Lower Dietary Intake of Plant Protein Is Associated with Genetic Risk of Diabetes-Related Traits in Urban Asian Indian Adults
Abstract
:1. Introduction
2. Methods
2.1. Study Participants
2.2. Anthropometric and Biochemical Measurements
2.3. Dietary Assessments
2.4. SNP Selection and GRS Construction
2.5. Genotyping
2.6. Statistical Analysis
3. Results
3.1. Characteristics of Study Participants
3.2. Association between Metabolic GRS and Metabolic Traits
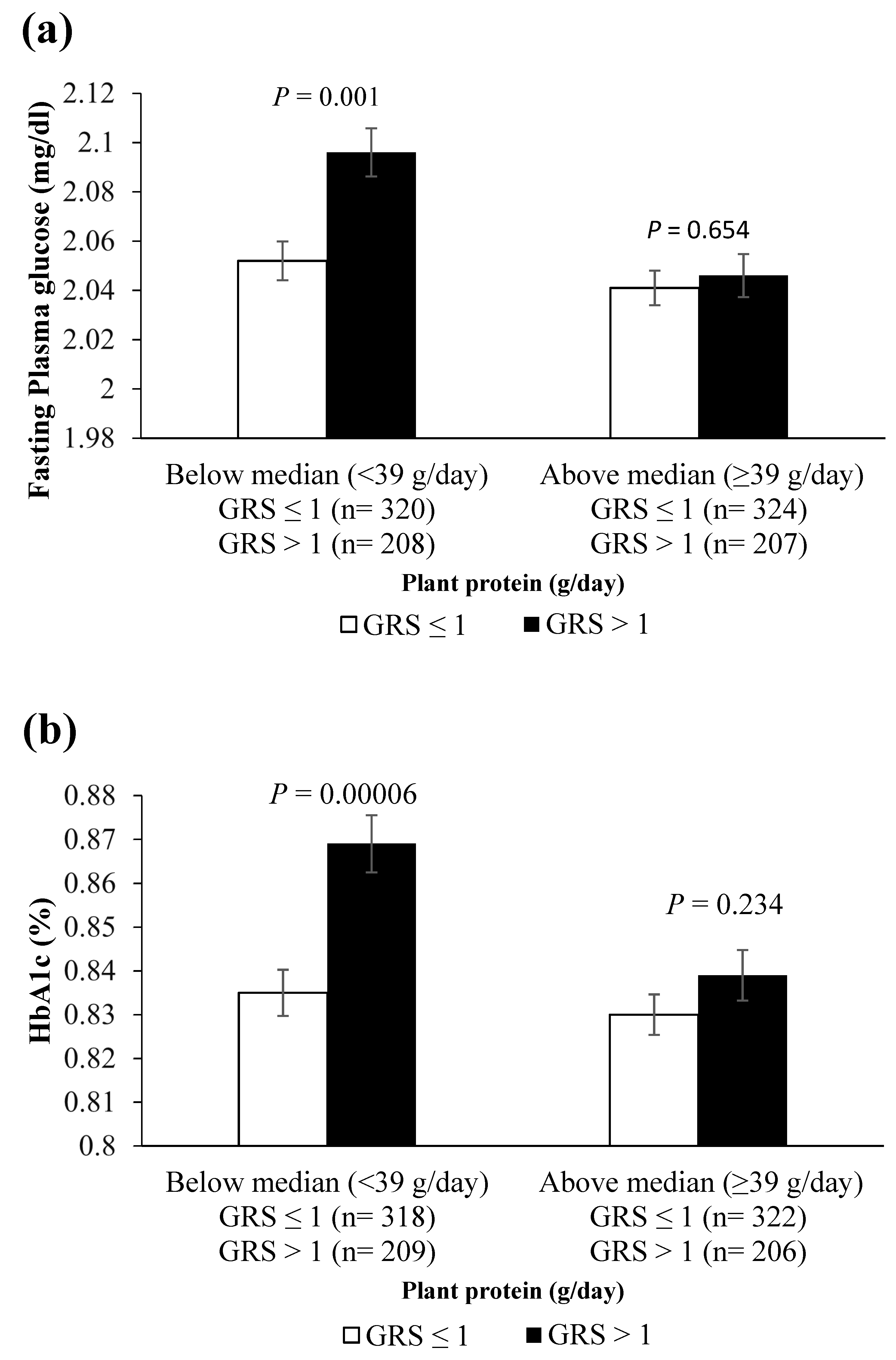
3.3. Interaction of 7-SNP and 3-SNP GRSs with Dietary Factors on Metabolic Traits
3.4. Sensitivity Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Khan, N.A.; Wang, H.; Anand, S.; Jin, Y.; Campbell, N.R.; Pilote, L.; Quan, H. Ethnicity and sex affect diabetes incidence and outcomes. Diabetes Care 2011, 34, 96–101. [Google Scholar] [CrossRef] [Green Version]
- Mohan, V.; Mathur, P.; Deepa, R.; Deepa, M.; Shukla, D.K.; Menon, G.R.; Anand, K.; Desai, N.G.; Joshi, P.P.; Mahanta, J.; et al. Urban rural differences in prevalence of self-reported diabetes in India—The WHO-ICMR Indian NCD risk factor surveillance. Diabetes Res. Clin. Pract. 2008, 80, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.C.; Malik, V.; Jia, W.; Kadowaki, T.; Yajnik, C.S.; Yoon, K.H.; Hu, F.B. Diabetes in Asia: Epidemiology, risk factors, and pathophysiology. JAMA 2009, 301, 2129–2140. [Google Scholar] [CrossRef] [PubMed]
- Mohan, V. Why are Indians more prone to diabetes? J. Assoc. Physicians India 2004, 52, 468–474. [Google Scholar]
- Ramachandran, A.; Ma, R.C.; Snehalatha, C. Diabetes in Asia. Lancet 2010, 375, 408–418. [Google Scholar] [CrossRef]
- Diabetes Atlas 9th Edition. Available online: https://www.diabetesatlas.org/en/ (accessed on 16 January 2021).
