Dietary Supplementation with Transgenic Camelina sativa Oil Containing 20:5n-3 and 22:6n-3 or Fish Oil Induces Differential Changes in the Transcriptome of CD3+ T Lymphocytes
Abstract
:1. Introduction
2. Methods
2.1. Preparation of Seed Oil from Transgenic C. sativa
2.2. Dietary Supplementation
2.3. T Cell Isolation and Culture
2.4. Analysis of Fatty Acid Composition by Gas Chromatography
2.5. Analysis of the T Cell Transcriptome by RNA Next-Generation Sequencing
2.6. Statistical Analysis
3. Results
3.1. Participant Characteristics
3.2. Provision of Fatty Acids from Test Oils
3.3. Effect of Dietary Supplementation on the Fatty Acid Composition of PBMCs
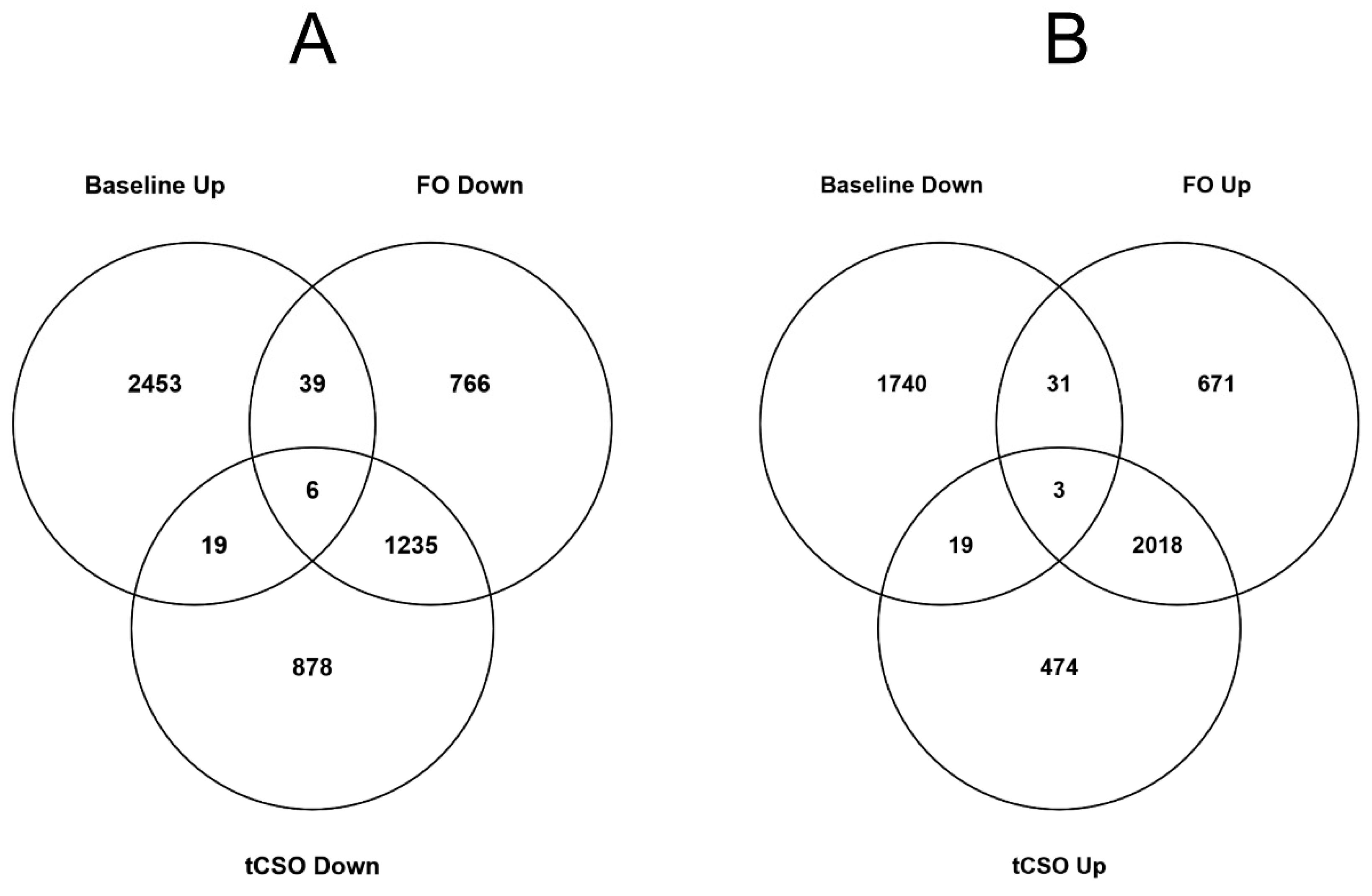
3.4. The Effect of Dietary Supplementation on the Number of Differentially Expressed Transcripts in Activated Compared to Unstimulated CD3+ T Cells
3.5. The Effect of Dietary Supplementation on the Most Differentially Expressed Transcripts in Activated Compared to Unstimulated CD3+ T Cells
3.6. Effect of Dietary Supplementation on Differentially Expressed Pathways in Activated CD3+ T Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclosures
References
- Mitchell, D.C.; Niu, S.L.; Litman, B.J. Optimization of receptor-G protein coupling by bilayer lipid composition I: Kinetics of rhodopsin-transducin binding. J. Biol. Chem. 2001, 276, 42801–42806. [Google Scholar] [CrossRef] [Green Version]
- Molloy, C.; Doyle, L.W.; Makrides, M.; Anderson, P.J. Docosahexaenoic acid and visual functioning in preterm infants: A review. Neuropsychol. Rev. 2012, 22, 425–437. [Google Scholar] [CrossRef]
- Burdge, G.C.; Rodway, H.; Kohler, J.A.; Lillycrop, K.A. Effect of fatty acid supplementation on growth and differentiation of human IMR-32 neuroblastoma cells in vitro. J. Cell. Biochem. 2000, 80, 266–273. [Google Scholar] [CrossRef]
- Burdge, G.C.; Lillycrop, K.A. Fatty acids and epigenetics. Curr. Opin. Clin. Nutr. Metabol. Care 2014, 17, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Givens, D.I.G.; Gibbs, R.A. Very long chain n-3 polyunsaturated fatty acids in the food chain in the UK and the potential of animal-derived foods to increase intake. Nutr. Bullet. 2006, 31, 104–110. [Google Scholar] [CrossRef]
- Calder, P.C.; Grimble, R.F. Polyunsaturated fatty acids, inflammation and immunity. Eur. J. Clin. Nutr. 2002, 56 (Suppl. 3), S14–S19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Browning, L.M.; Walker, C.G.; Mander, A.P.; West, A.L.; Madden, J.; Gambell, J.M.; Young, S.; Wang, L.; Jebb, S.A.; Calder, P.C. Incorporation of eicosapentaenoic and docosahexaenoic acids into lipid pools when given as supplements providing doses equivalent to typical intakes of oily fish. Am. J. Clin. Nutr. 2012, 96, 748–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faber, J.; Berkhout, M.; Vos, A.P.; Sijben, J.W.; Calder, P.C.; Garssen, J.; van Helvoort, A. Supplementation with a fish oil-enriched, high-protein medical food leads to rapid incorporation of EPA into white blood cells and modulates immune responses within one week in healthy men and women. J. Nutr. 2011, 141, 964–970. [Google Scholar] [CrossRef] [Green Version]
- Miles, E.A.; Calder, P.C. Influence of marine n-3 polyunsaturated fatty acids on immune function and a systematic review of their effects on clinical outcomes in rheumatoid arthritis. Br. J. Nutr. 2012, 107 (Suppl. 2), S171–S184. [Google Scholar] [CrossRef] [Green Version]
