Human Microbiota Network: Unveiling Potential Crosstalk between the Different Microbiota Ecosystems and Their Role in Health and Disease
Abstract
:1. Introduction
2. Main Microbial Communities in the Human Body
2.1. Skin Microbiota
2.2. Oral Microbiota
2.3. Respiratory Tract Microbiota
2.4. Gut Microbiota
2.5. Genital Microbiota
2.6. Urinary Microbiota
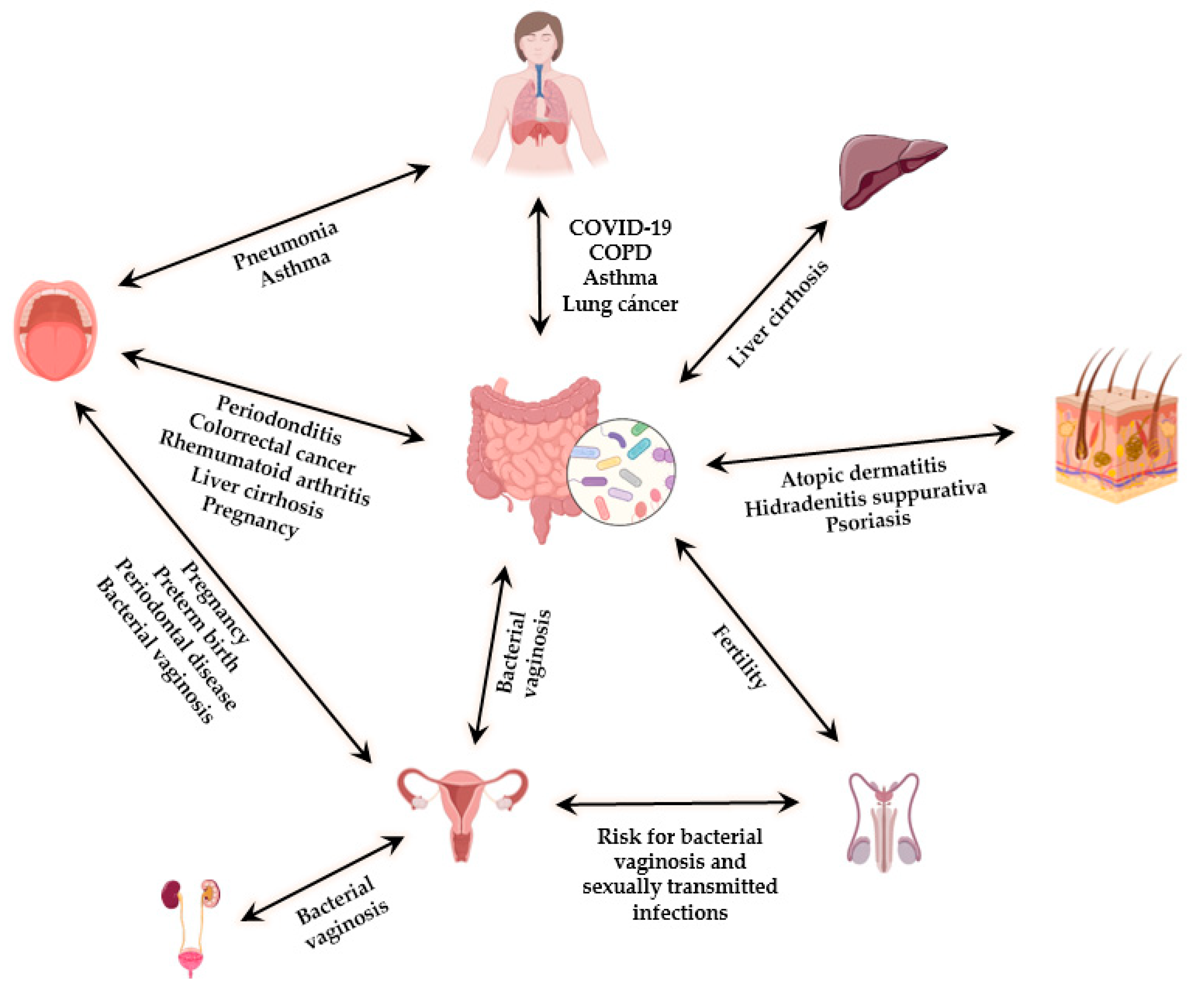
3. The Interplay between the Different Microbiotas
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lloyd-Price, J.; Abu-Ali, G.; Huttenhower, C. The healthy human microbiome. Genome Med. 2016, 8, 51. [Google Scholar] [CrossRef] [Green Version]
- Lederberg, B.J.; McCray, A.T. ‘Ome Sweet’ Omics—A Genealogical Treasury of Words. Scientist 2001, 15, 8. [Google Scholar]
- Villanueva-Millan, M.J.; Perez-Matute, P.; Oteo, J.A. Gut microbiota: A key player in health and disease. A review focused on obesity. J. Physiol. Biochem. 2015, 71, 509–525. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D. Human gut microbiome: Hopes, threats and promises. Gut 2018, 67, 1716–1725. [Google Scholar] [CrossRef] [PubMed]
- Vallianou, N.G.; Stratigou, T.; Tsagarakis, S. Metformin and gut microbiota: Their interactions and their impact on diabetes. Hormones 2019, 18, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davenport, E.R.; Sanders, J.G.; Song, S.J.; Amato, K.R.; Clark, A.G.; Knight, R. The human microbiome in evolution. BMC Biol. 2017, 15, 127. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Batto, J.; Bertalan, M.; Borruel, N.; Casellas, F.; Costea, P.I.; Hildebrand, F.; et al. Enterotypes of the human gut microbiome Manimozhiyan. Nature 2013, 3, 174–180. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Rajilić-Stojanović, M.; De Vos, W.M. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 2008, 57, 1605–1615. [Google Scholar] [CrossRef]
- Pascale, A.; Marchesi, N.; Marelli, C.; Coppola, A.; Luzi, L.; Govoni, S.; Giustina, A.; Gazzaruso, C. Microbiota and metabolic diseases. Endocrine 2018, 61, 357–371. [Google Scholar] [CrossRef]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, K.; Kassis, A.; Major, G.; Chou, C.J. Is the Gut Microbiota a New Factor Contributing to Obesity and Its Metabolic Disorders? J. Obes. 2012, 2012, 879151. [Google Scholar] [CrossRef] [Green Version]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; Fitzgerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [Green Version]
- de Goffau, M.C.; Lager, S.; Sovio, U.; Gaccioli, F.; Cook, E.; Peacock, S.J.; Parkhill, J.; Charnock-Jones, D.S.; Smith, G.C.S. Human placenta has no microbiome but can contain potential pathogens. Nature 2019, 572, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [Green Version]
- Bergström, A.; Skov, T.H.; Bahl, M.I.; Roager, H.M.; Christensen, L.B.; Ejlerskov, K.T.; Mølgaard, C.; Michaelsen, K.F.; Licht, T.R. Establishment of Intestinal Microbiota during Early Life: A Longitudinal, Explorative Study of a Large Cohort of Danish Infants Anders. Appl. Environ. Microbiol. 2014, 80, 2889–2900. [Google Scholar] [CrossRef] [Green Version]
- Rautava, S.; Luoto, R.; Salminen, S.; Isolauri, E. Microbial contact during pregnancy, intestinal colonization and human disease. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 565–576. [Google Scholar] [CrossRef]
- Wallace, J.G.; Gohir, W.; Sloboda, D.M. The impact of early life gut colonization on metabolic and obesogenic outcomes: What have animal models shown us? J. Dev. Orig. Health Dis. 2016, 7, 15–24. [Google Scholar] [CrossRef]
- Munyaka, P.M.; Khafipour, E.; Ghia, J.-E. External influence of early childhood establishment of gut microbiota and subsequent health implications. Front. Pediatr. 2014, 2, 109. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Zhang, M.; Wang, S.; Han, R.; Cao, Y.; Hua, W. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010, 4, 232–241. [Google Scholar] [CrossRef] [Green Version]
- Savage, N. Drugs and the microbiome can change each other in complex and little-understood ways. Nature 2020, 577, S10–S11. [Google Scholar] [CrossRef] [Green Version]