- Tandon, N.; Anjana, R.M.; Mohan, V.; Kaur, T.; Afshin, A.; Ong, K.; Mukhopadhyay, S.; Thomas, N.; Bhatia, E.; Krishnan, A.; et al. The increasing burden of diabetes and variations among the states of India: The Global Burden of Disease Study 1990–2016. Lancet Glob. Health 2018, 6, e1352–e1362. [Google Scholar] [CrossRef] [Green Version]
- Bodhini, D.; Gaal, S.; Shatwan, I.; Ramya, K.; Ellahi, B.; Surendran, S.; Sudha, V.; Anjana, M.R.; Mohan, V.; Lovegrove, J.A.; et al. Interaction between TCF7L2 polymorphism and dietary fat intake on high density lipoprotein cholesterol. PLoS ONE 2017, 12, e0188382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vimaleswaran, K.S.; Bodhini, D.; Lakshmipriya, N.; Ramya, K.; Anjana, R.M.; Sudha, V.; Lovegrove, J.A.; Kinra, S.; Mohan, V.; Radha, V. Erratum to: Interaction between FTO gene variants and lifestyle factors on metabolic traits in an Asian Indian population. Nutr. Metab. 2016, 13, 41. [Google Scholar] [CrossRef] [Green Version]
- Gassasse, Z.; Smith, D.; Finer, S.; Gallo, V. Association between urbanisation and type 2 diabetes: An ecological study. BMJ Glob. Health 2017, 2, e000473. [Google Scholar] [CrossRef] [Green Version]
- Ramachandran, A.; Snehalatha, C.; Latha, E.; Manoharan, M.; Vijay, V. Impacts of urbanisation on the lifestyle and on the prevalence of diabetes in native Asian Indian population. Diabetes Res. Clin. Pract. 1999, 44, 207–213. [Google Scholar] [CrossRef]
- Rampal, P. An Analysis of Protein Consumption in India Through Plant and Animal Sources. Food Nutr. Bull. 2018, 39, 564–580. [Google Scholar] [CrossRef] [PubMed]
- Sluijs, I.; Beulens, J.W.; van der, A.D.; Spijkerman, A.M.; Grobbee, D.E.; van der Schouw, Y.T. Dietary intake of total, animal, and vegetable protein and risk of type 2 diabetes in the European Prospective Investigation into Cancer and Nutrition (EPIC)-NL study. Diabetes Care 2010, 33, 43–48. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Manson, J.E.; Buring, J.E.; Liu, S. A prospective study of red meat consumption and type 2 diabetes in middle-aged and elderly women: The women’s health study. Diabetes Care 2004, 27, 2108–2115. [Google Scholar] [CrossRef] [Green Version]
- Ericson, U.; Sonestedt, E.; Gullberg, B.; Hellstrand, S.; Hindy, G.; Wirfält, E.; Orho-Melander, M. High intakes of protein and processed meat associate with increased incidence of type 2 diabetes. Br. J. Nutr. 2013, 109, 1143–1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Nielen, M.; Feskens, E.J.; Mensink, M.; Sluijs, I.; Molina, E.; Amiano, P.; Ardanaz, E.; Balkau, B.; Beulens, J.W.; Boeing, H.; et al. Dietary protein intake and incidence of type 2 diabetes in Europe: The EPIC-InterAct Case-Cohort Study. Diabetes Care 2014, 37, 1854–1862. [Google Scholar] [CrossRef] [Green Version]
- Pounis, G.D.; Tyrovolas, S.; Antonopoulou, M.; Zeimbekis, A.; Anastasiou, F.; Bountztiouka, V.; Metallinos, G.; Gotsis, E.; Lioliou, E.; Polychronopoulos, E.; et al. Long-term animal-protein consumption is associated with an increased prevalence of diabetes among the elderly: The Mediterranean Islands (MEDIS) study. Diabetes Metab. 2010, 36, 484–490. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, N.; Attia, J.; Oldmeadow, C.; Scott, R.J.; Holliday, E.G. The architecture of risk for type 2 diabetes: Understanding Asia in the context of global findings. Int. J. Endocrinol. 2014, 2014, 593982. [Google Scholar] [CrossRef]
- Raj, S.M.; Pei, A.; Foll, M.; Schlamp, F.; Excoffier, L.; Fuller, D.Q.; Kivisild, T.; Clark, A.G. Reconstruction of nine thousand years of agriculture-based diet and impact on human genetic diversity in Asia. bioRxiv 2019, 747709. [Google Scholar] [CrossRef]