- Miles, E.A.; Allen, E.; Calder, P.C. In vitro effects of eicosanoids derived from different 20-carbon Fatty acids on production of monocyte-derived cytokines in human whole blood cultures. Cytokine 2002, 20, 215–223. [Google Scholar] [CrossRef]
- Bagga, D.; Wang, L.; Farias-Eisner, R.; Glaspy, J.A.; Reddy, S.T. Differential effects of prostaglandin derived from omega-6 and omega-3 polyunsaturated fatty acids on COX-2 expression and IL-6 secretion. Proc. Natl. Acad. Sci. USA 2003, 100, 1751–1756. [Google Scholar] [CrossRef] [Green Version]
- Goldman, D.W.; Pickett, W.C.; Goetzl, E.J. Human neutrophil chemotactic and degranulating activities of leukotriene B5 (LTB5) derived from eicosapentaenoic acid. Biochem. Biophys. Res. Com. 1983, 117, 282–288. [Google Scholar] [CrossRef]
- Endres, S.; Ghorbani, R.; Kelley, V.E.; Georgilis, K.; Lonnemann, G.; van der Meer, J.W.; Cannon, J.G.; Rogers, T.S.; Klempner, M.S.; Weber, P.C.; et al. The effect of dietary supplementation with n-3 polyunsaturated fatty acids on the synthesis of interleukin-1 and tumor necrosis factor by mononuclear cells. N. Eng. J. Med. 1989, 320, 265–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, T.H.; Hoover, R.L.; Williams, J.D.; Sperling, R.I.; Ravalese, J., 3rd; Spur, B.W.; Robinson, D.R.; Corey, E.J.; Lewis, R.A.; Austen, K.F. Effect of dietary enrichment with eicosapentaenoic and docosahexaenoic acids on in vitro neutrophil and monocyte leukotriene generation and neutrophil function. N. Eng. J. Med. 1985, 312, 1217–1224. [Google Scholar] [CrossRef]
- Meydani, S.N.; Endres, S.; Woods, M.M.; Goldin, B.R.; Soo, C.; Morrill-Labrode, A.; Dinarello, C.A.; Gorbach, S.L. Oral (n-3) fatty acid supplementation suppresses cytokine production and lymphocyte proliferation: Comparison between young and older women. J. Nutr. 1991, 121, 547–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cleland, L.G.; Caughey, G.E.; James, M.J.; Proudman, S.M. Reduction of cardiovascular risk factors with longterm fish oil treatment in early rheumatoid arthritis. J. Rheumatol. 2006, 33, 1973–1979. [Google Scholar]
- Kremer, J.M.; Lawrence, D.A.; Jubiz, W.; DiGiacomo, R.; Rynes, R.; Bartholomew, L.E.; Sherman, M. Dietary fish oil and olive oil supplementation in patients with rheumatoid arthritis. Clinical and immunologic effects. Arthritis Rheum. 1990, 33, 810–820. [Google Scholar] [CrossRef] [PubMed]
- Sperling, R.I.; Weinblatt, M.; Robin, J.L.; Ravalese, J., 3rd; Hoover, R.L.; House, F.; Coblyn, J.S.; Fraser, P.A.; Spur, B.W.; Robinson, D.R.; et al. Effects of dietary supplementation with marine fish oil on leukocyte lipid mediator generation and function in rheumatoid arthritis. Arthritis Rheum. 1987, 30, 988–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Tempel, H.; Tulleken, J.E.; Limburg, P.C.; Muskiet, F.A.; van Rijswijk, M.H. Effects of fish oil supplementation in rheumatoid arthritis. Ann. Rheum. Dis. 1990, 49, 76–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calder, P.C. Omega-3 fatty acids and inflammatory processes: From molecules to man. Biochem. Soc. Trans. 2017, 45, 1105–1115. [Google Scholar] [CrossRef] [Green Version]
- Givens, D.I.; Gibbs, R.A. Current intakes of EPA and DHA in European populations and the potential of animal-derived foods to increase them. Proc. Nutr. Soc. 2008, 67, 273–280. [Google Scholar] [CrossRef] [Green Version]
- Scientific Advisory Committee on Nutrition Advice on Fish Consumption: Benefits and Risks; TSO: London, UK, 2004.