- Sekirov, I.; Russel, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut Microbiota in Health and Disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paun, A.; Yau, C.; Danska, J.S. The Influence of the Microbiome on Type 1 Diabetes. J. Immunol. 2017, 198, 590–595. [Google Scholar] [CrossRef] [PubMed]
- Codella, R.; Luzi, L.; Terruzzi, I. Exercise has the guts: How physical activity may positively modulate gut microbiota in chronic and immune-based diseases. Dig. Liver Dis. 2018, 50, 331–341. [Google Scholar] [CrossRef] [Green Version]
- Petriz, B.A.; Castro, A.P.; Almeida, J.A.; Gomes, C.P.; Fernandes, G.R.; Kruger, R.H.; Pereira, R.W.; Franco, O.L. Exercise induction of gut microbiota modifications in obese, non-obese and hypertensive rats. BMC Genom. 2014, 15, 511. [Google Scholar] [CrossRef] [Green Version]
- Karl, P.J.; Hatch, A.M.; Arcidiacono, S.M.; Pearce, S.C.; Pantoja-Feliciano, I.G.; Doherty, L.A.; Soares, J.W. Effects of Psychological, Environmental and Physical Stressors on the Gut Microbiota. Front. Microbiol. 2018, 9, 2013. [Google Scholar] [CrossRef] [Green Version]
- Smith, R.P.; Easson, C.; Lyle, S.M.; Kapoor, R.; Donnelly, C.P.; Davidson, E.J.; Parikh, E.; Lopez, J.V.; Tartar, J.L. Gut microbiome diversity is associated with sleep physiology in humans. PLoS ONE 2019, 14, e0222394. [Google Scholar] [CrossRef]
- Paulose, J.K.; Wright, J.M.; Patel, A.G.; Cassone, V.M. Human Gut Bacteria Are Sensitive to Melatonin and Express Endogenous Circadian Rhythmicity. PLoS ONE 2016, 11, e0146643. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-Bacterial Mutualism in the Human Intestine. Science 2005, 25, 1915–1920. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Indias, I.; Cardona, F.; Tinahones, F.J.; Queipo-Ortuño, I.M. Impact of the gut microbiota on the development of obesity and type 2 diabetes mellitus. Front. Microbiol. 2014, 5, 190. [Google Scholar] [CrossRef] [Green Version]
- Mowat, A.M.; Agace, W.W. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 2014, 14, 667–685. [Google Scholar] [CrossRef]
- The NIH HMP Working Group. The NIH Human Microbiome Project. Genome Res. 2009, 19, 2317–2323. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef] [PubMed]
- Ritari, J.; Salojärvi, J.; Lahti, L.; de Vos, W.M. Improved taxonomic assignment of human intestinal 16S rRNA sequences by a dedicated reference database. BMC Genom. 2015, 16, 1056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proctor, L.M.; Creasy, H.H.; Fettweis, J.M.; Lloyd-Price, J.; Mahurkar, A.; Zhou, W.; Buck, G.A.; Snyder, M.P.; Strauss, J.F.; Weinstock, G.M.; et al. The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.L.; Xu, S.Y.; Ren, Z.G.; Tao, L.; Jiang, J.W.; Zheng, S. Sen Application of metagenomics in the human gut microbiome. World J. Gastroenterol. 2015, 21, 803–814. [Google Scholar] [CrossRef]
- Morgan, X.C.; Huttenhower, C. Meta’omic Analytic Techniques for Studying the Intestinal Microbiome. Gastroenterology 2014, 146, 1437–1448. [Google Scholar] [CrossRef]
- Holling, C. Resilience and stability of ecological systems. Annu. Revis. Ecol. Evol. Syst. 1973, 4, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Backhed, F.; Fraser, C.M.; Ringel, Y.; Sanders, M.E.; Sartor, R.B.; Sherman, P.M.; Versalovic, J.; Young, V.; Finlay, B.B. Defining a Healthy Human Gut Microbiome: Current Concepts, Future Directions, and Clinical Applications. Cell Host Microbe 2012, 12, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Cabello-Olmo, M.; Araña, M.; Radichev, I.; Smith, P.; Huarte, E.; Barajas, M. New Insights into Immunotherapy Strategies for Treating Autoimmune Diabetes. Int. J. Mol. Sci. 2019, 20, 4789. [Google Scholar] [CrossRef] [Green Version]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nat. Publ. Gr. 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Chan, Y.K.; Estaki, M.; Gibson, D.L. Clinical Consequences of Diet-Induced Dysbiosis. Ann. Nutr. Metab. 2013, 63, 28–40. [Google Scholar] [CrossRef]