- Sanghera, D.K.; Ortega, L.; Han, S.; Singh, J.; Ralhan, S.K.; Wander, G.S.; Mehra, N.K.; Mulvihill, J.J.; Ferrell, R.E.; Nath, S.K.; et al. Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant risk. BMC Med. Genet. 2008, 9, 59. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, G.; Tabassum, R.; Mahajan, A.; Dwivedi, O.P.; Mahendran, Y.; Kaur, I.; Nigam, S.; Dubey, H.; Varma, B.; Madhu, S.V.; et al. Common variants of FTO and the risk of obesity and type 2 diabetes in Indians. J. Hum. Genet. 2011, 56, 720–726. [Google Scholar] [CrossRef] [Green Version]
- Ramya, K.; Radha, V.; Ghosh, S.; Majumder, P.P.; Mohan, V. Genetic variations in the FTO gene are associated with type 2 diabetes and obesity in south Indians (CURES-79). Diabetes Technol. Ther. 2011, 13, 33–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yajnik, C.S.; Janipalli, C.S.; Bhaskar, S.; Kulkarni, S.R.; Freathy, R.M.; Prakash, S.; Mani, K.R.; Weedon, M.N.; Kale, S.D.; Deshpande, J.; et al. FTO gene variants are strongly associated with type 2 diabetes in South Asian Indians. Diabetologia 2009, 52, 247–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kooner, J.S.; Saleheen, D.; Sim, X.; Sehmi, J.; Zhang, W.; Frossard, P.; Been, L.F.; Chia, K.S.; Dimas, A.S.; Hassanali, N.; et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat. Genet. 2011, 43, 984–989. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, G.; Spurgeon, C.J.; Tabassum, R.; Bhaskar, S.; Kulkarni, S.R.; Mahajan, A.; Chavali, S.; Kumar, M.V.; Prakash, S.; Dwivedi, O.P.; et al. Impact of common variants of PPARG, KCNJ11, TCF7L2, SLC30A8, HHEX, CDKN2A, IGF2BP2, and CDKAL1 on the risk of type 2 diabetes in 5164 Indians. Diabetes 2010, 59, 2068–2074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rees, S.D.; Hydrie, M.Z.; Shera, A.S.; Kumar, S.; O’Hare, J.P.; Barnett, A.H.; Basit, A.; Kelly, M.A. Replication of 13 genome-wide association (GWA)-validated risk variants for type 2 diabetes in Pakistani populations. Diabetologia 2011, 54, 1368–1374. [Google Scholar] [CrossRef] [Green Version]
- Humphries, S.E.; Gable, D.; Cooper, J.A.; Ireland, H.; Stephens, J.W.; Hurel, S.J.; Li, K.W.; Palmen, J.; Miller, M.A.; Cappuccio, F.P.; et al. Common variants in the TCF7L2 gene and predisposition to type 2 diabetes in UK European Whites, Indian Asians and Afro-Caribbean men and women. J. Mol. Med. 2006, 84, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Vasan, S.K.; Karpe, F.; Gu, H.F.; Brismar, K.; Fall, C.H.; Ingelsson, E.; Fall, T. FTO genetic variants and risk of obesity and type 2 diabetes: A meta-analysis of 28,394 Indians. Obesity 2014, 22, 964–970. [Google Scholar] [CrossRef]
- Chandak, G.R.; Janipalli, C.S.; Bhaskar, S.; Kulkarni, S.R.; Mohankrishna, P.; Hattersley, A.T.; Frayling, T.M.; Yajnik, C.S. Common variants in the TCF7L2 gene are strongly associated with type 2 diabetes mellitus in the Indian population. Diabetologia 2007, 50, 63–67. [Google Scholar] [CrossRef] [Green Version]
- Bodhini, D.; Radha, V.; Dhar, M.; Narayani, N.; Mohan, V. The rs12255372(G/T) and rs7903146(C/T) polymorphisms of the TCF7L2 gene are associated with type 2 diabetes mellitus in Asian Indians. Metabolism 2007, 56, 1174–1178. [Google Scholar] [CrossRef] [Green Version]
- Tong, Y.; Lin, Y.; Zhang, Y.; Yang, J.; Zhang, Y.; Liu, H.; Zhang, B. Association between TCF7L2 gene polymorphisms and susceptibility to type 2 diabetes mellitus: A large Human Genome Epidemiology (HuGE) review and meta-analysis. BMC Med. Genet. 2009, 10, 15. [Google Scholar] [CrossRef] [Green Version]
- Fuchsberger, C.; Flannick, J.; Teslovich, T.M.; Mahajan, A.; Agarwala, V.; Gaulton, K.J.; Ma, C.; Fontanillas, P.; Moutsianas, L.; McCarthy, D.J.; et al. The genetic architecture of type 2 diabetes. Nature 2016, 536, 41–47. [Google Scholar] [CrossRef] [Green Version]
