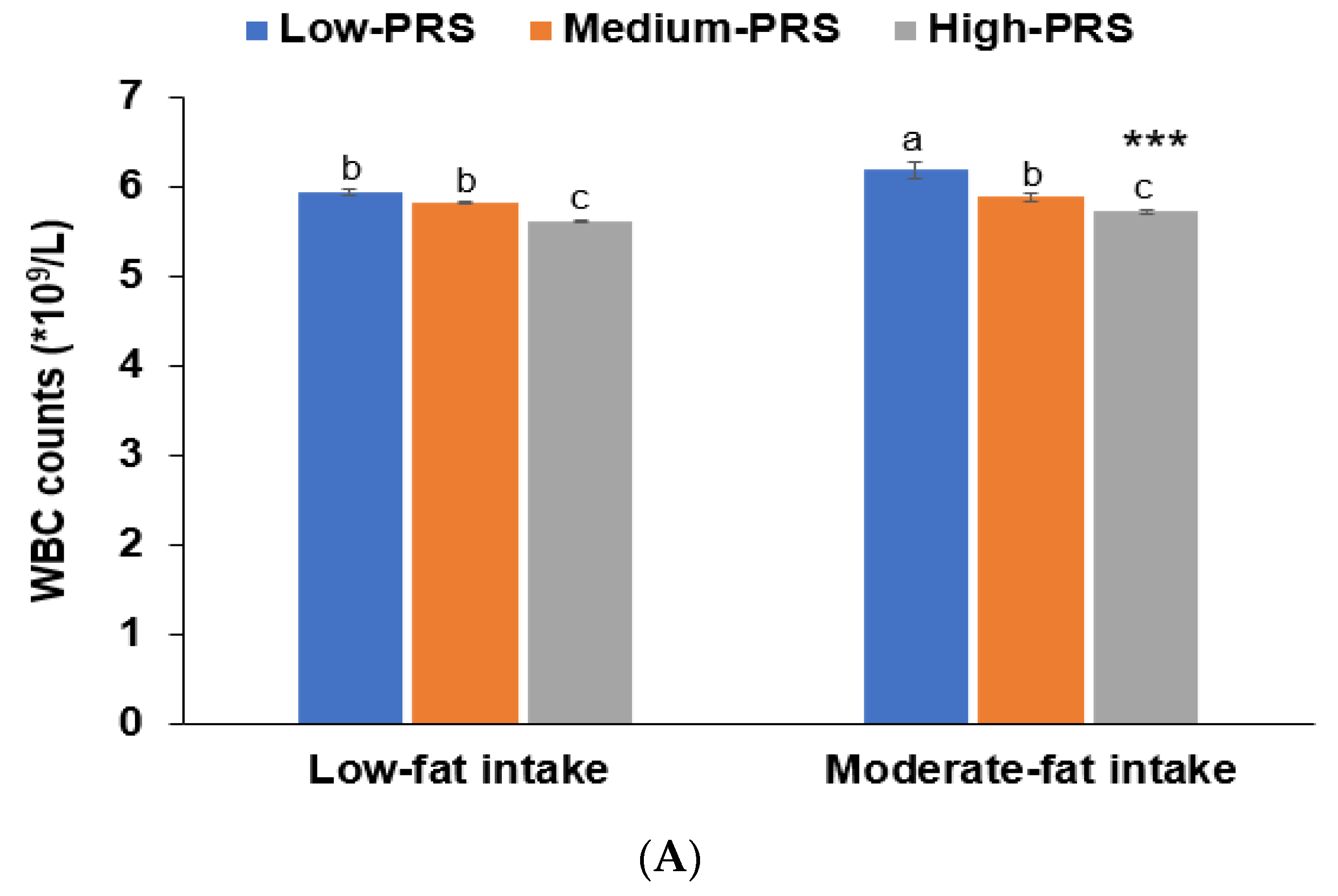
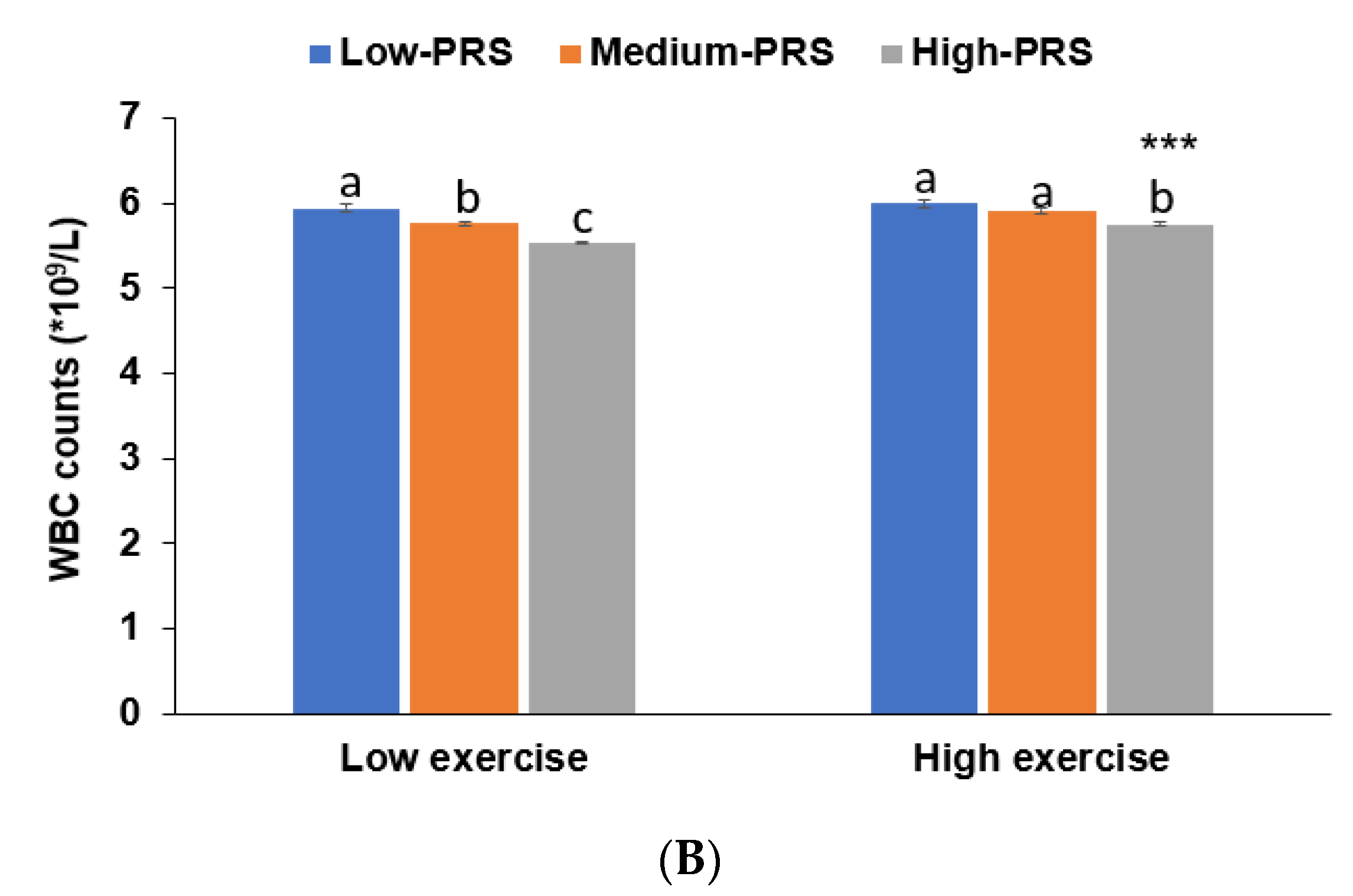
3.5. Interaction between PRS and Nutrient Intakes and a Low WBC Count Risk
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Zidar, D.A.; Al-Kindi, S.G.; Liu, Y.; Krieger, N.I.; Perzynski, A.T.; Osnard, M.; Nmai, C.; Anthony, D.D.; Lederman, M.M.; Freeman, M.L.; et al. Association of Lymphopenia With Risk of Mortality Among Adults in the US General Population. JAMA Netw. Open 2019, 2, e1916526. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Zhang, T. A Positive Association of Overactivated Immunity with Metabolic Syndrome Risk and Mitigation of Its Association by a Plant-Based Diet and Physical Activity in a Large Cohort Study. Nutrients 2021, 13, 2308. [Google Scholar] [CrossRef]
- Charles-Messance, H.; Sheedy, F.J. Train to Lose: Innate Immune Memory in Metaflammation. Mol. Nutr. Food Res. 2021, 65, e1900480. [Google Scholar] [CrossRef] [PubMed]
- Navegantes, K.C.; de Souza Gomes, R.; Pereira, P.A.T.; Czaikoski, P.G.; Azevedo, C.H.M.; Monteiro, M.C. Immune modula-tion of some autoimmune diseases: The critical role of macrophages and neutrophils in the innate and adaptive immunity. J. Transl. Med. 2017, 15, 36. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, C.P.; Gonzalez, M.C.; Aguilar-Salinas, C.A.; Nájera-Medina, O. Peripheral Lymphocytes, Obesity, and Metabolic Syndrome in Young Adults: An Immunometabolism Study. Metab. Syndr. Relat. Disord. 2018, 16, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Schloss, M.J.; Swirski, F.; Nahrendorf, M. Modifiable Cardiovascular Risk, Hematopoiesis, and Innate Immunity. Circ. Res. 2020, 126, 1242–1259. [Google Scholar] [CrossRef]
- Pietzner, M.; Kaul, A.; Henning, A.-K.; Kastenmüller, G.; Artati, A.; Lerch, M.M.; Adamski, J.; Nauck, M.; Friedrich, N. Com-prehensive metabolic profiling of chronic low-grade inflammation among generally healthy individuals. BMC Med. 2017, 15, 210. [Google Scholar] [CrossRef] [Green Version]
- Celerino da Silva, R.; Moura, R.R.; Victor Campos Coelho, A.; Arraes, L.C.; Brandão, L.A.C.; Crovella, S.; Lima Guimarães, R. HLA-C Single Nucleotide Polymorphism Associated with Increased Viral Load Level in HIV-1 Infected Individuals from Northeast Brazil. Curr. HIV Res. 2017, 15, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Harsløf, M.; Pedersen, K.M.; Nordestgaard, B.G.; Afzal, S. Low HDL (High-Density Lipoprotein) Cholesterol and High White Blood Cell Counts: A Mendelian Randomization Study. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 976–987. [Google Scholar] [CrossRef]