- Macia, L.; Thorburn, A.N.; Binge, L.C.; Marino, E.; Rogers, K.E.; Maslowski, K.M.; Vieira, A.T.; Kranich, J.; Mackay, C.R. Microbial influences on epithelial integrity and immune function as a basis for inflammatory diseases. Immunol. Rev. 2012, 245, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Ticinesi, A.; Tana, C.; Nouvenne, A. The intestinal microbiome and its relevance for functionality in older persons. Curr. Opin. Clin. Nutr. Metab. Care 2019, 22, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhou, J.M. The microbiota–gut–brain axis and its potential therapeutic role in autism spectrum disorder. Neuroscience 2016, 324, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Dhar, D.; Mohanty, A. Gut microbiota and Covid-19-possible link and implications. Virus Res. 2020, 285, 198018. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, J.; Li, F.; Shi, Y. Main Clinical Features of COVID-19 and Potential Prognostic and Therapeutic Value of the Microbiota in SARS-CoV-2 Infections. Front. Microbiol. 2020, 11, 1302. [Google Scholar] [CrossRef] [PubMed]
- Brubaker, L.; Wolfe, A.J. The Female Urinary Microbiota/Microbiome: Clinical and Research Implications. Rambam Maimonides Med. J. 2017, 8, e0015. [Google Scholar] [CrossRef]
- Mehta, S.D.; Zhao, D.; Green, S.J.; Agingu, W.; Otieno, F.; Bhaumik, R.; Bhaumik, D.; Bailey, R.C. The Microbiome Composition of a Man’s Penis Predicts Incident Bacterial Vaginosis in His Female Sex Partner with High Accuracy. Front. Cell. Infect. Microbiol. 2020, 10, 433. [Google Scholar] [CrossRef]
- Egert, M.; Simmering, R. The Microbiota of the Human Skin. In Microbiota of the Human Body: Implications in Health and Disease; Springer: Berlin/Heidelberg, Germany, 2016; Volume 902, pp. 61–81. ISBN 9783319312460. [Google Scholar]
- Lunjani, N.; Hlela, C.; O’Mahony, L. Microbiome and skin biology. Curr. Opin. Allergy Clin. Immunol. 2019, 19, 328–333. [Google Scholar] [CrossRef]
- Tlaskalova-Hogenova, H.; Stepankova, R.; Hudcovic, T.; Tuckova, L.; Cukrowska, B.; Lodinova-Zadnikova, R. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases and chronic inflammatory and autoimmune diseases. Immunol. Lett. 2004, 97–108. [Google Scholar] [CrossRef]
- Fournière, M.; Latire, T.; Souak, D.; Feuilloley, M.G.J.; Bedoux, G. Staphylococcus epidermidis and cutibacterium acnes: Two major sentinels of skin microbiota and the influence of cosmetics. Microorganisms 2020, 8, 1752. [Google Scholar] [CrossRef]
- Schommer, N.N.; Gallo, R.L. Structure and function of the human skin microbiome Nina. Physiol. Behav. 2013, 21, 660–668. [Google Scholar] [CrossRef]
- Blaser, M.J.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Estrada, I.; Gao, Z.; Clemente, J.C.; Costello, E.K.; Knight, R. Distinct cutaneous bacterial assemblages in a sampling of South American Amerindians and US residents. ISME J. 2013, 7, 85–95. [Google Scholar] [CrossRef]
- Chen, Y.E.; Fischbach, M.A.; Belkaid, Y. Skin microbiota-host interactions. Nature 2018, 553, 427–436. [Google Scholar] [CrossRef]
- Sanford, J.A.; Gallo, R.L. Functions of the skin microbiota in health and disease. Semin. Immunol. 2013, 25, 370–377. [Google Scholar] [CrossRef] [Green Version]
- Drago, L.; Zuccotti, G.V.; Romanò, C.L.; Goswami, K.; Villafañe, J.H.; Mattina, R.; Parvizi, J. Oral–Gut Microbiota and Arthritis: Is There an Evidence-Based Axis? J. Clin. Med. 2019, 8, 1753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, T.; Glatz, M.; Horiuchi, K.; Kawasaki, H.; Akiyama, H.; Kaplan, D.H.; Kong, H.H.; Amagai, M.; Nagao, K. Dysbiosis and Staphyloccus aureus Colonization Drives Inflammation in Atopic Dermatitis. Immunity 2015, 42, 756–766. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wang, X.; Li, H.; Ni, C.; Du, Z.; Yan, F. Human oral microbiota and its modulation for oral health. Biomed. Pharmacother. 2018, 99, 883–893. [Google Scholar] [CrossRef]