- Welch, A.A.; Shakya-Shrestha, S.; Lentjes, M.A.; Wareham, N.J.; Khaw, K.T. Dietary intake and status of n-3 polyunsaturated fatty acids in a population of fish-eating and non-fish-eating meat-eaters, vegetarians, and vegans and the precursor-product ratio of alpha-linolenic acid to long-chain n-3 polyunsaturated fatty acids: Results from the EPIC-Norfolk cohort. Am. J. Clin. Nutr. 2010, 92, 1040–1051. [Google Scholar] [PubMed]
- Ruiz-Lopez, N.; Haslam, R.P.; Napier, J.A.; Sayanova, O. Successful high-level accumulation of fish oil omega-3 long-chain polyunsaturated fatty acids in a transgenic oilseed crop. Plant J. 2014, 77, 198–208. [Google Scholar] [CrossRef] [Green Version]
- West, A.L.; Miles, E.A.; Lillycrop, K.A.; Han, L.; Napier, J.A.; Calder, P.C.; Burdge, G.C. Dietary supplementation with seed oil from transgenic Camelina sativa induces similar increments in plasma and erythrocyte DHA and EPA to fish oil in healthy humans. Br. J. Nutr. 2020, 124, 922–930. [Google Scholar] [CrossRef] [PubMed]
- West, A.L.; Miles, E.A.; Lillycrop, K.A.; Han, L.; Sayanova, O.; Napier, J.A.; Calder, P.C.; Burdge, G.C. Postprandial incorporation of EPA and DHA from transgenic Camelina sativa oil into blood lipids is equivalent to that from fish oil in healthy humans. Br. J. Nutr. 2019, 121, 1235–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miles, E.A.; Banerjee, T.; Dooper, M.M.; M’Rabet, L.; Graus, Y.M.; Calder, P.C. The influence of different combinations of gamma-linolenic acid, stearidonic acid and EPA on immune function in healthy young male subjects. Br. J. Nutr. 2004, 91, 893–903. [Google Scholar] [CrossRef] [Green Version]
- Folch, J.; Lees, M.; Sloane-Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Bio. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Burdge, G.C.; Wright, P.; Jones, A.E.; Wootton, S.A. A method for separation of phosphatidylcholine, triacylglycerol, non-esterified fatty acids and cholesterol esters from plasma by solid-phase extraction. Br. J. Nutr. 2000, 84, 781–787. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Scheffler, K.; Halpern, A.L.; Bekritsky, M.A.; Noh, E.; Kallberg, M.; Chen, X.; Kim, Y.; Beyter, D.; Krusche, P.; et al. Strelka2: Fast and accurate calling of germline and somatic variants. Nat. Methods 2018, 15, 591–594. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Sibbons, C.M.; Irvine, N.A.; Pérez-Mojica, J.E.; Calder, P.C.; Lillycrop, K.A.; Fielding, B.A.; Burdge, G.C. Polyunsaturated Fatty Acid Biosynthesis Involving Δ8 Desaturation and Differential DNA Methylation of FADS2 Regulates Proliferation of Human Peripheral Blood Mononuclear Cells. Front. Immunol. 2018, 9, 432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calder, P.C.; Yaqoob, P.; Newsholme, E.A. Triacylglycerol metabolism by lymphocytes and the effect of triacylglycerols on lymphocyte proliferation. Biochem. J. 1994, 298, 605–611. [Google Scholar] [CrossRef] [Green Version]
- Yaqoob, P.; Newsholme, E.A.; Calder, P.C. Fatty acid oxidation by lymphocytes. Biochem. Soc. Trans. 1994, 22, 116S. [Google Scholar] [CrossRef] [Green Version]
- Rode, H.N.; Szamel, M.; Schneider, S.; Resch, K. Phospholipid metabolism of stimulated lymphocytes. Preferential incorporation of polyunsaturated fatty acids into plasma membrane phospholipid upon stimulation with concanavalin A. Biochim. Biophys. Acta 1982, 688, 66–74. [Google Scholar] [CrossRef]
- Ferber, E.; De Pasquale, G.G.; Resch, K. Phospholipid metabolism of stimulated lymphocytes. Composition of phospholipid fatty acids. Biochim. Biophys. Acta 1975, 398, 364–376. [Google Scholar] [CrossRef]
- Anel, A.; Naval, J.; Gonzalez, B.; Torres, J.M.; Mishal, Z.; Uriel, J.; Pineiro, A. Fatty acid metabolism in human lymphocytes. I. Time-course changes in fatty acid composition and membrane fluidity during blastic transformation of peripheral blood lymphocytes. Biochim. Biophys. Acta. 1990, 1044, 323–331. [Google Scholar] [CrossRef]
- Calder, P.C.; Yaqoob, P.; Harvey, D.J.; Watts, A.; Newsholme, E.A. Incorporation of fatty acids by concanavalin A-stimulated lymphocytes and the effect on fatty acid composition and membrane fluidity. Biochem. J. 1994, 300, 509–518. [Google Scholar] [CrossRef] [Green Version]