- Kozuka, R.; Enomoto, M.; Sato-Matsubara, M.; Yoshida, K.; Motoyama, H.; Hagihara, A.; Fujii, H.; Uchida-Kobayashi, S.; Morikawa, H.; Tamori, A.; et al. Association between HLA-DQA1/DRB1 polymorphism and development of hepatocellular carcinoma during entecavir treatment. J. Gastroenterol. Hepatol. 2018, 34, 937–946. [Google Scholar] [CrossRef]
- Siedlinski, M.; Jozefczuk, E.; Xu, X.; Teumer, A.; Evangelou, E.; Schnabel, R.B.; Welsh, P.; Maffia, P.; Erdmann, J.; To-maszewski, M.; et al. White Blood Cells and Blood Pressure: A Mendelian Randomization Study. Circulation 2020, 141, 1307–1317. [Google Scholar] [CrossRef]
- Krebs, K.; Bovijn, J.; Zheng, N.; Lepamets, M.; Censin, J.C.; Jürgenson, T.; Särg, D.; Abner, E.; Laisk, T.; Luo, Y.; et al. Ge-nome-wide Study Identifies Association between HLA-B(∗)55:01 and Self-Reported Penicillin Allergy. Am. J. Hum. Genet. 2020, 107, 612–621. [Google Scholar] [CrossRef]
- Dmello, D.M.; Ariyanto, I.; Estiasari, R.; Halstrom, S.; Gaff, J.; Lee, S.; Price, P. Polymorphisms in IL10 may alter CD4 T-cell counts in Indonesian HIV patients beginning antiretroviral therapy. Hum. Immunol. 2017, 78, 387–390. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Jin, H.S.; Park, S. Protein and fat intake interacts with the haplotype of PTPN11_rs11066325, RPH3A_rs886477, and OAS3_rs2072134 to modulate serum HDL concentrations in middle-aged people. Clin. Nutr. 2020, 39, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Ahn, J.; Lee, B.-K. Self-rated Subjective Health Status Is Strongly Associated with Sociodemographic Factors, Lifestyle, Nutrient Intakes, and Biochemical Indices, but Not Smoking Status: KNHANES 2007-2012. J. Korean Med. Sci. 2015, 30, 1279–1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.; Han, B.G. Cohort Profile: The Korean Genome and Epidemiology Study (KoGES) Consortium. Int. J. Epidemiol. 2017, 46, 1350. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, D.S.; Kang, S. Carrying minor allele of FADS1 and haplotype of FADS1 and FADS2 increased the risk of met-abolic syndrome, and moderate but not low-fat diets lowered the risk in two Korean cohorts. Eur. J. Nutr. 2019, 58, 831–842. [Google Scholar] [CrossRef]
- Park, S.; Ahn, J.; Lee, B.K. Very low-fat diets may be associated with increased risk of metabolic syndrome in the adult pop-ulation. Clin. Nutr. 2016, 35, 1159–1167. [Google Scholar] [CrossRef]