- Koliarakis, I.; Messaritakis, I.; Nikolouzakis, T.K.; Hamilos, G.; Souglakos, J.; Tsiaoussis, J. Oral bacteria and intestinal dysbiosis in colorectal cancer. Int. J. Mol. Sci. 2019, 20, 4146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siqueira, J.F.; Rôças, I.N. The oral microbiota in health and disease: An overview of molecular findings. Methods Mol. Biol. 2017, 1537, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Huse, S.M.; Ye, Y.; Zhou, Y.; Fodor, A.A. A Core Human Microbiome as Viewed through 16S rRNA Sequence Clusters. PLoS ONE 2012, 7, e034242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamont, R.J.; Koo, H.; Hajishengallis, G. The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Microbiol. 2018, 16, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.; Segal, L.N. Looking higher: Is It Prime Time for the Oral–Lung Axis in HIV-related Lung Disease? Am. J. Respir. Crit. Care Med. 2020, 201, 402–403. [Google Scholar] [CrossRef]
- Lanaspa, M.; Bassat, Q.; Medeiros, M.M.; Muñoz-Almagro, C. Respiratory microbiota and lower respiratory tract disease. Expert Rev. Anti-Infect. Ther. 2017, 15, 703–711. [Google Scholar] [CrossRef]
- Dickson, R.P.; Erb-Downward, J.R.; Martinez, F.J.; Huffnagle, G.B. The Microbiome and the Respiratory Tract. Annu. Rev. Physiol. 2016, 78, 481–504. [Google Scholar] [CrossRef] [Green Version]
- Edouard, S.; Million, M.; Bachar, D.; Dubourg, G.; Michelle, C.; Ninove, L.; Charrel, R.; Raoult, D. The nasopharyngeal microbiota in patients with viral respiratory tract infections is enriched in bacterial pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1725–1733. [Google Scholar] [CrossRef]
- Man, W.H.; De Steenhuijsen Piters, W.A.A.; Bogaert, D. The microbiota of the respiratory tract: Gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017, 15, 259–270. [Google Scholar] [CrossRef]
- Huffnagle, G.; Dickson, R.; Lukacs, N. The respiratory tract microbiome and lung inflammation: A two-way street. Physiol. Behav. 2017, 10, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Marques, T.M.; Cryan, J.F.; Shanahan, F.; Fitzgerald, G.F.; Ross, R.P.; Dinan, T.G.; Stanton, C. Gut microbiota modulation and implications for host health: Dietary strategies to influence the gut-brain axis. Innov. Food Sci. Emerg. Technol. 2014, 22, 239–247. [Google Scholar] [CrossRef]
- Walter, J.; Ley, R. The human gut microbiome: Ecology and recent evolutionary changes. Annu. Rev. Microbiol. 2011, 65, 411–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conlon, M.A.; Bird, A.R. The impact of diet and lifestyle on gut microbiota and human health. Nutrients 2015, 7, 17–44. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Howitt, M.R.; Garrett, W.S. Exploring host—Microbiota interactions in animal models and humans. Genes Dev. 2013, 27, 701–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cotillard, A.; Kennedy, S.P.; Kong, L.C.; Prifti, E.; Pons, N.; Le Chatelier, E.; Almeida, M.; Quinquis, B.; Levenez, F.; Galleron, N.; et al. Dietary intervention impact on gut microbial gene richness. Nature 2013, 500, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Kolodziejczyk, A.A.; Zheng, D.; Elinav, E. Diet–microbiota interactions and personalized nutrition. Nat. Rev. Microbiol. 2019, 17, 742–753. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.X.; Wang, Y.P. Gut microbiota-brain axis. Chin. Med. J. 2016, 129, 2373–2380. [Google Scholar] [CrossRef]
- Cryan, J.F.; O’riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The microbiota-gut-brain axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef]
- Quigley, E.M.M. Microbiota-Brain-Gut Axis and Neurodegenerative Diseases. Curr. Neurol. Neurosci. Rep. 2017, 17, 94. [Google Scholar] [CrossRef] [PubMed]
- Fox, C.; Eichelberger, K. Maternal microbiome and pregnancy outcomes. Fertil. Steril. 2015, 104, 1358–1363. [Google Scholar] [CrossRef] [Green Version]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.K.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108, 4680–4687. [Google Scholar] [CrossRef] [Green Version]
- Van De Wijgert, J.H.H.M.; Borgdorff, H.; Verhelst, R.; Crucitti, T.; Francis, S.; Verstraelen, H.; Jespers, V. The vaginal microbiota: What have we learned after a decade of molecular characterization? PLoS ONE 2014, 9, e105998. [Google Scholar] [CrossRef] [Green Version]
- Lopes dos Santos Santiago, G.; Cools, P.; Verstraelen, H.; Trog, M.; Missine, G.; El Aila, N.; Verhelst, R.; Tency, I.; Claeys, G.; Temmerman, M.; et al. Longitudinal Study of the Dynamics of Vaginal Microflora during Two Consecutive Menstrual Cycles. PLoS ONE 2011, 6, e028180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stapleton, A.E. The vaginal microbiota and urinary tract infection. Urin. Tract Infect. Mol. Pathog. Clin. Manag. 2016, 79–86. [Google Scholar] [CrossRef] [Green Version]