- Morris, A.P.; Voight, B.F.; Teslovich, T.M.; Ferreira, T.; Segrè, A.V.; Steinthorsdottir, V.; Strawbridge, R.J.; Khan, H.; Grallert, H.; Mahajan, A.; et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012, 44, 981–990. [Google Scholar] [PubMed]
- Scott, R.A.; Scott, L.J.; Mägi, R.; Marullo, L.; Gaulton, K.J.; Kaakinen, M.; Pervjakova, N.; Pers, T.H.; Johnson, A.D.; Eicher, J.D.; et al. An Expanded Genome-Wide Association Study of Type 2 Diabetes in Europeans. Diabetes 2017, 66, 2888–2902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahajan, A.; Taliun, D.; Thurner, M.; Robertson, N.R.; Torres, J.M.; Rayner, N.W.; Payne, A.J.; Steinthorsdottir, V.; Scott, R.A.; Grarup, N.; et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018, 50, 1505–1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Udler, M.S.; McCarthy, M.I.; Florez, J.C.; Mahajan, A. Genetic Risk Scores for Diabetes Diagnosis and Precision Medicine. Endocr. Rev. 2019, 40, 1500–1520. [Google Scholar] [CrossRef] [Green Version]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef] [Green Version]
- Ordovas, J.M.; Ferguson, L.R.; Tai, E.S.; Mathers, J.C. Personalised nutrition and health. Bmj 2018, 361, bmj–k2173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietrich, S.; Jacobs, S.; Zheng, J.S.; Meidtner, K.; Schwingshackl, L.; Schulze, M.B. Gene-lifestyle interaction on risk of type 2 diabetes: A systematic review. Obes. Rev. 2019, 20, 1557–1571. [Google Scholar] [CrossRef]
- Wang, T.; Huang, T.; Zheng, Y.; Rood, J.; Bray, G.A.; Sacks, F.M.; Qi, L. Genetic variation of fasting glucose and changes in glycemia in response to 2-year weight-loss diet intervention: The POUNDS LOST trial. Int. J. Obes. 2016, 40, 1164–1169. [Google Scholar] [CrossRef] [Green Version]
- Eriksen, R.; Gibson, R.; Aresu, M.; Heard, A.; Chan, Q.; Evangelou, E.; Gao, H.; Elliott, P.; Frost, G. Gene-diet quality interactions on haemoglobin A1c and type 2 diabetes risk: The Airwave Health Monitoring Study. Endocrinol. Diabetes Metab. 2019, 2, e00074. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.S.; Kim, B.C.; Daily, J.W.; Park, S. High genetic risk scores for impaired insulin secretory capacity doubles the risk for type 2 diabetes in Asians and is exacerbated by Western-type diets. Diabetes Metab. Res. Rev. 2018, 34, e2944. [Google Scholar] [CrossRef]
- Qi, L.; Cornelis, M.C.; Zhang, C.; van Dam, R.M.; Hu, F.B. Genetic predisposition, Western dietary pattern, and the risk of type 2 diabetes in men. Am. J. Clin. Nutr. 2009, 89, 1453–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.S.; Li, K.; Huang, T.; Chen, Y.; Xie, H.; Xu, D.; Sun, J.; Li, D. Genetic Risk Score of Nine Type 2 Diabetes Risk Variants that Interact with Erythrocyte Phospholipid Alpha-Linolenic Acid for Type 2 Diabetes in Chinese Hans: A Case-Control Study. Nutrients 2017, 9, 376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deepa, M.; Pradeepa, R.; Rema, M.; Mohan, A.; Deepa, R.; Shanthirani, S.; Mohan, V. The Chennai Urban Rural Epidemiology Study (CURES)-study design and methodology (urban component) (CURES-I). J. Assoc. Physicians India 2003, 51, 863–870. [Google Scholar]
- Alberti, K.G.; Zimmet, P.Z. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: Diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet. Med. 1998, 15, 539–553. [Google Scholar] [CrossRef]
- Regional Office for the Western Pacific. The Asia-Pacific Perspective. Redefining Obesity and Its Treatment. Sydney: Health Communications Australia. Available online: https://apps.who.int/iris/handle/10665/206936 (accessed on 5 June 2021).
