A Synbiotic Formulation Comprising Bacillus subtilis DSM 32315 and L-Alanyl-L-Glutamine Improves Intestinal Butyrate Levels and Lipid Metabolism in Healthy Humans
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Vitro Colonic Screening Platform
2.1.1. Testing of Compounds in I-Screen
2.1.2. DNA Isolation
2.1.3. V4 16S rRNA Gene Sequencing
2.1.4. Short Chain Fatty Acid and Lactate Analysis
2.1.5. Data Analysis
2.2. Human Study
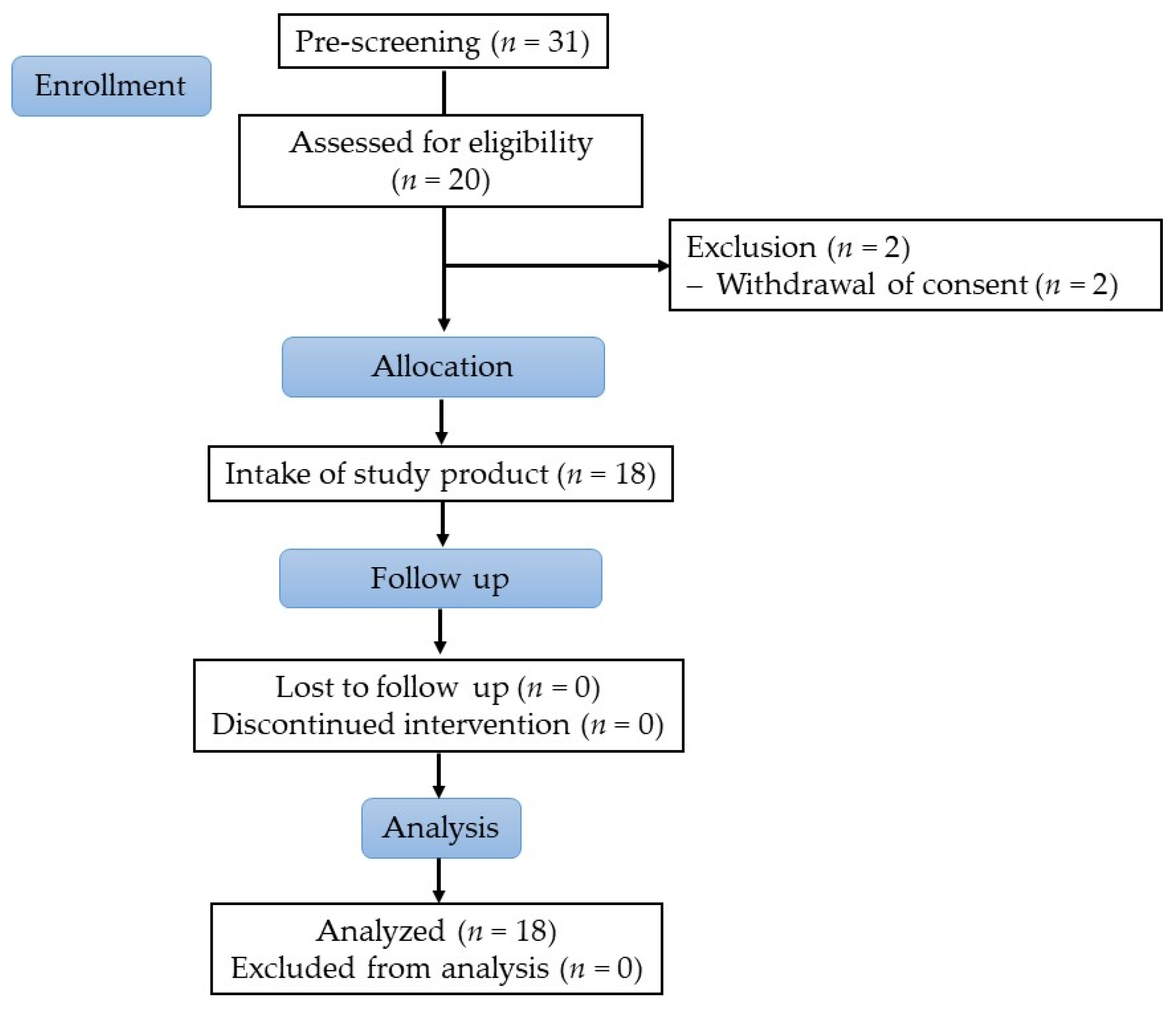
2.2.1. Study Subjects
2.2.2. Study Design
2.2.3. Study Product