- Prince, A.L.; Chu, D.M.; Seferovic, M.D.; Antony, K.M.; Ma, J.; Aagaard, K.M. The Perinatal Microbiome and Pregnancy: Moving Beyond the Vaginal Microbiome. Cold Spring Harb. Perspect. Med. 2015, 5, a023051. [Google Scholar] [CrossRef] [PubMed]
- Marangoni, A.; Laghi, L.; Zagonari, S.; Patuelli, G.; Zhu, C.; Foschi, C.; Morselli, S.; Pedna, M.F.; Sambri, V. New Insights into Vaginal Environment during Pregnancy. Front. Mol. Biosci. 2021, 8, 656844. [Google Scholar] [CrossRef] [PubMed]
- Younes, J.A.; Lievens, E.; Hummelen, R.; van der Westen, R.; Reid, G.; Petrova, M.I. Women and Their Microbes: The Unexpected Friendship. Trends Microbiol. 2018, 26, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Machado, D.; Castro, J.; Palmeira-de-Oliveira, A.; Martinez-de-Oliveira, J.; Cerca, N. Bacterial vaginosis biofilms: Challenges to current therapies and emerging solutions. Front. Microbiol. 2016, 6, 1528. [Google Scholar] [CrossRef] [Green Version]
- Marrazzo, J.M.; Fiedler, T.L.; Srinivasan, S.; Thomas, K.K.; Liu, C.; Ko, D.; Xie, H.; Saracino, M.; Fredricks, D.N. Extravaginal Reservoirs of Vaginal Bacteria as Risk Factors for Incident Bacterial Vaginosis. J. Infect. Dis. 2012, 205, 1580–1588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendling, W.; Palmeira-de-Oliveira, A.; Biber, S.; Prasauskas, V. An update on the role of Atopobium vaginae in bacterial vaginosis: What to consider when choosing a treatment? A mini review. Arch. Gynecol. Obstet. 2019, 300, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Balle, C.; Esra, R.; Havyarimana, E.; Jaumdally, S.Z.; Lennard, K.; Konstantinus, I.N.; Barnabas, S.L.; Happel, A.U.; Gill, K.; Pidwell, T.; et al. Relationship between the oral and vaginal microbiota of South African adolescents with high prevalence of bacterial vaginosis. Microorganisms 2020, 8, 1004. [Google Scholar] [CrossRef] [PubMed]
- Machado, A.; Cerca, N. Influence of Biofilm Formation by Gardnerella vaginalis and Other Anaerobes on Bacterial Vaginosis. J. Infect. Dis. 2015, 212, 1856–1861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chee, W.J.Y.; Chew, S.Y.; Than, L.T.L. Vaginal microbiota and the potential of Lactobacillus derivatives in maintaining vaginal health. Microb. Cell Fact. 2020, 19, 203. [Google Scholar] [CrossRef] [PubMed]
- D’Enfert, C.; Kaune, A.K.; Alaban, L.R.; Chakraborty, S.; Cole, N.; Delavy, M.; Kosmala, D.; Marsaux, B.; Fróis-Martins, R.; Morelli, M.; et al. The impact of the Fungus-Host-Microbiota interplay upon Candida albicans infections: Current knowledge and new perspectives. FEMS Microbiol. Rev. 2021, 45, fuaa060. [Google Scholar] [CrossRef]
- Onywera, H.; Williamson, A.L.; Cozzuto, L.; Bonnin, S.; Mbulawa, Z.Z.A.; Coetzee, D.; Ponomarenko, J.; Meiring, T.L. The penile microbiota of Black South African men: Relationship with human papillomavirus and HIV infection. BMC Microbiol. 2020, 20, 78. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.M.; Prodger, J.L.; Tobian, A.A.R.; Abraham, A.G.; Kigozi, G.; Hungate, B.A.; Aziz, M.; Nalugoda, F.; Sariya, S.; Serwadda, D.; et al. Penile anaerobic dysbiosis as a risk factor for HIV infection. MBio 2017, 8, e00996-17. [Google Scholar] [CrossRef] [Green Version]
- Pearce, M.M.; Hilt, E.E.; Rosenfeld, A.B.; Zilliox, M.J.; Thomas-White, K.; Fok, C.; Kliethermes, S.; Schreckenberger, P.C.; Brubaker, L.; Gai, X.; et al. The Female Urinary Microbiome: A Comparison of Women with and without Urgency Urinary Incontinence. MBio 2014, 5, e01283-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Govender, Y.; Gabriel, I.; Minassian, V.; Fichorova, R. The Current Evidence on the Association between the Urinary Microbiome and Urinary Incontinence in Women. Front. Cell. Infect. Microbiol. 2019, 9, 133. [Google Scholar] [CrossRef] [Green Version]