- Shires, S.E.; Kelleher, J.; Trejdosiewicz, L.K. Effects of linoleic acid and mitogenic stimulation on the fatty acid composition of human lymphocytes. Biochim. Biophys. Acta 1989, 1002, 74–78. [Google Scholar] [CrossRef]
- Myhrstad, M.C.; Narverud, I.; Telle-Hansen, V.H.; Karhu, T.; Lund, D.B.; Herzig, K.H.; Makinen, M.; Halvorsen, B.; Retterstol, K.; Kirkhus, B.; et al. Effect of the fat composition of a single high-fat meal on inflammatory markers in healthy young women. Br. J. Nutr. 2011, 106, 1826–1835. [Google Scholar] [CrossRef] [Green Version]
- Naeini, Z.; Toupchian, O.; Vatannejad, A.; Sotoudeh, G.; Teimouri, M.; Ghorbani, M.; Nasli-Esfahani, E.; Koohdani, F. Effects of DHA-enriched fish oil on gene expression levels of p53 and NF-kappaB and PPAR-gamma activity in PBMCs of patients with T2DM: A randomized, double-blind, clinical trial. Nutr. Metab. Cardiovasc. Dis. 2020, 30, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Rundblad, A.; Holven, K.B.; Bruheim, I.; Myhrstad, M.C.; Ulven, S.M. Effects of fish and krill oil on gene expression in peripheral blood mononuclear cells and circulating markers of inflammation: A randomised controlled trial. J. Nutr. Sci. 2018, 7, e10. [Google Scholar] [CrossRef] [Green Version]
- Myhrstad, M.C.; Ulven, S.M.; Gunther, C.C.; Ottestad, I.; Holden, M.; Ryeng, E.; Borge, G.I.; Kohler, A.; Bronner, K.W.; Thoresen, M.; et al. Fish oil supplementation induces expression of genes related to cell cycle, endoplasmic reticulum stress and apoptosis in peripheral blood mononuclear cells: A transcriptomic approach. J. Intern. Med. 2014, 276, 498–511. [Google Scholar] [CrossRef] [Green Version]
- Bouwens, M.; van de Rest, O.; Dellschaft, N.; Bromhaar, M.G.; de Groot, L.C.; Geleijnse, J.M.; Muller, M.; Afman, L.A. Fish-oil supplementation induces antiinflammatory gene expression profiles in human blood mononuclear cells. Am. J. Clin. Nutr. 2009, 90, 415–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polus, A.; Zapala, B.; Razny, U.; Gielicz, A.; Kiec-Wilk, B.; Malczewska-Malec, M.; Sanak, M.; Childs, C.E.; Calder, P.C.; Dembinska-Kiec, A. Omega-3 fatty acid supplementation influences the whole blood transcriptome in women with obesity, associated with pro-resolving lipid mediator production. Biochim. Biophys. Acta 2016, 1861, 1746–1755. [Google Scholar] [CrossRef] [Green Version]
- Fabriek, B.O.; Dijkstra, C.D.; van den Berg, T.K. The macrophage scavenger receptor CD163. Immunobiology 2005, 210, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Hahn, B.H.; Bischoff, D.S. Effects of Peptide-Induced Immune Tolerance on Murine Lupus. Front. Immunol. 2021, 12, 662901. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.S.; Gumperz, J.E.; Brenner, M.B. Understanding the function of CD1-restricted T cells. Nat. Immunol. 2003, 4, 517–523. [Google Scholar] [CrossRef]
- Sousa, I.G.; Simi, K.C.R.; do Almo, M.M.; Bezerra, M.A.G.; Doose, G.; Raiol, T.; Stadler, P.F.; Hoffmann, S.; Maranhao, A.Q.; Brigido, M.M. Gene expression profile of human T cells following a single stimulation of peripheral blood mononuclear cells with anti-CD3 antibodies. BMC Genom. 2019, 20, 593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara, M.; Fukagawa, T. Dynamics of kinetochore structure and its regulations during mitotic progression. Cell. Mol. Life Sci. 2020, 77, 2981–2995. [Google Scholar] [CrossRef] [PubMed]
- Sabat, R.; Grutz, G.; Warszawska, K.; Kirsch, S.; Witte, E.; Wolk, K.; Geginat, J. Biology of interleukin-10. Cytokine Growth Factor Rev. 2010, 21, 331–344. [Google Scholar] [CrossRef] [Green Version]
- Huber, S.; Gagliani, N.; Esplugues, E.; O’Connor, W., Jr.; Huber, F.J.; Chaudhry, A.; Kamanaka, M.; Kobayashi, Y.; Booth, C.J.; Rudensky, A.Y.; et al. Th17 cells express interleukin-10 receptor and are controlled by Foxp3(-) and Foxp3+ regulatory CD4+ T cells in an interleukin-10-dependent manner. Immunity 2011, 34, 554–565. [Google Scholar] [CrossRef] [Green Version]