- Sudha, V.; Radhika, G.; Sathya, R.M.; Ganesan, A.; Mohan, V. Reproducibility and validity of an interviewer-administered semi-quantitative food frequency questionnaire to assess dietary intake of urban adults in southern India. Int. J. Food Sci. Nutr. 2006, 57, 481–493. [Google Scholar] [CrossRef]
- Fretts, A.M.; Follis, J.L.; Nettleton, J.A.; Lemaitre, R.N.; Ngwa, J.S.; Wojczynski, M.K.; Kalafati, I.P.; Varga, T.V.; Frazier-Wood, A.C.; Houston, D.K.; et al. Consumption of meat is associated with higher fasting glucose and insulin concentrations regardless of glucose and insulin genetic risk scores: A meta-analysis of 50,345 Caucasians. Am. J. Clin. Nutr. 2015, 102, 1266–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Khor, C.C.; Fan, J.; Lv, J.; Yu, C.; Guo, Y.; Bian, Z.; Yang, L.; Millwood, I.Y.; Walters, R.G.; et al. Genetic risk, adherence to a healthy lifestyle, and type 2 diabetes risk among 550,000 Chinese adults: Results from 2 independent Asian cohorts. Am. J. Clin. Nutr. 2020, 111, 698–707. [Google Scholar] [CrossRef] [Green Version]
- Scuteri, A.; Sanna, S.; Chen, W.M.; Uda, M.; Albai, G.; Strait, J.; Najjar, S.; Nagaraja, R.; Orru, M.; Usala, G.; et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007, 3, e115. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Z.; Song, Y.; Zhou, D.; Zhang, D.; Zhao, T.; Chen, Z.; Yu, L.; Yang, Y.; Feng, G.; et al. Meta-analysis added power to identify variants in FTO associated with type 2 diabetes and obesity in the Asian population. Obesity 2010, 18, 1619–1624. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Zhu, Y.; Lü, B.; Xu, F.; Li, X.; Lai, M. TCF7L2 gene polymorphisms and type 2 diabetes risk: A comprehensive and updated meta-analysis involving 121,174 subjects. Mutagenesis 2013, 28, 25–37. [Google Scholar] [CrossRef] [Green Version]
- Tönjes, A.; Zeggini, E.; Kovacs, P.; Böttcher, Y.; Schleinitz, D.; Dietrich, K.; Morris, A.P.; Enigk, B.; Rayner, N.W.; Koriath, M.; et al. Association of FTO variants with BMI and fat mass in the self-contained population of Sorbs in Germany. Eur. J. Hum. Genet. EJHG 2010, 18, 104–110. [Google Scholar] [CrossRef] [Green Version]
- Prabhakaran, D.; Jeemon, P.; Sharma, M.; Roth, G.A.; Johnson, C.; Harikrishnan, S.; Gupta, R.; Pandian, J.D.; Naik, N.; Roy, A. The changing patterns of cardiovascular diseases and their risk factors in the states of India: The Global Burden of Disease Study 1990–2016. Lancet Glob. Health 2018, 6, e1339–e1351. [Google Scholar] [CrossRef] [Green Version]
- Gupta, V.; Vinay, D.G.; Sovio, U.; Rafiq, S.; Kranthi Kumar, M.V.; Janipalli, C.S.; Evans, D.; Mani, K.R.; Sandeep, M.N.; Taylor, A.; et al. Association study of 25 type 2 diabetes related Loci with measures of obesity in Indian sib pairs. PLoS ONE 2013, 8, e53944. [Google Scholar]
- Wheeler, E.; Leong, A.; Liu, C.T.; Hivert, M.F.; Strawbridge, R.J.; Podmore, C.; Li, M.; Yao, J.; Sim, X.; Hong, J.; et al. Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis. PLoS Med. 2017, 14, e1002383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janipalli, C.S.; Kumar, M.V.; Vinay, D.G.; Sandeep, M.N.; Bhaskar, S.; Kulkarni, S.R.; Aruna, M.; Joglekar, C.V.; Priyadharshini, S.; Maheshwari, N.; et al. Analysis of 32 common susceptibility genetic variants and their combined effect in predicting risk of Type 2 diabetes and related traits in Indians. Diabet. Med. 2012, 29, 121–127. [Google Scholar] [CrossRef]
- Anand, S.S.; Meyre, D.; Pare, G.; Bailey, S.D.; Xie, C.; Zhang, X.; Montpetit, A.; Desai, D.; Bosch, J.; Mohan, V.; et al. Genetic information and the prediction of incident type 2 diabetes in a high-risk multiethnic population: The EpiDREAM genetic study. Diabetes Care 2013, 36, 2836–2842. [Google Scholar] [CrossRef] [Green Version]
- Alsulami, S.; Aji, A.S.; Ariyasra, U.; Sari, S.R.; Tasrif, N.; Yani, F.F.; Lovegrove, J.A.; Sudji, I.R.; Lipoeto, N.I.; Vimaleswaran, K.S. Interaction between the genetic risk score and dietary protein intake on cardiometabolic traits in Southeast Asian. Genes Nutr. 2020, 15, 19. [Google Scholar] [CrossRef]
- Czajkowski, P.; Adamska-Patruno, E.; Bauer, W.; Fiedorczuk, J.; Krasowska, U.; Moroz, M.; Gorska, M.; Kretowski, A. The Impact of FTO Genetic Variants on Obesity and Its Metabolic Consequences is Dependent on Daily Macronutrient Intake. Nutrients 2020, 12, 3255. [Google Scholar] [CrossRef]
- Li, S.X.; Imamura, F.; Schulze, M.B.; Zheng, J.; Ye, Z.; Agudo, A.; Ardanaz, E.; Aune, D.; Boeing, H.; Dorronsoro, M.; et al. Interplay between genetic predisposition, macronutrient intake and type 2 diabetes incidence: Analysis within EPIC-InterAct across eight European countries. Diabetologia 2018, 61, 1325–1332. [Google Scholar] [CrossRef] [Green Version]