- Nelson, D.E.; van der Pol, B.; Dong, Q.; Revanna, K.V.; Fan, B.; Easwaran, S.; Sodergren, E.; Weinstock, G.M.; Diao, L.; Fortenberry, D.J. Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. PLoS ONE 2010, 5, e014116. [Google Scholar] [CrossRef]
- Dong, Q.; Nelson, D.E.; Toh, E.; Diao, L.; Gao, X.; Fortenberry, J.D.; van Der Pol, B. The microbial communities in male first catch urine are highly similar to those in paired urethral swab specimens. PLoS ONE 2011, 6, e019709. [Google Scholar] [CrossRef] [Green Version]
- Datcu, R. Characterization of the vaginal microflora in health and disease. Dan. Med. J. 2014, 61, 1–24. [Google Scholar]
- Shrestha, E.; White, J.R.; Yu, S.H.; Kulac, I.; Ertunc, O.; De Marzo, A.M.; Yegnasubramanian, S.; Mangold, L.A.; Partin, A.W.; Sfanos, K.S. Profiling the Urinary Microbiome in Men with Positive versus Negative Biopsies for Prostate Cancer. J. Urol. 2018, 199, 161–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundy, S.D.; Sangwan, N.; Parekh, N.V.; Selvam, M.K.P.; Gupta, S.; McCaffrey, P.; Bessoff, K.; Vala, A.; Agarwal, A.; Sabanegh, E.S.; et al. Functional and Taxonomic Dysbiosis of the Gut, Urine, and Semen Microbiomes in Male Infertility [Formula presented]. Eur. Urol. 2021, 79, 826–836. [Google Scholar] [CrossRef] [PubMed]
- Reddel, S.; Del Chierico, F.; Quagliariello, A.; Giancristoforo, S.; Vernocchi, P.; Russo, A.; Fiocchi, A.; Rossi, P.; Putignani, L.; El Hachem, M. Gut microbiota profile in children affected by atopic dermatitis and evaluation of intestinal persistence of a probiotic mixture. Sci. Rep. 2019, 9, 4996. [Google Scholar] [CrossRef] [Green Version]
- Hidalgo-Cantabrana, C.; Gómez, J.; Delgado, S.; Requena-López, S.; Queiro-Silva, R.; Margolles, A.; Coto, E.; Sánchez, B.; Coto-Segura, P. Gut microbiota dysbiosis in a cohort of patients with psoriasis. Br. J. Dermatol. 2019, 181, 1287–1295. [Google Scholar] [CrossRef]
- Li, B.Z.; Zhou, H.Y.; Guo, B.; Chen, W.J.; Tao, J.H.; Cao, N.W.; Chu, X.J.; Meng, X. Dysbiosis of oral microbiota is associated with systemic lupus erythematosus. Arch. Oral Biol. 2020, 113, 104708. [Google Scholar] [CrossRef] [PubMed]
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014, 513, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Hufnagl, K.; Pali-Schöll, I.; Roth-Walter, F.; Jensen-Jarolim, E. Dysbiosis of the gut and lung microbiome has a role in asthma. Semin. Immunopathol. 2020, 42, 75–93. [Google Scholar] [CrossRef] [Green Version]
- Donati Zeppa, S.; Agostini, D.; Piccoli, G.; Stocchi, V.; Sestili, P. Gut Microbiota Status in COVID-19: An Unrecognized Player? Front. Cell. Infect. Microbiol. 2020, 10, 742. [Google Scholar] [CrossRef]
- Walton, G.E.; Gibson, G.R.; Hunter, K.A. Mechanisms linking the human gut microbiome to prophylactic and treatment strategies for COVID-19. Br. J. Nutr. 2021, 126, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, H.; Lin, Y.; Pan, J.; Su, J. The Gut Microbiota and Respiratory Diseases: New Evidence. J. Immunol. Res. 2020, 2020, 2340670. [Google Scholar] [CrossRef] [PubMed]
- Budden, K.F.; Gellatly, S.L.; Wood, D.L.A.; Cooper, M.A.; Morrison, M.; Hugenholtz, P.; Hansbro, P.M. Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 2017, 15, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Li, S.; Wang, N.; Tan, H.Y.; Zhang, Z.; Feng, Y. The Cross-Talk between Gut Microbiota and Lungs in Common Lung Diseases. Front. Microbiol. 2020, 11, 301. [Google Scholar] [CrossRef]
- Bauer, T.M.; Schwacha, H.; Steinbrückner, B.; Brinkmann, F.E.; Ditzen, A.K.; Aponte, J.J.; Pelz, K.; Berger, D.; Kist, M.; Blum, H.E. Small Intestinal Bacterial Overgrowth in Human Cirrhosis Is Associated with Systemic Endotoxemia. Am. J. Gastroenterol. 2002, 97, 2364–2370. [Google Scholar] [CrossRef] [PubMed]
- Gottschick, C.; Deng, Z.L.; Vital, M.; Masur, C.; Abels, C.; Pieper, D.H.; Wagner-Döbler, I. The urinary microbiota of men and women and its changes in women during bacterial vaginosis and antibiotic treatment. Microbiome 2017, 5, 99. [Google Scholar] [CrossRef] [Green Version]
- Komesu, Y.M.; Dinwiddie, D.L.; Richter, H.E.; Lukacz, E.S.; Sung, V.W.; Siddiqui, N.Y.; Zyczynski, H.M.; Ridgeway, B.; Rogers, R.G.; Arya, L.A.; et al. Defining the relationship between vaginal and urinary microbiomes. Am. J. Obs. Gynecol. 2020, 222, 154.e1–154.e10. [Google Scholar] [CrossRef] [PubMed]