- Chehimi, M.; Ward, R.; Pestel, J.; Robert, M.; Pesenti, S.; Bendridi, N.; Michalski, M.C.; Laville, M.; Vidal, H.; Eljaafari, A. Omega-3 Polyunsaturated Fatty Acids Inhibit IL-17A Secretion through Decreased ICAM-1 Expression in T Cells Co-Cultured with Adipose-Derived Stem Cells Harvested from Adipose Tissues of Obese Subjects. Mol. Nutr. Food Res. 2019, 63, e1801148. [Google Scholar] [CrossRef] [PubMed]
- Shoda, H.; Yanai, R.; Yoshimura, T.; Nagai, T.; Kimura, K.; Sobrin, L.; Connor, K.M.; Sakoda, Y.; Tamada, K.; Ikeda, T.; et al. Dietary Omega-3 Fatty Acids Suppress Experimental Autoimmune Uveitis in Association with Inhibition of Th1 and Th17 Cell Function. PLoS ONE 2015, 10, e0138241. [Google Scholar] [CrossRef]
- Mansoori, A.; Sotoudeh, G.; Djalali, M.; Eshraghian, M.R.; Keramatipour, M.; Nasli-Esfahani, E.; Shidfar, F.; Alvandi, E.; Toupchian, O.; Koohdani, F. Effect of DHA-rich fish oil on PPARgamma target genes related to lipid metabolism in type 2 diabetes: A randomized, double-blind, placebo-controlled clinical trial. J. Clin. Lipidol. 2015, 9, 770–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoile, S.P.; Clarke-Harris, R.; Huang, R.C.; Calder, P.C.; Mori, T.A.; Beilin, L.J.; Lillycrop, K.A.; Burdge, G.C. Supplementation with N-3 Long-Chain Polyunsaturated Fatty Acids or Olive Oil in Men and Women with Renal Disease Induces Differential Changes in the DNA Methylation of FADS2 and ELOVL5 in Peripheral Blood Mononuclear Cells. PLoS ONE 2014, 9, e109896. [Google Scholar] [CrossRef] [PubMed]
- Perez-Mojica, J.E.; Lillycrop, K.A.; Cooper, C.; Calder, P.C.; Burdge, G.C. Docosahexaenoic acid and oleic acid induce altered DNA methylation of individual CpG loci in Jurkat T cells. Prostaglandins Leukot. Essent. Fatty Acids 2020, 158, 102128. [Google Scholar] [CrossRef]
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef] [Green Version]


| Whole Group | Males | Females | RNAseq Subgroup * | ||||
|---|---|---|---|---|---|---|---|
| All (Baseline) | FO Group | tCSO Group | ‡t Test | ||||
| n | 31 | 13 | 18 | 16 | 8 | 8 | p |
| Age (years) | 44.6 ± 3.4 | 46.6 ± 5.3 | 43.1 ± 4.9 | 44.4 ± 5.5 | 40.1 ± 7.9 | 46.1 ± 7.6 | 0.595 |
| BMI (kg/m2) | 24.5 ± 0.5 | 25.2 ± 0.8 | 23.9 ± 0.6 | 26.6 ± 5.5 | 23.2 ± 1.3 | 24.3 ± 0.7 | 0.468 |
| Systolic bp (mm Hg) | 118 ± 2 | 123 ± 2 | 114 ± 3.5 | 106 ± 8 | 117 ± 5.3 | 111.6 ± 5.8 | 0.522 |
| Diastolic bp (mm Hg) | 69 ± 2 | 73 ± 2 | 66 ± 2 | 65 ± 2 | 66.9 ± 2.8 | 65.3 ± 2.9 | 0.713 |
| Glucose (mmol/L) | 5.1 ± 0.1 | 4.9 ± 0.1 | 5.3 ± 0.2 | 5.4 ± 0.2 | 5.1 ± 0.2 | 5.6 ± 0.4 | 0.258 |
| Cholesterol (mmol/L) | 4.9 ± 0.2 | 4.8 ± 0.2 | 5.0 ± 0.3 | 5.2 ± 0.3 | 5.2 ± 0.4 | 4.9 ± 0.5 | 0.665 |
| Fatty Acid | Daily Provision (mg/day) | Cumulative Provision (g over 56 days) | ||
|---|---|---|---|---|
| FO | tCSO | FO | tCSO | |
| 14:0 | 100 | 3 | 5.6 | 0.2 |
| 16:0 | 232 | 156 | 13.0 | 8.7 |
| 18:0 | 45 | 130 | 2.5 | 7.3 |
| 20:0 | 3 | 68 | 0.1 | 3.8 |
| 24:0 | 2 | 24 | 0.1 | 1.3 |
| Total SFAs | 381 | 380 | 21.4 | 21.3 |
| 16:1n-7 | 139 | 5 | 7.8 | 0.3 |
| 18:1n-9 | 224 | 139 | 12.6 | 7.8 |
| 18:1n-7 | 63 | 37 | 3.5 | 2.1 |
| 20:1n-9 | 102 | 144 | 5.7 | 8.0 |
| Total MUFAs | 527 | 325 | 29.5 | 18.2 |
| 18:2n-6 | 24 | 468 | 1.4 | 26.2 |
| 18:3n-6 | 3 | 73 | 0.2 | 4.1 |
| 20:2n-6 | 4 | 20 | 0.2 | 1.1 |
| 20:3n-6 | 3 | 21 | 0.2 | 1.2 |
| 20:4n-6 | 15 | 67 | 0.9 | 3.8 |
| Total n-6 PUFAs | 50 | 650 | 2.8 | 36.4 |
| 18:3n-3 | 16 | 321 | 0.9 | 18.0 |
| 20:4n-3 | 96 | 66 | 5.4 | 3.7 |
| 20:5n-3 | 247 | 254 | 13.8 | 14.2 |
| 22:5n-3 | 29 | 155 | 1.6 | 8.7 |
| 22:6n-3 | 199 | 202 | 11.1 | 11.3 |
| 20:5n-3 + 22:6n-3 | 445 | 456 | 24.9 | 25.5 |
| Total n-3 PUFAs | 587 | 998 | 33 | 56 |
| Total fatty acids | 1545 | 2352 | 86.5 | 131.7 |
| Proportion of Total Fatty Acids | ANOVA | |||||||
|---|---|---|---|---|---|---|---|---|
| % | Time | Time × Test Oil | ||||||
| FO | tCSO | F | p | F | p | |||
| Start | End | Start | End | |||||
| 14:0 | 0.5 ± 0.1 | 0.6 ± 0.1 | 0.7 ± 0.1 | 0.5 ± 0.1 | 0.82 | 0.35 | 0.25 | 0.88 |
| 16:0 | 19.8 ± 0.9 | 19.9 ± 1.2 | 21.3 ± 1.2 | 18.6 ± 1.2 | 3.34 | 0.08 | 4.14 | 0.05 |