- Ahn, Y.; Kwon, E.; Shim, J.E.; Park, M.K.; Joo, Y.; Kimm, K.; Park, C.; Kim, D.H. Validation and reproducibility of food fre-quency questionnaire for Korean genome epidemiologic study. Eur. J. Clin. Nutr. 2007, 61, 1435–1441. [Google Scholar] [CrossRef]
- Park, S.; Kang, S. A Western-style diet interacts with genetic variants of the LDL receptor to hyper-LDL cholesterolemia in Korean adults. Public Health Nutr. 2021, 24, 2964–2974. [Google Scholar] [CrossRef]
- Van Woudenbergh, G.J.; Theofylaktopoulou, D.; Kuijsten, A.; Ferreira, I.; Van Greevenbroek, M.M.; Van Der Kallen, C.J.; Schalkwijk, C.G.; Da Stehouwer, C.; Ocké, M.C.; Nijpels, G.; et al. Adapted dietary inflammatory index and its association with a summary score for low-grade inflammation and markers of glucose metabolism: The Cohort study on Diabetes and Atherosclerosis Maastricht (CODAM) and the Hoorn study. Am. J. Clin. Nutr. 2013, 98, 1533–1542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, S.; Kim, Y.J.; Han, S.; Hwang, M.Y.; Shin, D.M.; Park, M.Y.; Lu, Y.; Yoon, K.; Jang, H.-M.; Kim, Y.K.; et al. The Korea Biobank Array: Design and Identification of Coding Variants Associated with Blood Biochemical Traits. Sci. Rep. 2019, 9, 1382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabbee, N.; Speed, T.P. A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics 2005, 22, 7–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jyothi, K.U.; Reddy, B.M. Gene–gene and gene–environment interactions in the etiology of type 2 diabetes mellitus in the population of Hyderabad, India. Meta Gene 2015, 5, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.-W.; Kim, S.H.; Zhang, X.; Park, S. Interactions among the variants of insulin-related genes and nutrients increase the risk of type 2 diabetes. Nutr. Res. 2018, 51, 82–92. [Google Scholar] [CrossRef]
- Monserrat-Mesquida, M.; Quetglas-Llabrés, M.; Capó, X.; Bouzas, C.; Mateos, D.; Pons, A.; Tur, J.A.; Sureda, A. Metabolic Syndrome Is Associated with Oxidative Stress and Proinflammatory State. Antioxidants 2020, 9, 236. [Google Scholar] [CrossRef] [Green Version]
- Mahdiani, A.; Kheirandish, M.; Bonakdaran, S. Correlation Between White Blood Cell Count and Insulin Resistance in Type 2 Diabetes. Curr. Diabetes Rev. 2018, 15, 62–66. [Google Scholar] [CrossRef]
- Hartl, S.; Breyer, M.-K.; Burghuber, O.C.; Ofenheimer, A.; Schrott, A.; Urban, M.H.; Agusti, A.; Studnicka, M.; Wouters, E.F.; Breyer-Kohansal, R. Blood eosinophil count in the general population: Typical values and potential confounders. Eur. Respir. J. 2020, 55, 1901874. [Google Scholar] [CrossRef]