- Shobana, R.; Snehalatha, C.; Latha, E.; Vijay, V.; Ramachandran, A. Dietary profile of urban south Indians and its relations with glycaemic status. Diabetes Res. Clin. Pract. 1998, 42, 181–186. [Google Scholar] [CrossRef]
- Joshi, S.R.; Bhansali, A.; Bajaj, S.; Banzal, S.S.; Dharmalingam, M.; Gupta, S.; Mukhopadhyay, S.; Shah, P.R.; Sahay, R.; Sarkar, S.; et al. Results from a dietary survey in an Indian T2DM population: A STARCH study. BMJ Open 2014, 4, e005138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, E.T.; de Koning, L.; Kanaya, A.M. Higher protein intake is associated with diabetes risk in South Asian Indians: The Metabolic Syndrome and Atherosclerosis in South Asians Living in America (MASALA) study. J. Am. Coll. Nutr. 2010, 29, 130–135. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.Y.; Zhang, Z.L.; Wang, P.Y.; Qin, L.Q. Effects of high-protein diets on body weight, glycaemic control, blood lipids and blood pressure in type 2 diabetes: Meta-analysis of randomised controlled trials. Br. J. Nutr. 2013, 110, 781–789. [Google Scholar] [CrossRef]
- Hession, M.; Rolland, C.; Kulkarni, U.; Wise, A.; Broom, J. Systematic review of randomized controlled trials of low-carbohydrate vs. low-fat/low-calorie diets in the management of obesity and its comorbidities. Obes. Rev. 2009, 10, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Santesso, N.; Akl, E.A.; Bianchi, M.; Mente, A.; Mustafa, R.; Heels-Ansdell, D.; Schünemann, H.J. Effects of higher- versus lower-protein diets on health outcomes: A systematic review and meta-analysis. Eur. J. Clin. Nutr. 2012, 66, 780–788. [Google Scholar] [CrossRef]
- Malik, V.S.; Li, Y.; Tobias, D.K.; Pan, A.; Hu, F.B. Dietary Protein Intake and Risk of Type 2 Diabetes in US Men and Women. Am. J. Epidemiol. 2016, 183, 715–728. [Google Scholar] [CrossRef]
- Viguiliouk, E.; Stewart, S.E.; Jayalath, V.H.; Ng, A.P.; Mirrahimi, A.; de Souza, R.J.; Hanley, A.J.; Bazinet, R.P.; Blanco Mejia, S.; Leiter, L.A.; et al. Effect of Replacing Animal Protein with Plant Protein on Glycemic Control in Diabetes: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Nutrients 2015, 7, 9804–9824. [Google Scholar] [CrossRef]
- Tian, S.; Xu, Q.; Jiang, R.; Han, T.; Sun, C.; Na, L. Dietary Protein Consumption and the Risk of Type 2 Diabetes: A Systematic Review and Meta-Analysis of Cohort Studies. Nutrients 2017, 9, 982. [Google Scholar]
- Satija, A.; Bhupathiraju, S.N.; Rimm, E.B.; Spiegelman, D.; Chiuve, S.E.; Borgi, L.; Willett, W.C.; Manson, J.E.; Sun, Q.; Hu, F.B. Plant-Based Dietary Patterns and Incidence of Type 2 Diabetes in US Men and Women: Results from Three Prospective Cohort Studies. PLoS Med. 2016, 13, e1002039. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Zuurmond, M.G.; van der Schaft, N.; Nano, J.; Wijnhoven, H.A.H.; Ikram, M.A.; Franco, O.H.; Voortman, T. Plant versus animal based diets and insulin resistance, prediabetes and type 2 diabetes: The Rotterdam Study. Eur. J. Epidemiol. 2018, 33, 883–893. [Google Scholar] [CrossRef] [Green Version]
- Alhazmi, A.; Stojanovski, E.; McEvoy, M.; Garg, M.L. Macronutrient intake and type 2 diabetes risk in middle-aged Australian women. Results from the Australian Longitudinal Study on Women’s Health. Public Health Nutr. 2014, 17, 1587–1594. [Google Scholar] [CrossRef] [Green Version]
- Nanri, A.; Mizoue, T.; Kurotani, K.; Goto, A.; Oba, S.; Noda, M.; Sawada, N.; Tsugane, S. Low-carbohydrate diet and type 2 diabetes risk in Japanese men and women: The Japan Public Health Center-Based Prospective Study. PLoS ONE 2015, 10, e0118377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.; Mathur, V. Structural changes in demand for food in India. J. Agric. Econ. 1996, 51, 664–673. [Google Scholar]
- National Nutrition Monitoring Bureau (1984) Report on Urban Population (1975–1980). Available online: https://www.nin.res.in/downloads/Report_Of_Urban_Population_75-80.pdf (accessed on 16 April 2021).
- Swaminathan, S.; Vaz, M.; Kurpad, A.V. Protein intakes in India. Br. J. Nutr. 2012, 108, S50–S58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Indian Council of Medical Research (ICMR). Dietary Guidelines for Indians. Available online: https://www.nin.res.in/downloads/DietaryGuidelinesforNINwebsite.pdf (accessed on 5 June 2021).
- National Family Health Survey (NFHS-4). International Institute of Population Science (IIPS). Available online: http://rchiips.org/nfhs/factsheet_nfhs-4.shtml (accessed on 5 June 2021).
- Indian Council of Social Science Research (ICSSR). Available online: http://www.icssrdataservice.in/datarepository/index.php/catalog/91 (accessed on 5 June 2021).