| 16:1n-7 | 0.3 ± 0.0 | 0.3 ± 0.0 | 0.3 ± 0.0 | 0.3 ± 0.0 | 0.04 | 0.85 | 0.42 | 0.52 |
| 18:0 | 27.7 ± 0.7 | 27.4 ± 0.5 | 27.6 ± 0.7 | 27.0 ± 0.8 | 0.83 | 0.37 | 1.21 | 0.73 |
| 18:1n-9 | 14.8 ± 0.5 | 15.1 ± 0.4 | 13.8 ± 0.5 | 15.3 ± 0.5 | 5.98 | 0.02 | 2.47 | 0.12 |
| 18:1n-7 | 1.4 ± 0.1 | 1.3 ± 0.0 | 1.3 ± 0.0 | 1.2 ± 0.1 | 6.22 | 0.02 | 0.01 | 0.91 |
| 18:2n-6 | 6.7 ± 0.3 | 6.1 ± 0.2 | 6.2 ± 0.2 | 6.0 ± 0.2 | 5.35 | 0.02 | 1.36 | 0.24 |
| 18:3n-6 | 0.5 ± 0.0 | 0.4 ± 0.0 | 0.5 ± 0.1 | 0.3 ± 0.0 | 3.895 | 0.04 | 0.42 | 0.52 |
| 18:3n-3 | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.2 ± 0.1 | 1.1 ± 0.1 | 0.45 | 0.51 | 0.70 | 0.79 |
| 20:0 | 0.6 ± 0.1 | 0.6 ± 0.1 | 0.6 ± 0.1 | 0.5 ± 0.1 | 0.79 | 0.39 | 1.22 | 0.27 |
| 20:1n-9 | 1.2 ± 0.1 | 1.2 ± 0.1 | 1.3 ± 0.1 | 1.1 ± 0.1 | 1.65 | 0.20 | 1.35 | 0.25 |
| 20:2n-6 | 0.6 ± 0.1 | 0.6 ± 0.1 | 0.6 ± 0.1 | 0.6 ± 0.1 | 0.001 | 0.98 | 0.55 | 0.46 |
| 20:3n-6 | 1.8 ± 0.1 | 1.8 ± 0.1 | 1.9 ± 0.1 | 1.7 ± 0.1 | 1.1 | 0.31 | 4.88 | 0.07 |
| 20:4n-6 | 16.5 ± 0.9 | 16.6 ± 0.9 | 16.3 ± 1.1 | 18.4 ± 1.1 | 2.49 | 0.12 | 2.42 | 0.13 |
| 20:5n-3 | 0.8 ± 0.1 | 0.9 ± 0.1 | 0.8 ± 0.1 | 0.9 ± 0.1 | 4.51 | 0.04 | 0.07 | 0.80 |
| 22:5n-3 | 1.9 ± 0.1 | 1.9 ± 0.1 | 1.7 ± 0.1 | 2.2 ± 0.1 | 12.22 | 0.001 | 5.35 | 0.02 |
| 22:6n-3 | 2.1 ± 0.1 | 2.2 ± 0.1 | 1.9 ± 0.1 | 2.2 ± 0.1 | 4.20 | 0.04 | 1.13 | 0.30 |
| Difference in mRNA Expression in Concanavalin A-Stimulated Cells and Unstimulated Cells | ||||||||
|---|---|---|---|---|---|---|---|---|
| Baseline | Post tCSO Supplementation | Post FO Supplementation | ||||||
| n | 16 | 8 | 8 | |||||
| Gene | log2 Fold Change | Adjusted p | Gene | log2 Fold Change | Adjusted p | Gene | log2 Fold Change | Adjusted p |
| Up-regulated genes | ||||||||
| CXCL9 | 8.97 | 1.16 × 10−32 | KIF20A | 8.71 | 6.13 × 10−73 | CSF2 | 8.50 | 2.48 × 10−26 |
| IL17A | 8.40 | 1.14 × 10−15 | IL17F | 8.68 | 9.23 × 10−21 | GJB2 | 8.13 | 1.15 × 10−12 |
| IL17F | 7.84 | 6.67 × 10−21 | ZBED2 | 8.60 | 5.08 × 10−45 | CAVIN3 | 8.03 | 1.17 × 10−17 |
| GJB2 | 7.80 | 6.24 × 10−26 | SPC25 | 8.60 | 3.61 × 10−34 | RIBC2 | 7.91 | 1.46 × 10−17 |
| CCL17 | 7.69 | 5.03 × 10−18 | IL22 | 8.57 | 3.52 × 10−17 | IL17F | 7.91 | 2.68 × 10−17 |
| ZBED2 | 7.63 | 3.78 × 10−28 | IL17A | 8.50 | 2.94 × 10−12 | CBSL | 7.84 | 2.09 × 10−11 |
| CXCL10 | 7.42 | 1.20 × 10−18 | XIRP1 | 8.40 | 6.14 × 10−26 | MCM10 | 7.76 | 2.56 × 10−29 |
| IL5 | 7.27 | 1.62 × 10−15 | PKMYT1 | 8.32 | 2.78 × 10−68 | IL5 | 7.57 | 2.19 × 10−07 |
| HIST1H2BH | 7.20 | 4.41 × 10−29 | IL5 | 8.23 | 5.14 × 10−13 | CCL1 | 7.53 | 6.48 × 10−16 |
| IFNG | 7.19 | 3.46 × 10−27 | UHRF1 | 8.17 | 1.65 × 10−140 | CDC25C | 7.49 | 3.57 × 10−17 |
| EBI3 | 7.17 | 1.53 × 10−46 | KIF18B | 8.13 | 5.09 × 10−88 | TYMS | 7.47 | 3.65 × 10−20 |
| IL22 | 7.15 | 1.04 × 10−15 | CCNB2 | 8.11 | 5.83 × 10−67 | CDC25A | 7.40 | 1.26 × 10−20 |
| SPC25 | 7.14 | 1.82 × 10−19 | KIF1A | 8.08 | 8.82 × 10−22 | CDC20 | 7.33 | 3.20 × 10−17 |
| FLJ21408 | 7.13 | 2.02 × 10−20 | RRM2 | 8.02 | 2.36 × 10−49 | KIF1A | 7.33 | 1.63 × 10−16 |
| KIF1A | 7.06 | 8.69 × 10−23 | TYMS | 8.02 | 4.02 × 10−78 | FGF2 | 7.29 | 9.95 × 10−16 |
| IL21 | 7.02 | 4.12 × 10−21 | BIRC5 | 7.85 | 5.30 × 10−73 | IL24 | 7.29 | 1.20 × 10−11 |
| MCM10 | 7.01 | 7.03 × 10−23 | CBSL | 7.82 | 1.59 × 10−15 | BIRC5 | 7.28 | 5.84 × 10−16 |
| MYBL2 | 7.01 | 1.29 × 10−28 | LINC01132 | 7.80 | 4.72 × 10−23 | ANXA3 | 7.24 | 3.10 × 10−09 |
| TYMS | 6.97 | 1.64 × 10−25 | GNG4 | 7.76 | 2.32 × 10−33 | PBK | 7.24 | 2.41 × 10−16 |
| DLGAP5 | 6.96 | 7.44 × 10−25 | STC2 | 7.69 | 6.13 × 10−20 | SMIM11A | 7.19 | 0.012 |
| Down-regulated genes | ||||||||
| TREM2 | −6.45 | 3.34 × 10−13 | RNASE1 | −8.15 | 5.49 × 10−23 | SMN1 | −26.27 | 5.38 × 10−17 |
| FAM198B | −6.01 | 2.85 × 10−10 | TREM2 | −6.77 | 1.31 × 10−12 | EEF1E1_BLOC1S5 | −23.38 | 1.23 × 10−13 |
| GPNMB | −5.90 | 2.93 × 10−21 | HSFX2 | −6.76 | 0.04 | LOC102723996 | −23.19 | 2.00 × 10−13 |
| RNASE1 | −5.84 | 2.34 × 10−07 | CNTNAP2 | −6.53 | 4.28 × 10−10 | RNASE1 | −7.60 | 8.65 × 10−05 |
| APOC2 | −5.67 | 3.95 × 10−07 | OLR1 | −6.27 | 5.65 × 10−07 | TREM2 | −6.96 | 2.91 × 10−07 |
| KCNJ5 | −5.63 | 1.03 × 10−07 | LINC00891 | −6.26 | 0.002 | GPNMB | −6.17 | 8.35 × 10−15 |