- Chen, T.; Chen, H.; Xiao, H.; Tang, H.; Xiang, Z.; Wang, X.; Wang, X.; Zou, H. Comparison of the Value of Neutrophil to High-Density Lipoprotein Cholesterol Ratio and Lymphocyte to High-Density Lipoprotein Cholesterol Ratio for Predict-ing Metabolic Syndrome Among a Population in the Southern Coast of China. Diabetes Metab. Syndr. Obes. Targets Ther. 2020, 13, 597–605. [Google Scholar] [CrossRef] [Green Version]
- Mosaad, Y.M. Clinical Role of Human Leukocyte Antigen in Health and Disease. Scand. J. Immunol. 2015, 82, 283–306. [Google Scholar] [CrossRef]
- Luckey, D.; Weaver, E.A.; Osborne, D.G.; Billadeau, D.D.; Taneja, V. Immunity to Influenza is dependent on MHC II poly-morphism: Study with 2 HLA transgenic strains. Sci. Rep. 2019, 9, 19061. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, A.; David, J.K.; Maden, S.K.; Wood, M.A.; Weeder, B.R.; Nellore, A.; Thompson, R.F. Human Leukocyte Antigen Susceptibility Map for Severe Acute Respiratory Syndrome Coronavirus 2. J. Virol. 2020, 94, 00510–00520. [Google Scholar] [CrossRef] [Green Version]
- Reiner, A.P.; Lettre, G.; Nalls, M.A.; Ganesh, S.K.; Mathias, R.; Austin, M.A.; Déan, E.; Arepalli, S.; Britton, A.; Chen, Z.; et al. Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: The Continental Origins and Genetic Epidemiology Network (COGENT). PLoS Genet. 2011, 7, e1002108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bencomo-Alvarez, A.E.; Rubio, A.J.; Olivas, I.M.; Gonzalez, M.A.; Ellwood, R.; Fiol, C.R.; Eide, C.A.; Lara, J.J.; Barreto-Vargas, C.; Jave-Suarez, L.F.; et al. Proteasome 26S subunit, non-ATPases 1 (PSMD1) and 3 (PSMD3), play an oncogenic role in chronic myeloid leukemia by stabilizing nuclear factor-kappa B. Oncogene 2021, 40, 2697–2710. [Google Scholar] [CrossRef]
- Yau, W.W.; Yen, P.M. Thermogenesis in Adipose Tissue Activated by Thyroid Hormone. Int. J. Mol. Sci. 2020, 21, 3020. [Google Scholar] [CrossRef]
- Moran, C.; Chatterjee, K. Resistance to thyroid hormone due to defective thyroid receptor alpha. Best Pract. Res. Clin. Endocrinol. Metab. 2015, 29, 647–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Artemniak-Wojtowicz, D.; Witkowska-Sędek, E.; Borowiec, A.; Pyrżak, B. Peripheral blood picture and aminotransferase activity in children with newly diagnosed Graves’ disease at baseline and after the initiation of antithyroid drug therapy. Cent. Eur. J. Immunol. 2019, 44, 132–137. [Google Scholar] [CrossRef]