2.2.4. Sampling and Assessments
DNA Extraction and Sequencing from Stool Samples
Bioinformatic Processing and Data Cleaning
2.2.5. Safety
2.2.6. Statistical Analysis
Clinical Parameters
Biostatistical Analyses of the Microbial Community Data
3. Results
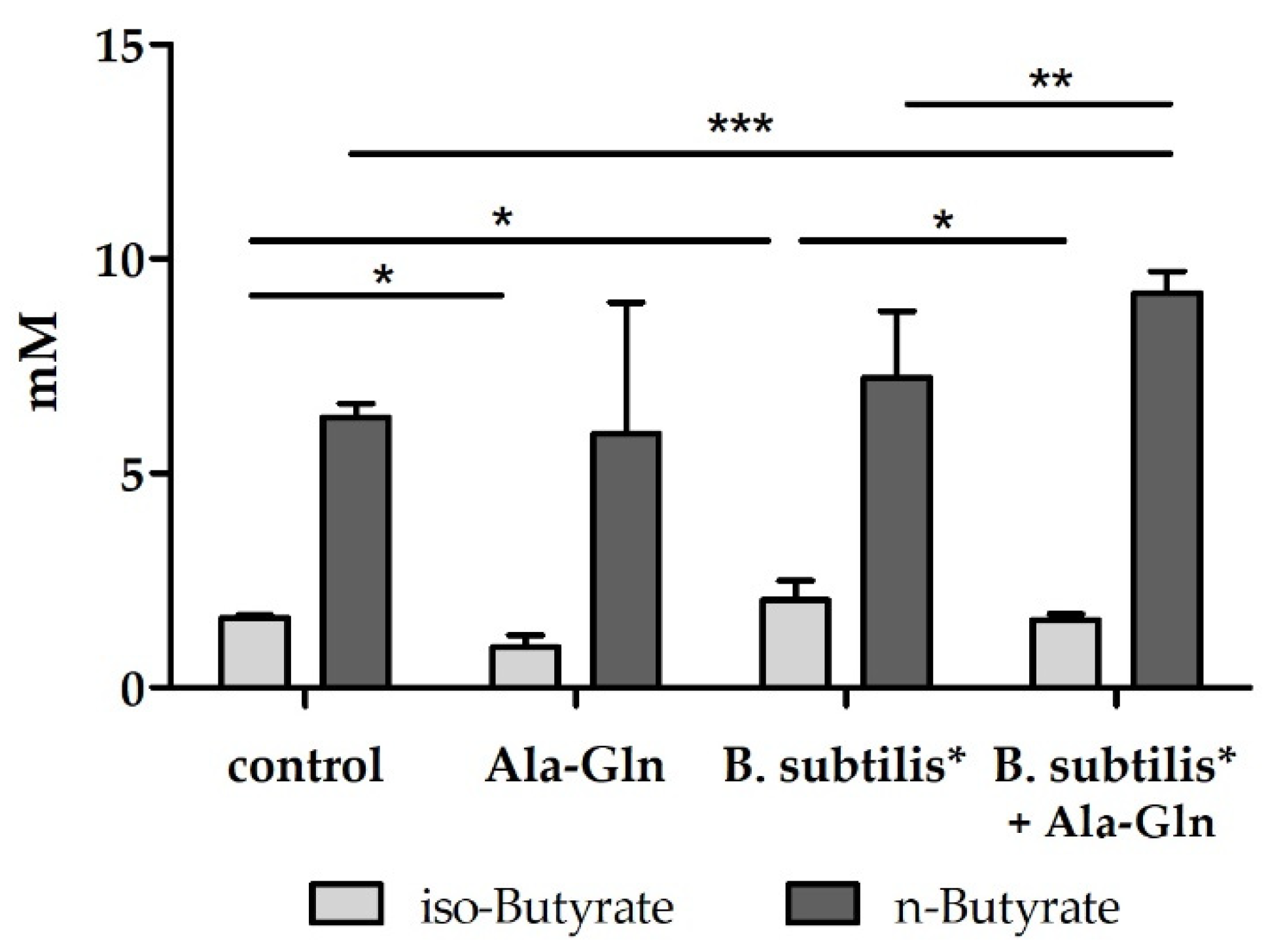