| A2M | −5.58 | 9.60 × 10−17 | F13A1 | −6.24 | 1.54 × 10−11 | LILRB5 | −5.99 | 6.70 × 10−06 |
| RAB42 | −5.42 | 4.82 × 10−08 | GPNMB | −6.15 | 5.01 × 10−28 | CD163 | −5.98 | 1.05 × 10−12 |
| LILRB5 | −5.23 | 2.02 × 10−07 | C1QA | −6.09 | 2.01 × 10−07 | CNTNAP2 | −5.91 | 4.45 × 10−09 |
| FCN1 | −5.10 | 2.49 × 10−10 | FPR3 | −6.06 | 6.52 × 10−16 | SLCO2B1 | −5.88 | 2.22 × 10−08 |
| DCANP1 | −5.08 | 1.11 × 10−08 | FAM198B | −6.00 | 1.72 × 10−10 | FAM198B | −5.83 | 0.0003 |
| STAB1 | −5.01 | 1.04 × 10−13 | GTF2IP4 | −5.85 | 0.02 | DCANP1 | −5.69 | 1.10 × 10−05 |
| ATP6V0D2 | −4.98 | 6.54 × 10−07 | STAB1 | −5.80 | 4.66 × 10−21 | MSR1 | −5.66 | 0.0001 |
| CNTNAP2 | −4.91 | 2.72 × 10−15 | HNMT | −5.76 | 4.85 × 10−08 | CLEC10A | −5.62 | 6.39 × 10−10 |
| CLEC9A | −4.84 | 5.86 × 10−07 | A2M | −5.74 | 5.33 × 10−15 | F13A1 | −5.57 | 1.77 × 10−05 |
| F13A1 | −4.79 | 2.04 × 10−09 | CLEC10A | −5.66 | 5.39 × 10−10 | DNASE1L3 | −5.55 | 1.88 × 10−06 |
| APOE | −4.72 | 1.20 × 10−16 | DCANP1 | −5.56 | 4.06 × 10−06 | THBD | −5.53 | 0.001 |
| HS3ST2 | −4.72 | 0.0002 | LILRB5 | −5.37 | 1.51 × 10−07 | FABP4 | −5.42 | 0.002 |
| CD1D | −4.54 | 2.05 × 10−05 | PDK4 | −5.28 | 7.81 × 10−11 | LOC387810 | −5.41 | 0.0001 |
| NLRC4 | −4.44 | 1.59 × 10−05 | CLEC7A | −5.24 | 5.99 × 10−12 | UMODL1-AS1 | −5.32 | 3.56 × 10−05 |
| Pathway | −log(p) | Ratio | No Overlap with Dataset (Number, %) |
|---|---|---|---|
| Baseline | |||
| Kinetochore Metaphase Signalling Pathway | 14.4 | 0.43 | 51/101 (50%) |
| Hepatic Fibrosis/Hepatic Stellate Cell Activation | 11.5 | 0.31 | 115/186 (62%) |
| Cardiac Hypertrophy Signalling (Enhanced) | 11.1 | 0.22 | 343/497 (69%) |
| Th1 and Th2 Activation Pathway | 11 | 0.31 | 103/171 (60%) |
| STAT3 Pathway | 10.6 | 0.33 | 82/135 (61%) |
| HMGB1 Signalling | 10.0 | 0.30 | 105/165 (64%) |
| IL-10 Signalling | 9.6 | 0.41 | 37/70 (53%) |
| Role of BRCA1 in DNA Damage Response | 9.4 | 0.39 | 46/80 (57%) |
| Hepatic Cholestasis | 8.5 | 0.27 | 122/186 (66%) |
| IL-17 Signalling | 8.4 | 0.27 | 123/187 (66%) |
| Airway Pathology in Chronic Obstructive Pulmonary Disease | 8.0 | 0.31 | 66/118 (56%) |
| GADD45 Signalling | 8.0 | 0.68 | 5/19 (26%) |
| Molecular Mechanisms of Cancer | 7.9 | 0.22 | 280/400 (70%) |
| Breast Cancer Regulation by Stathmin1 | 7.8 | 0.20 | 431/590 (73%) |
| Cell Cycle Control of Chromosomal Replication | 7.7 | 0.41 | 30/56 (54%) |
| Aryl Hydrocarbon Receptor Signalling | 7.6 | 0.29 | 88/143 (62%) |
| Tumor Microenvironment Pathway | 7.5 | 0.27 | 113/176 (64%) |
| Hereditary Breast Cancer Signalling | 7.4 | 0.29 | 93/140 (66%) |
| Role of Cytokines in Mediating Communication between Immune Cells | 7.3 | 0.41 | 27/54 (50%) |
| Erythropoietin Signalling Pathway | 7.3 | 0.27 | 114/173 (66%) |
| Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis | 7.2 | 0.22 | 220/314 (70%) |
| PI3K/AKT Signalling | 6.9 | 0.26 | 129/184 (70%) |
| Th2 Pathway | 6.7 | 0.28 | 87/136 (64%) |
| Cell Cycle: G2/M DNA Damage Checkpoint Regulation | 6.7 | 0.41 | 26/49 (53%) |
| Oestrogen-Mediated S-phase Entry | 6.7 | 0.54 | 11/26 (42%) |
| Post-FO supplementation | |||
| Kinetochore Metaphase Signalling Pathway | 14.6 | 0.42 | 6/101 (6%) |
| STAT3 Pathway | 12.1 | 0.34 | 15/135 (11%) |
| Th1 and Th2 Activation Pathway | 10.3 | 0.29 | 21/171 (12%) |
| IL-10 Signalling | 10.2 | 0.41 | 9/70 (13%) |
| Cardiac Hypertrophy Signalling (Enhanced) | 9.8 | 0.21 | 103/497 (21%) |
| Hepatic Fibrosis/Hepatic Stellate Cell Activation | 9.4 | 0.27 | 60/186 (32%) |
| Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses | 9.3 | 0.29 | 35/154 (23%) |
| HMGB1 Signalling | 9.3 | 0.29 | 27/165 (16%) |
| Altered T Cell and B Cell Signalling in Rheumatoid Arthritis | 9.2 | 0.36 | 19/90 (21%) |
| IL-17 Signalling | 8.8 | 0.27 | 60/187 (32%) |
| Airway Pathology in Chronic Obstructive Pulmonary Disease | 8.7 | 0.31 | 46/118 (39%) |
| Hepatic Cholestasis | 8.4 | 0.26 | 50/186 (27%) |
| Cell Cycle Control of Chromosomal Replication | 8.2 | 0.41 | 0/56 (0%) |
| Oestrogen-Mediated S-phase Entry | 8.0 | 0.58 | 0/26 (0%) |