- Ahmed, S.S.; Mohammed, A.A. Effects of thyroid dysfunction on hematological parameters: Case controlled study. Ann. Med. Surg. 2020, 57, 52–55. [Google Scholar] [CrossRef] [PubMed]
- Ozdemir, O.; Mete, E.; Catal, F.; Ozol, D. Food Intolerances and Eosinophilic Esophagitis in Childhood. Dig. Dis. Sci. 2008, 54, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.-Y.; Park, S. Associations of Polygenetic Variants at the 11q23 Locus and Their Interactions with Macronutrient In-take for the risk of 3GO, a Combination of Hypertension, Hyperglycemia, and Dyslipidemia. J. Pers. Med. 2021, 11, 207. [Google Scholar] [CrossRef]
- Zhou, J.-Y.; Song, M.Y.; Park, S. Carbohydrate and sodium intake and physical activity interact with genetic risk scores of four genetic variants mainly related to lipid metabolism to modulate metabolic syndrome risk in Korean middle-aged adults. Br. J. Nutr. 2019, 122, 919–927. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Berube, J.; McNamara, M.; Saksena, S.; Hartman, M.; Arshad, T.; Bornheimer, S.J.; O’Gorman, M. Lymphocyte Subset Counts in COVID-19 Patients: A Meta-Analysis. Cytom. Part A 2020, 97, 772–776. [Google Scholar] [CrossRef]
- Chi, Q.; Dai, X.; Jiang, X.; Zhu, L.; Du, J.; Chen, Y.; Zheng, J.; Huang, J. Differential diagnosis for suspected cases of corona-virus disease 2019: A retrospective study. BMC Infect. Dis. 2020, 20, 679. [Google Scholar] [CrossRef] [PubMed]
| Low (<4) (n = 4176) | Normal (4≤ <6.2) (n = 23,911) | High (<6.2) (n = 12,640) | Adjusted ORs (95% CI) of Low-WBC 16 | |
|---|---|---|---|---|
| Age 1 (years) | 54.1 (53.9–54.3) 14,b | 53.9 (53.8–54.0) b | 53.2 (53.1–53.3) a,*** | 1.134 (1.040–1.237) |
| Gender (N, male %) | 742 (17.8) 15 | 7432 (31.1) | 5682 (44.9) +++ | 0.614 (0.534–0.706) |
| Cancer (N, Yes %) | 283 (6.8) | 981 (4.1) | 423 (3.4) +++ | 1.467 (1.219–1.765) |
| Serum hs-CRP 2 (ng/mL) | 0.097 (0.083–0.113) c | 0.111 (0.105–0.117) b | 0.209 (0.201–0.217) a,*** | 0.542 (0.416–0.706) |
| Metabolic syndrome (N, Yes %) | 239 (5.7) | 2860 (12.0) | 2625 (20.7) +++ | 0.458 (0.385–0.545) |
| BMI 3 (kg/m2) | 23.0 (22.9–23.1) c | 23.8 (23.7–23.8) b | 24.3 (24.3–24.4) a,*** | 0.561 (0.510–0.618) |
| Fat mass 4 (%) | 1749 (41.9) | 11848 (49.5) | 6835(53.9) *** | 0.541 (0.498–0.588) |
| Waist circumference 5 (cm) | 79.8 (79.6–79.9) c | 80.3 (80.3–80.4) b | 80.8 (80.7–80.9) a,*** | 0.548 (0.485–0.618) |
| Plasma glucose 6 (mg/dL) | 92.7 (92.1–93.3) c | 94.7 (94.4–94.9) b | 97.3 (97.0–97.7) a,*** | 0.464 (0.386–0.557) |
| HbA1c 7 (%) | 5.56 (5.58–5.59) c | 5.67 (5.66–5.68) b | 5.85 (5.84–5.87) a,*** | 0.376 (0.299–0.473) |