- Mazorra-Alonso, M.; Tomás, G.; Soler, J.J. Microbially mediated chemical ecology of animals: A review of its role in conspecific communication, parasitism and predation. Biology 2021, 10, 274. [Google Scholar] [CrossRef]
- Ñahui Palomino, R.A.; Vanpouille, C.; Costantini, P.E.; Margolis, L. Microbiota–host communications: Bacterial extracellular vesicles as a common language. PLoS Pathog. 2021, 17, e1009508. [Google Scholar] [CrossRef]
- Cuesta, C.M.; Guerri, C.; Ureña, J. Role of Microbiota-Derived Extracellular Vesicles in Gut-Brain Communication. Int. J. Mol. Sci. 2021, 22, 4235. [Google Scholar] [CrossRef]
- Tripathi, A.; Debelius, J.; Brenner, D.A.; Karin, M.; Schnabl, B.; Knight, R.; Diego, S.; Jolla, L.; Diego, S.; Jolla, L.; et al. The gut-liver axis and the intersection with the microbiome Anupriya. Nat. Rev. Gastriebterik. Hepatol. 2018, 15, 397–411. [Google Scholar] [CrossRef]
- Morais, L.H.; Schreiber, H.L.; Mazmanian, S.K. The gut microbiota–brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 2021, 19, 241–255. [Google Scholar] [CrossRef]
- De Pessemier, B.; Grine, L.; Debaere, M.; Maes, A.; Paetzold, B.; Callewaert, C. Gut–skin axis: Current knowledge of the interrelationship between microbial dysbiosis and skin conditions. Microorganisms 2021, 9, 353. [Google Scholar] [CrossRef]
- Gaeckle, N.T.; Pragman, A.A.; Pendleton, K.M.; Baldomero, A.K.; Criner, G.J. The oral-lung axis: The impact of oral health on lung health. Respir. Care 2020, 65, 1211–1220. [Google Scholar] [CrossRef]
- Dang, A.T.; Marsland, B.J. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 2019, 12, 843–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szántó, M.; Dózsa, A.; Antal, D.; Szabó, K.; Kemény, L.; Bai, P. Targeting the gut-skin axis—Probiotics as new tools for skin disorder management? Exp. Dermatol. 2019, 28, 1210–1218. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Luo, X.; Tang, J.; Mo, Q.; Zhong, H.; Zhang, H.; Feng, F. A bridge for short-chain fatty acids to affect inflammatory bowel disease, type 1 diabetes, and non-alcoholic fatty liver disease positively: By changing gut barrier. Eur. J. Nutr. 2020, 60, 2317–2330. [Google Scholar] [CrossRef]
- O’Neill, C.A.; Monteleone, G.; McLaughlin, J.T.; Paus, R. The gut-skin axis in health and disease: A paradigm with therapeutic implications. BioEssays 2016, 38, 1167–1176. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Z.; Koo, H.; Chen, Q.; Zhou, X.; Liu, Y.; Simon-Soro, A. Potential implications of SARS-CoV-2 oral infection in the host microbiota. J. Oral Microbiol. 2021, 13, 1853451. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients with COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955.e8. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.Y.; Zhang, F.; Liu, Q.; Li, A.Y.L.; Chung, A.C.K.; Cheung, C.P.; Tso, E.Y.K.; Fung, K.S.C.; et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Lansbury, L.; Lim, B.; Baskaran, V.; Lim, W.S. Co-infections in people with COVID-19: A systematic review and meta-analysis. J. Infect. 2020, 81, 266–275. [Google Scholar] [CrossRef]
- Krone, C.L.; van de Groep, K.; Trzciński, K.; Sanders, E.A.M.; Bogaert, D. Immunosenescence and pneumococcal disease: An imbalance in host-pathogen interactions. Lancet Respir. Med. 2014, 2, 141–153. [Google Scholar] [CrossRef]
- Pace, C.C.; McCullough, G.H. The Association Between Oral Microorgansims and Aspiration Pneumonia in the Institutionalized Elderly: Review and Recommendations. Dysphagia 2010, 25, 307–322. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Chen, F.; Huang, Z.; Ma, L.; Chen, L.; Pan, Y.; Xu, J.; Kim, S.; Kinane, D.; Koo, H.; et al. Meeting report: A close look at oral biofilms and microbiomes. Int. J. Oral Sci. 2018, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Dumas, A.; Bernard, L.; Poquet, Y.; Lugo-Villarino, G.; Neyrolles, O. The role of the lung microbiota and the gut–lung axis in respiratory infectious diseases. Cell. Microbiol. 2018, 20, e12966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pu, C.Y.; Seshadri, M.; Manuballa, S.; Yendamuri, S. The Oral Microbiome and Lung Diseases. Curr. Oral Health Rep. 2020, 7, 79–86. [Google Scholar] [CrossRef]