| Role of BRCA1 in DNA Damage Response | 7.9 | 0.35 | 2/80 (3%) |
| Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis | 7.9 | 0.22 | 62/314 (20%) |
| Aryl Hydrocarbon Receptor Signalling | 7.8 | 0.28 | 26/143 (18%) |
| Erythropoietin Signalling Pathway | 7.6 | 0.26 | 28/173 (16%) |
| Diff. Reg. of Cytokine Production in Macrophages and T Helper Cells by IL-17A and IL-17F | 7.5 | 0.67 | 1/18 (6%) |
| Molecular Mechanisms of Cancer | 7.4 | 0.20 | 45/400 (11%) |
| Crosstalk between Dendritic Cells and Natural Killer Cells | 7.4 | 0.33 | 11/89 (12%) |
| TREM1 Signalling | 7.3 | 0.35 | 13/75 (17%) |
| GADD45 Signalling | 7.1 | 0.63 | 0/19 (0%) |
| Role of Cytokines in Mediating Communication between Immune Cells | 7.0 | 0.39 | 25/54 (46%) |
| Protein Ubiquitination Pathway | 6.9 | 0.22 | 27/273 (10%) |
| Post tCSO supplementation | |||
| Kinetochore Metaphase Signalling Pathway | 14.4 | 0.43 | 6/101 (6%) |
| IL-10 Signalling | 11.1 | 0.44 | 9/70 (13%) |
| Th1 and Th2 Activation Pathway | 11 | 0.31 | 19/171 (11%) |
| Hepatic Fibrosis/Hepatic Stellate Cell Activation | 10.5 | 0.30 | 50/186 (27%) |
| Airway Pathology in Chronic Obstructive Pulmonary Disease | 10.4 | 0.35 | 42/118 (36%) |
| Hepatic Cholestasis | 9.9 | 0.29 | 46/186 (25%) |
| Diff. Reg. of Cytokine Production in Macrophages and T Helper Cells by IL-17A and IL-17F | 9.8 | 0.78 | 1/18 (6%) |
| Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses | 9.6 | 0.31 | 31/154 (20%) |
| STAT3 Pathway | 9.4 | 0.32 | 11/135 (8%) |
| IL-17 Signalling | 8.9 | 0.28 | 55/187 (29%) |
| Differential Regulation of Cytokine Production in Intestinal Epithelial Cells by IL-17A and IL-17F | 8.7 | 0.65 | 4/23 (17%) |
| Granulocyte Adhesion and Diapedesis | 8.2 | 0.28 | 67/173 (39%) |
| Role of Cytokines in Mediating Communication between Immune Cells | 8.0 | 0.43 | 22/54 (41%) |
| Role of BRCA1 in DNA Damage Response | 8.0 | 0.36 | 1/80 (1%) |
| HMGB1 Signalling | 8.0 | 0.28 | 24/165 (15%) |
| Altered T Cell and B Cell Signalling in Rheumatoid Arthritis | 7.9 | 0.34 | 20/90 (22%) |
| Airway Inflammation in Asthma | 7.9 | 0.53 | 3/32 (9%) |
| Cardiac Hypertrophy Signalling (Enhanced) | 7.8 | 0.20 | 92/497 (19%) |
| Systemic Lupus Erythematosus in B Cell Signalling Pathway | 7.8 | 0.24 | 43/275 (16%) |
| Th2 Pathway | 7.7 | 0.29 | 15/136 (11%) |
| Aryl Hydrocarbon Receptor Signalling | 7.6 | 0.29 | 23/143 (16%) |
| Cell Cycle: G2/M DNA Damage Checkpoint Regulation | 7.5 | 0.43 | 0/49 (0%) |
| PI3K/AKT Signalling | 7.3 | 0.26 | 12/184 (7%) |
| Erythropoietin Signalling Pathway | 7.3 | 0.27 | 23/173 (13%) |
| Role of Hypercytokinemia/Hyperchemokinemia in the Pathogenesis of Influenza | 7.2 | 0.34 | 27/86 (31%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
West, A.L.; Miles, E.A.; Han, L.; Lillycrop, K.A.; Napier, J.A.; Calder, P.C.; Burdge, G.C. Dietary Supplementation with Transgenic Camelina sativa Oil Containing 20:5n-3 and 22:6n-3 or Fish Oil Induces Differential Changes in the Transcriptome of CD3+ T Lymphocytes. Nutrients 2021, 13, 3116. https://doi.org/10.3390/nu13093116
West AL, Miles EA, Han L, Lillycrop KA, Napier JA, Calder PC, Burdge GC. Dietary Supplementation with Transgenic Camelina sativa Oil Containing 20:5n-3 and 22:6n-3 or Fish Oil Induces Differential Changes in the Transcriptome of CD3+ T Lymphocytes. Nutrients. 2021; 13(9):3116. https://doi.org/10.3390/nu13093116
Chicago/Turabian StyleWest, Annette L., Elizabeth A. Miles, Lihua Han, Karen A. Lillycrop, Johnathan A. Napier, Philip C. Calder, and Graham C. Burdge. 2021. "Dietary Supplementation with Transgenic Camelina sativa Oil Containing 20:5n-3 and 22:6n-3 or Fish Oil Induces Differential Changes in the Transcriptome of CD3+ T Lymphocytes" Nutrients 13, no. 9: 3116. https://doi.org/10.3390/nu13093116
APA StyleWest, A. L., Miles, E. A., Han, L., Lillycrop, K. A., Napier, J. A., Calder, P. C., & Burdge, G. C. (2021). Dietary Supplementation with Transgenic Camelina sativa Oil Containing 20:5n-3 and 22:6n-3 or Fish Oil Induces Differential Changes in the Transcriptome of CD3+ T Lymphocytes. Nutrients, 13(9), 3116. https://doi.org/10.3390/nu13093116