| Serum total cholesterol 8 (mg/dL) | 193 (192–194) c | 198 (197–198) b | 199 (199–200) a,*** | 0.670 (0.606–0.740 |
| Serum HDL 9 (mg/dL) | 56.9 (56.5–57.3) a | 54.8 (54.6–54.9) b | 53.3 (53.1–53.5) c,*** | 0.725 (0.658–0.800) |
| Serum LDL 10 (mg/dL) | 115 (114–116) b | 119 (118–119) a | 118 (117–119) a,*** | 0.662 (0.588–0.744) |
| Serum TG 11 (mg/dL) | 106 (103–109) c | 122 (121–123) b | 140 (139–142) a,*** | 0.516 (0.464–0.574) |
| SBP (mmHg) 12 | 120.8 (120.4–121.3) c | 122.4 (122.2–122.6) b | 123.7 (123.4–123.9) a,*** | 0.817 (0.746–0.895) |
| DBP (mmHg) 13 | 74.3 (74.0–74.6) c | 75.3 (75.1–75.4) b | 76.0 (75.8–76.1) a,*** | 0.799 (0.674–0.946) |
| KERRYPNX | Low (<4) (n = 4176) | Normal (4≤ <6.2) (n = 23,911) | High (<6.2) (n = 12,640) | Adjusted ORs (95% CI) of Low-WBC 3 |
|---|---|---|---|---|
| Smoking (N, %) | ||||
| Non-smoker | 3630 (87.0) 1 | 18,216 (76.4) | 7700 (61.2) +++ | 1 |
| Former-smoker | 412 (9.90) | 3902 (16.4) | 2236 (17.8) | 0.352 (0.271–0.458) |
| Smoker | 121 (2.91) | 1723 (7.23) | 2656 (21.1) | 0.298 (0.230–0.387) ### |
| Regular exercise 4 (%) | 2430 (58.3) | 13,354 (56.0) | 6453 (51.2) +++ | 1.262 (1.165–1.367) ### |
| Alcohol intake 5 (≥20g/week) | 1413 (33.8) | 14,234 (42.5) | 1498 (48.7) *** | 0.849 (0.768–0.936) ### |
| Coffee 6 (cups/week) | 3.5 (3.4–3.6) c | 3.7 (3.7–3.8) b | 4.0 (3.9–4.0) a,*** | 0.856 (0.790–0.928) ### |
| Energy intake 7 (%EER) | 94.7 (93.7–95.6) 2,c | 96.2 (95.9–96.6) b | 95.4 (94.3–96.5) a,b,** | 0.949 (0.876–1.028) |
| CHO intake 8 (energy %) | 71.7 (71.5–71.9) a | 71.7 (71.6–71.8) a | 71.4 (71.3–71.5) b,* | 0.983 (0.880–1.097) |
| Protein intake 9 (energy %) | 13.4 (13.3–13.5) | 13.4 (13.3–13.4) | 13.4 (13.4–13.5) | 0.993 (0.918–1.075) |
| Fat intake 10 (energy %) | 13.9 (13.7–14.1) a | 13.9 (13.9–14.0) a | 14.1 (14.0–14.2) b,** | 0.952 (0.875–1.036) |
| Vitamin D 11 (ug/day) | 6.48 (6.34–6.61) a | 6.39 (6.34–6.45) a | 6.23 (6.15–6.31) b,** | 1.080 (0.975–1.197) |
| Anti-inflammation index (scores) 12 | 1933 (1891–1975) | 1926 (1908–1943) | 1918 (1894–1943) | 1.010 (0.917–1.113) |
| Korean balanced diet 13 (<66th per, N, %) | 1176 (28.2) | 7337 (30.7) | 4245 (33.5) +++ | 1.034 (0.893–1.198) |
| Plant-based diet 13 (N, %) | 1625 (38.9) | 8153 (34.1) | 3611 (28.5) +++ | 1.231 (1.041–1.456) |
| Western-style diet 13 (N, %) | 1181 (28.3) | 7728 (32.3) | 4676 (37.0) +++ | 1.032 (0.818–1.303) |
| Rice-main diet 13 (N, %) | 1417 (33.9) | 7672 (32.1) | 4205 (33.2) + | 1.079 (0.976–1.192) |