- Sowmya, N.; Lakshmipriya, N.; Arumugam, K.; Venkatachalam, S.; Vijayalakshmi, P.; Ruchi, V.; Geetha, G.; Anjana, R.M.; Mohan, V.; Krishnaswamy, K.; et al. Comparison of dietary profile of a rural south Indian population with the current dietary recommendations for prevention of non-communicable diseases (CURES 147). Indian J. Med Res. 2016, 144, 112–119. [Google Scholar] [PubMed]
- Agrawal, S.; Ebrahim, S. Association between legume intake and self-reported diabetes among adult men and women in India. BMC Public Health 2013, 13, 706. [Google Scholar] [CrossRef] [Green Version]
- Lakshmi Priya, N.; Gayathri, R.; Sudha, V.; Geetha, G.; Gayathri, N.; Shilpa, B.; Shanthi Rani, C.; Kamala, K.; Anjana, R.; Ranjit, U.; et al. Prospective associations between a food-based Indian Diet Quality Score and type 2 diabetes risk among South Indian adults (CURES-154). J. Diabetol. 2020, 11, 115–124. [Google Scholar]
- Radhika, G.; Sathya, R.M.; Ganesan, A.; Saroja, R.; Vijayalakshmi, P.; Sudha, V.; Mohan, V. Dietary profile of urban adult population in South India in the context of chronic disease epidemiology (CURES-68). Public Health Nutr. 2011, 14, 591–598. [Google Scholar] [CrossRef] [Green Version]
- Anjana, R.M.; Deepa, M.; Pradeepa, R.; Mahanta, J.; Narain, K.; Das, H.K.; Adhikari, P.; Rao, P.V.; Saboo, B.; Kumar, A.; et al. Prevalence of diabetes and prediabetes in 15 states of India: Results from the ICMR-INDIAB population-based cross-sectional study. Lancet Diabetes Endocrinol. 2017, 5, 585–596. [Google Scholar] [CrossRef]
- Deepa, M.; Anjana, R.M.; Manjula, D.; Narayan, K.M.; Mohan, V. Convergence of prevalence rates of diabetes and cardiometabolic risk factors in middle and low income groups in urban India: 10-year follow-up of the Chennai Urban Population Study. J. Diabetes Sci. Technol. 2011, 5, 918–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodarzi, M.O. Genetics of obesity: What genetic association studies have taught us about the biology of obesity and its complications. Lancet Diabetes Endocrinol. 2018, 6, 223–236. [Google Scholar] [CrossRef]
| Total | NGT Controls | T2D Cases | p Value | ||||
|---|---|---|---|---|---|---|---|
| n | n | n | |||||
| Sex | 0.807 ** | ||||||
| Men (%) | 591 | 56 | 278 | 56 | 313 | 55 | |
| Women (%) | 471 | 44 | 218 | 44 | 253 | 45 | |
| Age (years) | 1062 | 45 ± 12 | 496 | 38 ± 10 | 566 | 51 ± 11 | 1.160 × 10−71 * |
| Diabetes duration | - | - | - | - | 566 | 5.20 ± 5.29 | - |
| Anti-diabetic medication | - | - | - | - | 164 | 15.4% | - |
| BMI (kg/m2) | 1061 | 24.6 ± 4.56 | 496 | 23.5 ± 4.64 | 565 | 25.5 ± 4.30 | 1.480 × 10−12 * |
| WC (cm) | 1022 | 87 ± 12 | 479 | 83 ± 12 | 543 | 91 ± 10 | 5.692 × 10−33 * |
| HBA1C (%) | 1056 | 7.3 ± 2.4 | 492 | 5.6 ± 0.47 | 564 | 8.8 ± 2.4 | 1.480 × 10−14 * |
| FPG (mg/dL) | 1060 | 126 ± 64 | 495 | 85 ± 8 | 565 | 162 ± 69 | 1.392 × 10−127 * |
| Fasting Insulin (μIU/mL) | 699 | 9 ± 7 | 448 | 8 ± 6 | 251 | 12 ± 7 | 6.386 × 10−101 * |
| Energy (kcal/day) | 1062 | 2536 ± 805 | 496 | 2685 ± 708 | 566 | 2406 ± 861 | 8.773 × 10−9 * |
| Protein (%) | 1062 | 11 ± 1 | 496 | 11.27 ± 1.17 | 566 | 11.45 ± 1.23 | 0.014 * |
| Animal protein (g/day) | 1062 | 22 ± 12 | 496 | 25 ± 13 | 566 | 19 ± 11 | 3.787 × 10−14 * |
| Plant protein (g/day) | 1062 | 40 ± 14 | 496 | 42 ± 15 | 566 | 39 ± 13 | 0.006 * |
| Fat (%) | 1062 | 23 ± 5 | 496 | 24 ± 5 | 566 | 23 ± 5 | 0.113 * |
| Carbohydrate (%) | 1062 | 65 ± 6 | 496 | 64 ± 6 | 566 | 65 ± 6 | 0.003 * |
| Dietary fibre (g) | 1062 | 32 ± 11 | 496 | 32 ± 10 | 566 | 31 ± 12 | 0.150 * |
| Total SFA (g) | 1062 | 24 ± 10 | 496 | 27 ± 10 | 566 | 22 ± 10 | 2.295 × 10−12 * |