3.1. Effects of Amino Acids, Peptides and Bacillus Strains in an In Vitro Colonic Screening Platform
3.2. Human Study
3.2.1. Subject Characteristics
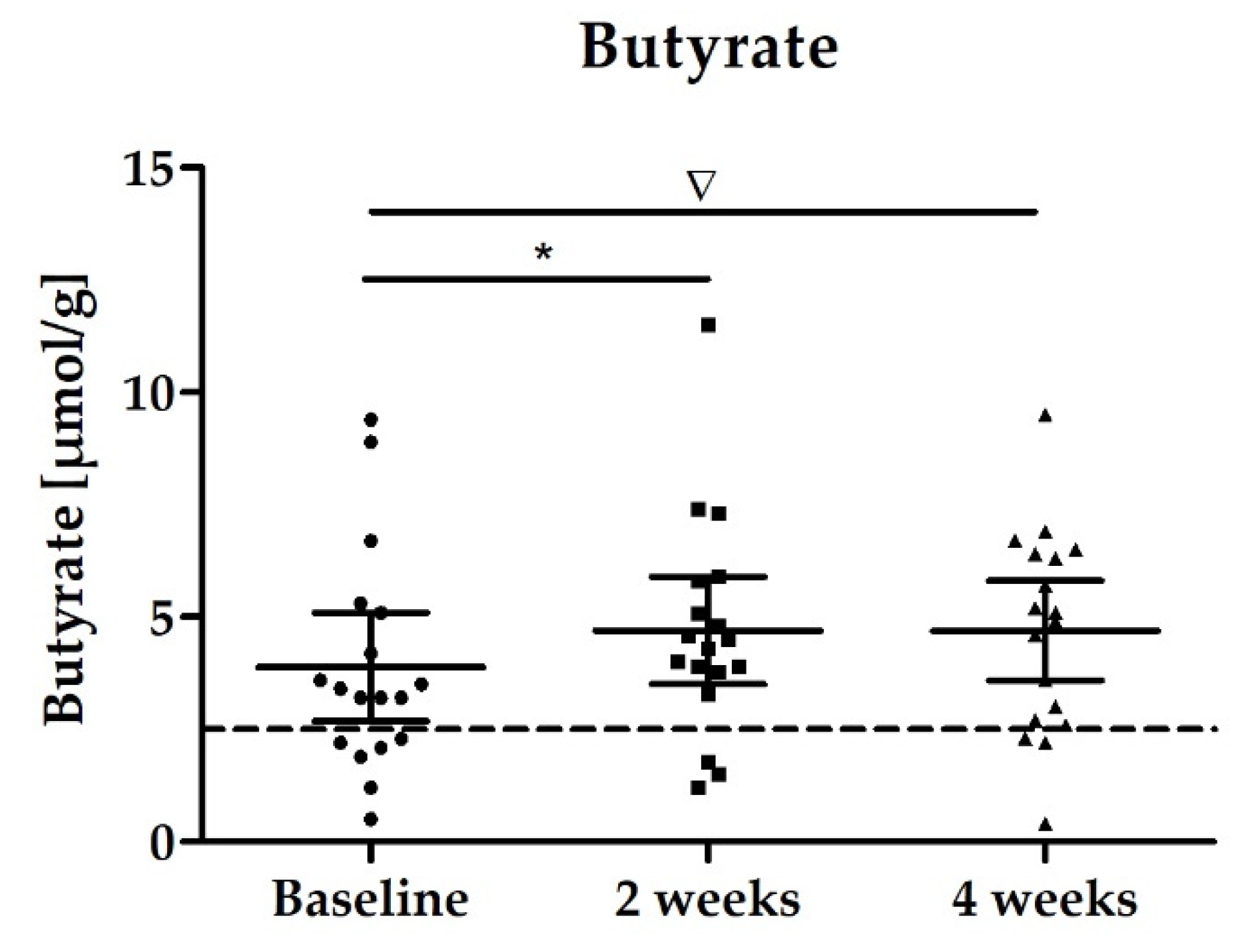
3.2.2. Stool Biomarker: SCFAs
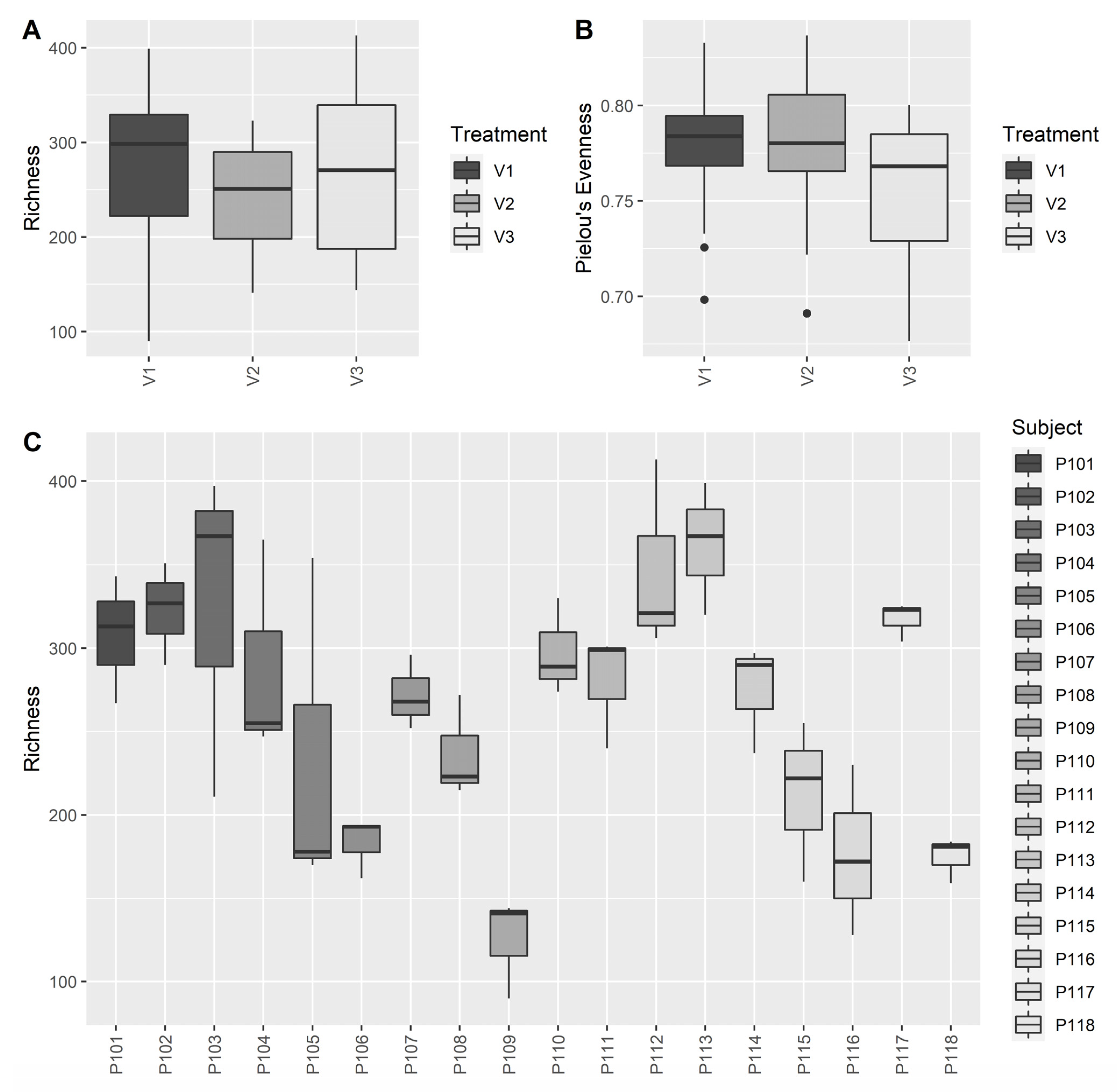