| Chr.1 | SNP 2 | Position | Mi 3 | Ma 4 | OR 5 (95% CI) 6 | p-Value Adjusted 7 | OR 8 (95% CI) | p-Value Adjusted 9 | MAF 10 | HWE 11 | Gene | Functional Consequence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | rs80157389 | 136546733 | C | G | 0.75 (0.69–0.81) | 1.90 × 10−13 | 0.73 (0.59–0.904) | 0.004 | 0.179 | 0.79 | LCT | intron |
| 6 | rs2308575 | 31239057 | T | C | 0.82 (0.77–0.88) | 4.90 × 10−8 | 0.78 (0.54–0.96) | 0.029 | 0.202 | 0.349 | HLA-C | missense |
| 6 | rs34791928 | 31781398 | T | C | 0.81 (0.72–0.92) | 7.10 × 10−4 | 0.75 (0.50–0.97) | 0.043 | 0.062 | 0.887 | HSPA1A | near-gene-5 |
| 6 | rs532162239 | 32558725 | T | C | 0.85 (0.80–0.90) | 3.30 × 10−8 | 0.76 (0.59–0.93) | 0.015 | 0.346 | 0.523 | HLA-DRB1 | upstream |
| 6 | rs112181319 | 33039694 | T | G | 0.86 (0.78–0.94) | 9.50 × 10−4 | 0.73 (0.55–0.95) | 0.012 | 0.107 | 0.546 | HLA-DPA1 | intron |
| 6 | rs3097649 | 33056962 | T | C | 1.10 (1.04–1.16) | 9.00 × 10−5 | 1.16 (1.01–1.32) | 0.043 | 0.363 | 0.95 | HLA-DPB1 | utr-3 |
| 6 | rs3176337 | 36648920 | A | C | 0.86 (0.81–0.92) | 4.90 × 10−6 | 0.77 (0.60–0.97) | 0.035 | 0.245 | 0.697 | CDKN1A | intron |
| 7 | rs445 | 92408370 | T | C | 1.18 (1.12–1.25) | 8.61 × 10−9 | 1.21 (1.02–1.40) | 0.002 | 0.327 | 0.888 | CDK6 | intron |
| 17 | rs9898547 | 38136026 | T | G | 1.23 (1.16–1.29) | 2.40 × 10−13 | 1.45 (1.24–1.68) | 2.2 × 10−6 | 0.399 | 0.475 | PSMD3 | near-gene-5 |
| 19 | rs7502539 | 38219005 | A | G | 1.18 (1.12–1.25) | 3.60 × 10−9 | 1.21 (1.03–1.42) | 0.017 | 0.347 | 0.669 | THRA | near-gene-5 |
| Adjusted for Gender and Age | Adjusted for Gender, Age, Residence Area, BMI, and Serum CRP | |||||||
|---|---|---|---|---|---|---|---|---|
| Model | TRBA | TEBA | p-Value | CVC | TRBA | TEBA | p-Value | CVC |
| PSMD3_rs9898547 | 0.5270 | 0.5247 | 10 (0.0010) | 9/10 | 0.5270 | 0.5225 | 10 (0.0010) | 6/10 |
| Model 1 plus LCT_rs80157389 | 0.5391 | 0.5381 | 10 (0.0010) | 10/10 | 0.5392 | 0.5383 | 10 (0.0010) | 10/10 |
| Model 2 plus HLA-C_rs2308575 | 0.5421 | 0.5334 | 10 (0.0010) | 5/10 | 0.5425 | 0.5328 | 10 (0.0010) | 4/10 |
| Model 2 plus HLA-DRB1 _rs532162239 HLA_DPB1 _rs3097649 | 0.5486 | 0.5351 | 10 (0.0010) | 7/10 | 0.5494 | 0.5349 | 10 (0.0010) | 8/10 |
| Model 4 plus CDKN1A_rs3176337 | 0.5589 | 0.5273 | 10 (0.0010) | 6/10 | 0.5597 | 0.5304 | 10 (0.0010) | 7/10 |
| Model 5 plus HLA-C_rs2308575 | 0.5758 | 0.5177 | 10 (0.0010) | 5/10 | 0.5768 | 0.5208 | 10 (0.0010) | 5/10 |
| Model 6 plus THRA_rs7502539 | 0.6028 | 0.5259 | 10 (0.0010) | 10/10 | 0.6040 | 0.5291 | 10 (0.0010) | 10/10 |