- Ray, K. The oral–gut axis in IBD. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 532. [Google Scholar] [CrossRef] [PubMed]
- Petricevic, L.; Domig, K.J.; Nierscher, F.J.; Krondorfer, I.; Janitschek, C.; Kneifel, W.; Kiss, H. Characterisation of the oral, vaginal and rectal Lactobacillus flora in healthy pregnant and postmenopausal women. Eur. J. Obstet. Gynecol. Reprod. Biol. 2012, 160, 93–99. [Google Scholar] [CrossRef] [PubMed]
| Microbiota Niche | Predominant Taxonomic Groups | Associated-Diseases or Conditions and Characteristic Microbiota Composition (If Available) | Potential Communication with Other Microbiota Niches/Organs |
|---|---|---|---|
| Skin microbiota | Actinobacteria, Firmicutes, and Proteobacteria phyla [50]. Corynebacterium, Propionibacterium, Staphylococcus, and Streptococcus genera [50,51]. | ||
| Oral microbiota | Actinobacteria, Bacteroidetes, Firmicutes, Proteobacterias, and Synergistetes phyla [62] Fusobacterium, Gemmela, Veillonella, Streptococcus and Granulicatella genera [63] Saliva: Streptococcus, Veillonella, and Prevotella spp. [60] Soft tissues: Streptococcus salivarus, Rothia, and Eubacterium spp. [60]. Tooth: Corynebacterium, Actinomyces, Spirochaetes, Fusobacteria, Actinobacteria, Proteobacteria, and Bacteroidetes spp. [60] |
|
|
| Respiratory tract microbiota | Bacteroidetes, Actinobacteria, and Firmicutes [109] Streptococcus, Haemophilus, Moraxella, Staphylococcus, and Veillonella spp. [68]. Upper respiratory tract (nasal cavity, nasopharynx, and oropharynx): Staphylococcus, Propionibacterium, Corynebacterium, Streptococuus, Moraxella, Haemophillus, Prevotella, and Veillonella spp. [68]. Lower respiratory tract (trachea and lung’s bronchial trees): Prevotella, Veillonella, Streptococcus, and Tropheryma spp. [68]. | ||
| Gut microbiota | Bacteroidetes and Firmicutes phyla [10]. Bifidobacterium, Lactobacillus, Bacteroides, Clostridium, Escherichia, Streptococcus and Ruminococcus spp. [74]. |
| |
| Vaginal microbiota | Lactobacillus spp., (L. crispatus, L. iners, L. gasseri, and L. jensenii), Atopobium, Dialister, Gardnerella, Megasphaera, Prevotella, Peptoniphilus, Veinovella, Lachnospiraceae, Streptococcus, Staphylococcus and Gemella [82,83]. | ||
| Penile microbiota | Corynebacteriaceae, Prevotellaceae, Clostridiales, Porphyromonadaceae, and Staphylococcaceae families [96]. Corynebacterium spp. [96] |
| |
| Female urinary microbiota | Lactobacillus spp. [48]. |
| |
| Male urinary microbiota | Firmicutes, Actinobacteria, Fusobacteria, Proteobacteria, and Bacteroidetes [100]. Lactobacillus (L. iners), Aerococcus, Anaerococcus, Prevotella, Gemella, Veillonella, Sneathia, Corynebacterium, Staphylococcus and Streptococcus spp. [100,103]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez, J.E.; Vargas, A.; Pérez-Sánchez, T.; Encío, I.J.; Cabello-Olmo, M.; Barajas, M. Human Microbiota Network: Unveiling Potential Crosstalk between the Different Microbiota Ecosystems and Their Role in Health and Disease. Nutrients 2021, 13, 2905. https://doi.org/10.3390/nu13092905
Martínez JE, Vargas A, Pérez-Sánchez T, Encío IJ, Cabello-Olmo M, Barajas M. Human Microbiota Network: Unveiling Potential Crosstalk between the Different Microbiota Ecosystems and Their Role in Health and Disease. Nutrients. 2021; 13(9):2905. https://doi.org/10.3390/nu13092905
Chicago/Turabian StyleMartínez, Jose E., Augusto Vargas, Tania Pérez-Sánchez, Ignacio J. Encío, Miriam Cabello-Olmo, and Miguel Barajas. 2021. "Human Microbiota Network: Unveiling Potential Crosstalk between the Different Microbiota Ecosystems and Their Role in Health and Disease" Nutrients 13, no. 9: 2905. https://doi.org/10.3390/nu13092905
APA StyleMartínez, J. E., Vargas, A., Pérez-Sánchez, T., Encío, I. J., Cabello-Olmo, M., & Barajas, M. (2021). Human Microbiota Network: Unveiling Potential Crosstalk between the Different Microbiota Ecosystems and Their Role in Health and Disease. Nutrients, 13(9), 2905. https://doi.org/10.3390/nu13092905