| Total MUFA (g) | 1062 | 20 ± 8 | 496 | 21 ± 8 | 566 | 18 ± 8 | 3.943 × 10−9 * |
| Total PUFA (g) | 1062 | 18 ± 10 | 496 | 19 ± 9 | 566 | 18 ± 10 | 0.184 * |
| Physical activity level | |||||||
| Sedentary | 695 | 71% | 335 | 73% | 360 | 70% | 0.001 ** |
| Moderate | 223 | 23% | 110 | 24% | 113 | 22% | |
| Vigorously active | 58 | 6% | 13 | 3% | 45 | 8% | |
| Smoking | 0.206 ** | ||||||
| Non-smokers | 865 | 81.5% | 396 | 79.8% | 469 | 82.9% | |
| Smokers | 197 | 18.5% | 100 | 20.2% | 97 | 17.1% | |
| Alcohol consumption | |||||||
| Non-alcoholics | 793 | 74.7% | 358 | 72.2% | 435 | 76.9% | 0.080 ** |
| Alcoholics | 269 | 25.3% | 138 | 27.8% | 131 | 23.1% | |
| 7-SNP GRS | 3-SNP GRS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | GRS < 6 | n | GRS ≥ 6 | p Value | n | GRS ≤ 1 | n | GRS > 1 | p Value * | |
| BMI (kg/m2) | 526 | 24.5 ± 0.2 | 535 | 24.7 ± 0.2 | 0.572 | 645 | 24.7 ± 0.2 | 416 | 24.5 ± 0.2 | 0.572 |
| WC (cm) | 508 | 86.7 ± 0.5 | 514 | 87.4 ± 0.5 | 0.668 | 620 | 87.0 ± 0.47 | 402 | 88.0 ± 0.57 | 0.010 |
| HBA1C (%) | 524 | 7.1 ± 0.1 | 532 | 7.4 ± 0.1 | 0.935 | 640 | 7.0 ± 0.1 | 416 | 7.7 ± 0.1 | 0.000066 |
| FPG (mg/dL) | 526 | 119.9 ± 2.6 | 534 | 131.6 ± 2.9 | 0.181 | 644 | 120.0 ± 2.35 | 416 | 135.0 ± 3.39 | 0.002 |
| Fasting insulin (μIU/mL) | 373 | 9.5 ± 0.4 | 326 | 9.4 ± 0.3 | 0.767 | 419 | 10.0 ± 0.36 | 280 | 9.0 ± 0.33 | 0.171 |
| GRS | OR | 95% CI for OR | p Value * | |
|---|---|---|---|---|
| Lower | Upper | |||
| 7-SNP GRS | 2.083 | 1.496 | 2.898 | 0.0000134 |
| 3-SNP GRS | 1.559 | 1.121 | 2.170 | 0.008 |
| 7-SNP GRS | 3-SNP GRS | |||||
|---|---|---|---|---|---|---|
| Protein | Fat | Carbohydrate | Protein | Fat | Carbohydrate | |
| (% of TEI) | (% of TEI) | (% of TEI) | (% of TEI) | (% of TEI) | (% of TEI) | |
| BMI (kg/m2) | 0.176 | 0.388 | 0.195 | 0.36 | 0.653 | 0.805 |
| WC (cm) | 0.852 | 0.786 | 0.892 | 0.638 | 0.958 | 0.914 |
| HBA1C (%) | 0.032 | 0.629 | 0.618 | 0.007 | 0.677 | 0.756 |
| FPG (mg/dL) | 0.249 | 0.489 | 0.507 | 0.011 | 0.367 | 0.231 |
| Fasting insulin (μIU/mL) | 0.952 | 0.085 | 0.04 | 0.299 | 0.567 | 0.999 |
| T2D | 0.956 | 0.214 | 0.152 | 0.764 | 0.508 | 0.365 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alsulami, S.; Bodhini, D.; Sudha, V.; Shanthi Rani, C.S.; Pradeepa, R.; Anjana, R.M.; Radha, V.; Lovegrove, J.A.; Gayathri, R.; Mohan, V.; et al. Lower Dietary Intake of Plant Protein Is Associated with Genetic Risk of Diabetes-Related Traits in Urban Asian Indian Adults. Nutrients 2021, 13, 3064. https://doi.org/10.3390/nu13093064
Alsulami S, Bodhini D, Sudha V, Shanthi Rani CS, Pradeepa R, Anjana RM, Radha V, Lovegrove JA, Gayathri R, Mohan V, et al. Lower Dietary Intake of Plant Protein Is Associated with Genetic Risk of Diabetes-Related Traits in Urban Asian Indian Adults. Nutrients. 2021; 13(9):3064. https://doi.org/10.3390/nu13093064
Chicago/Turabian StyleAlsulami, Sooad, Dhanasekaran Bodhini, Vasudevan Sudha, Coimbatore Subramanian Shanthi Rani, Rajendra Pradeepa, Ranjit Mohan Anjana, Venkatesan Radha, Julie A. Lovegrove, Rajagopal Gayathri, Viswanathan Mohan, and et al. 2021. "Lower Dietary Intake of Plant Protein Is Associated with Genetic Risk of Diabetes-Related Traits in Urban Asian Indian Adults" Nutrients 13, no. 9: 3064. https://doi.org/10.3390/nu13093064
APA StyleAlsulami, S., Bodhini, D., Sudha, V., Shanthi Rani, C. S., Pradeepa, R., Anjana, R. M., Radha, V., Lovegrove, J. A., Gayathri, R., Mohan, V., & Vimaleswaran, K. S. (2021). Lower Dietary Intake of Plant Protein Is Associated with Genetic Risk of Diabetes-Related Traits in Urban Asian Indian Adults. Nutrients, 13(9), 3064. https://doi.org/10.3390/nu13093064