| Model 7 plus HLA-DPA1_rs112181319 | 0.6254 | 0.5239 | 10 (0.0010) | 10/10 | 0.6260 | 0.5248 | 10 (0.0010) | 10/10 |
| Model 8 plus HSPA1A_rs34791928 | 0.6447 | 0.5189 | 9 (0.0107) | 10/10 | 0.6452 | 0.5215 | 10 (0.0010) | 10/10 |
| Model 9 plus CDK6_rs445 | 0.6559 | 0.5198 | 10 (0.0010) | 10/10 | 0.6561 | 0.5218 | 10 (0.0010) | 10/10 |
| Low-PRS (n = 2719) | Medium-PRS (n = 11150) | High-PRS (n = 26,899) | Gene-Nutrient Interaction p-Value | |
|---|---|---|---|---|
| Low energy 1 High energy | 1 | 1.401(1.074–1.829) 1.625(1.158–2.280) | 2.130(1.657–2.739) 2.104(1.522–2.909) | 0.3184 |
| Low CHO 2 High CHO | 1 | 2.020 (1.132–3.606) 1.412 (1.128–1.767) | 2.659 (1.525–4.635) 2.038 (1.648–2.521) | 0.3799 |
| Low protein 3 High protein | 1 | 1.316(0.992–1.745) 1.547(1.276–1.875) | 1.879(1.439–2.453) 1.718(1.256–2.349) | 0.6677 |
| Low fat 4 High fat | 1 | 1.656 (1.165–1.819) 1.227(0.939–2.177) | 2.085 (1.688–2.575) 2.638(1.307–4.184) | 0.0170 |
| Low KBD 5 High KBD | 1 | 1.325 (1.044–1.682) 1.603 (1.232–2.086) | 1.928 (1.539–2.415) 2.285 (1.780–2.935) | 0.2819 |
| Low PBD 5 High PBD | 1 | 1.266 (0.979–1.638) 1.454 (1.171–1.805) | 1.924 (1.510–2.451) 2.089 (1.702–2.564) | 0.2670 |
| Low WSD 5 High WSD | 1 | 1.434 (1.117–1.839) 1.428 (1.114–1.829) | 2.118 (1.673–2.681) 1.937 (1.531–2.449) | 0.1327 |
| Low RMD 5 High RMD | 1 | 1.533 (1.175–2.001) 1.468 (1.156–1.917) | 2.210 (1.716–2.846) 2.126 (1.672–2.703) | 0.4678 |
| Low exercise 6 High exercise | 1 | 1.287 (0.949–1.746) 1.651 (1.238–2.202) | 1.799 (1.349–2.398) 2.371 (1.803–3.120) | 0.0482 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.; Kang, S. Association between Polygenetic Risk Scores of Low Immunity and Interactions between These Scores and Moderate Fat Intake in a Large Cohort. Nutrients 2021, 13, 2849. https://doi.org/10.3390/nu13082849
Park S, Kang S. Association between Polygenetic Risk Scores of Low Immunity and Interactions between These Scores and Moderate Fat Intake in a Large Cohort. Nutrients. 2021; 13(8):2849. https://doi.org/10.3390/nu13082849
Chicago/Turabian StylePark, Sunmin, and Suna Kang. 2021. "Association between Polygenetic Risk Scores of Low Immunity and Interactions between These Scores and Moderate Fat Intake in a Large Cohort" Nutrients 13, no. 8: 2849. https://doi.org/10.3390/nu13082849
APA StylePark, S., & Kang, S. (2021). Association between Polygenetic Risk Scores of Low Immunity and Interactions between These Scores and Moderate Fat Intake in a Large Cohort. Nutrients, 13(8), 2849. https://doi.org/10.3390/nu13082849