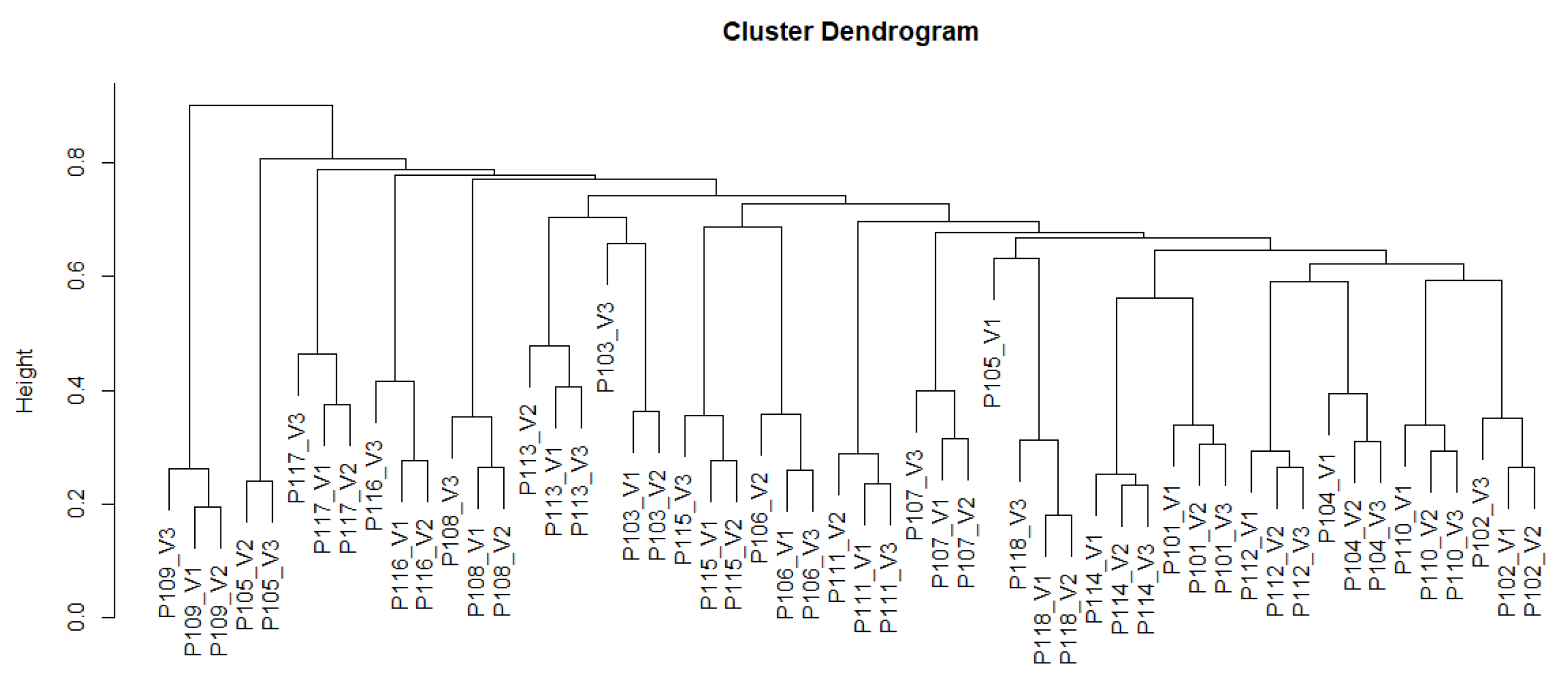
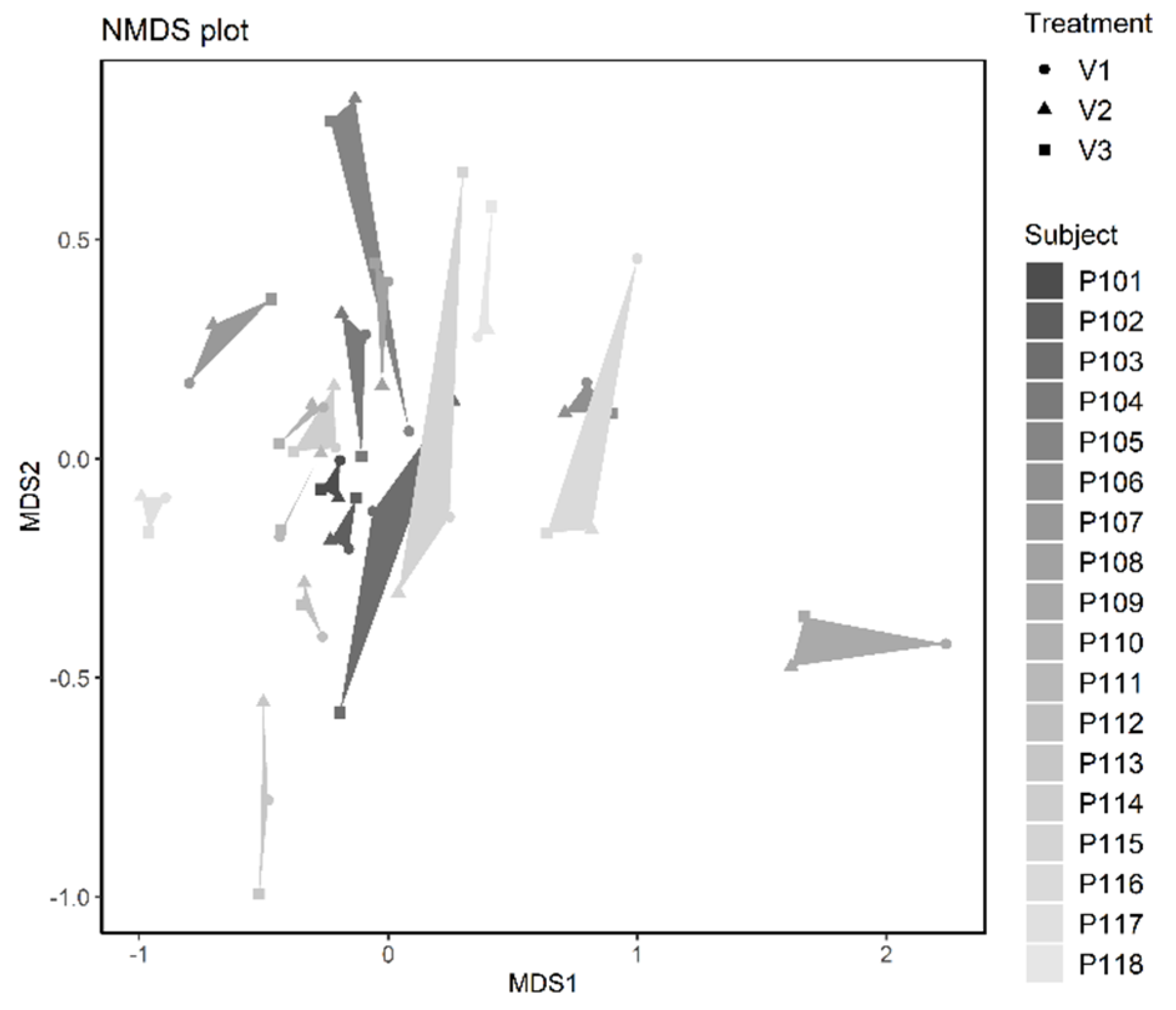
3.2.3. Microbiome Analysis
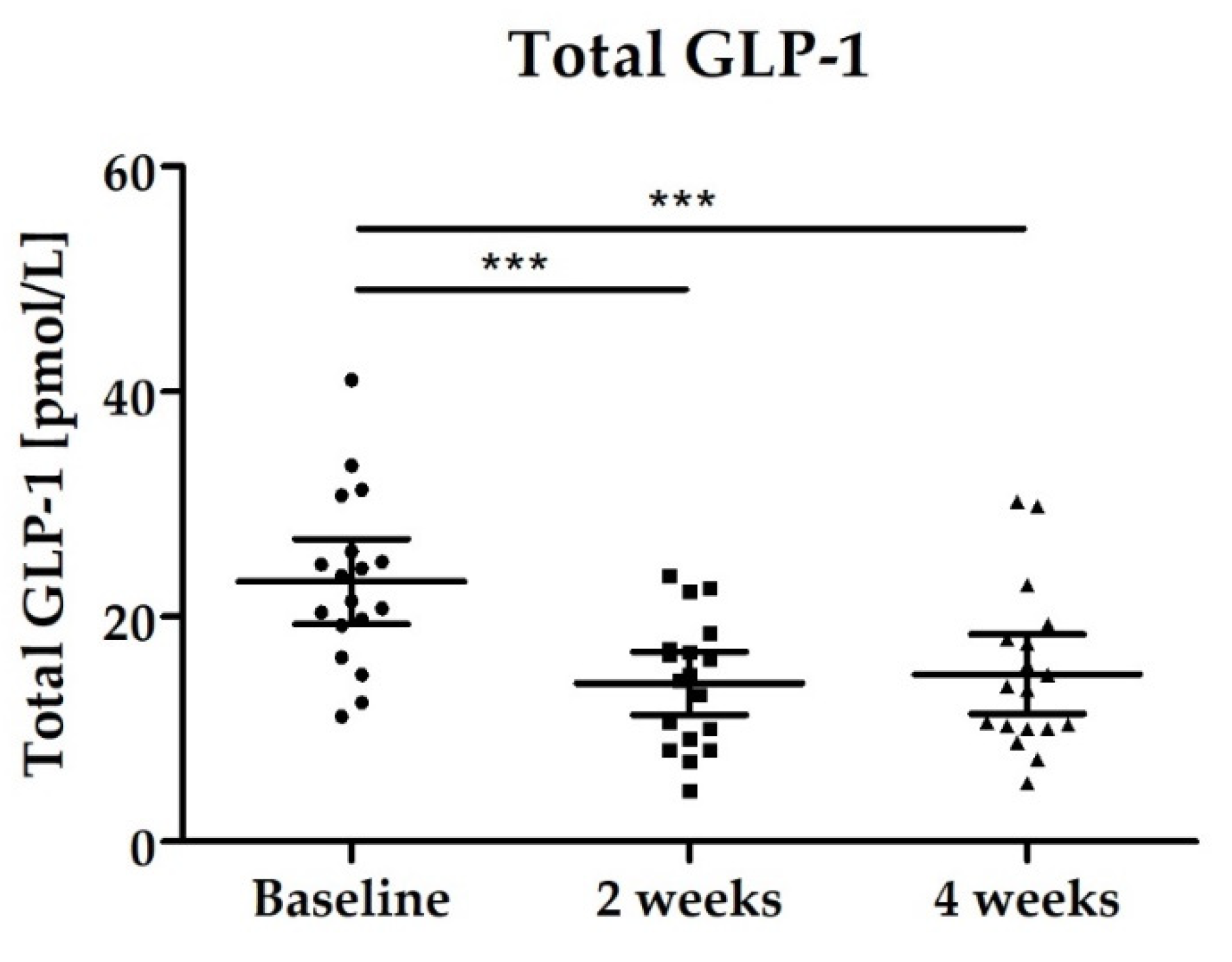
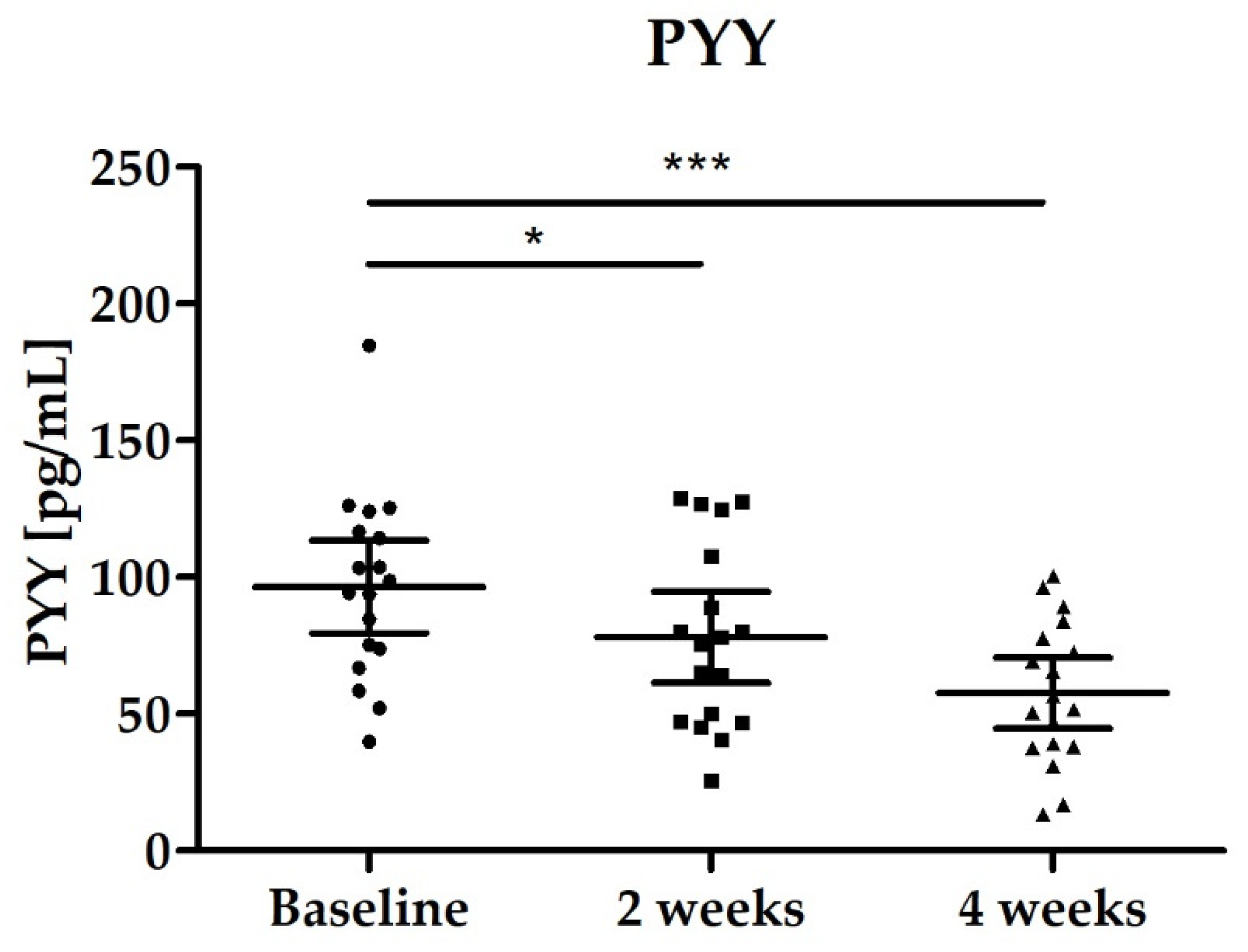
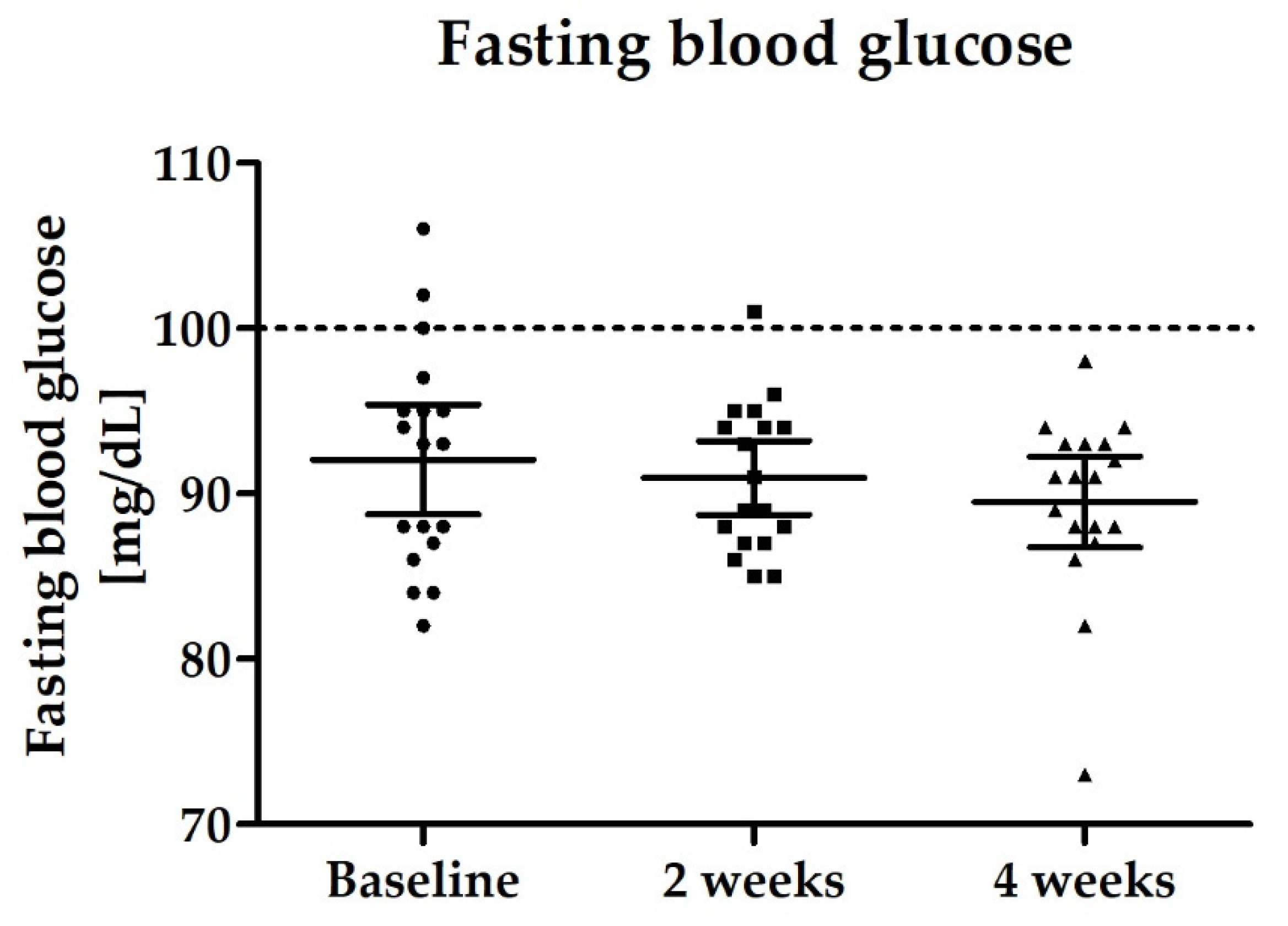
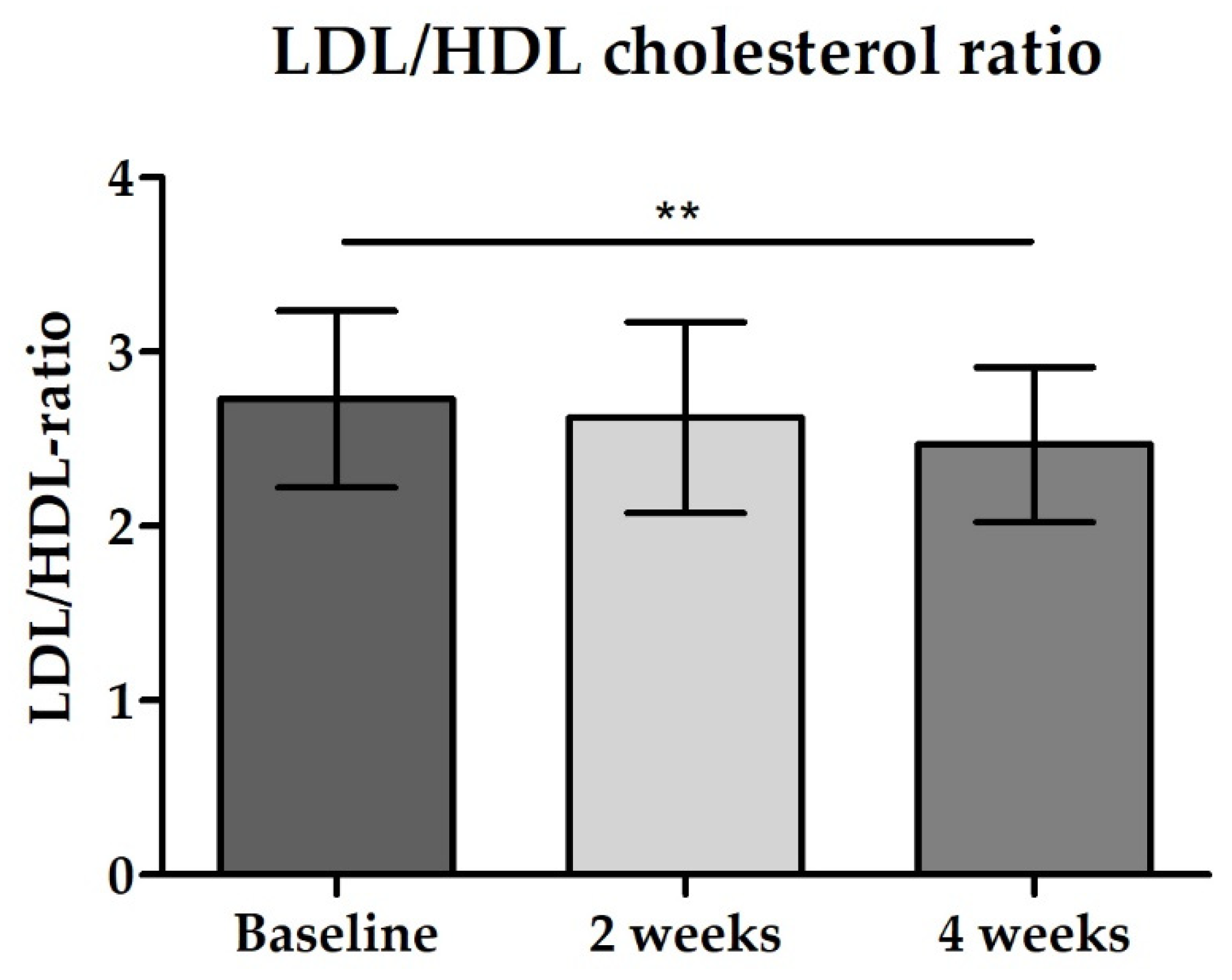
3.2.4. Blood Biomarkers: Total GLP-1, PYY, Fasting Blood Glucose and Lipid Status
3.2.5. Safety Assessment and Compliance
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gasaly, N.; Hermoso, M.A.; Gotteland, M. Butyrate and the Fine-Tuning of Colonic Homeostasis: Implication for Inflammatory Bowel Diseases. Int. J. Mol. Sci. 2021, 22, 3061. [Google Scholar] [CrossRef]
- Boets, E.; Gomand, S.V.; Deroover, L.; Preston, T.; Vermeulen, K.; De Preter, V.; Hamer, H.M.; Van den Mooter, G.; De Vuyst, L.; Courtin, C.M.; et al. Systemic availability and metabolism of colonic-derived short-chain fatty acids in healthy subjects: A stable isotope study. J. Physiol. 2017, 595, 541–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nogal, A.; Valdes, A.M.; Menni, C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes 2021, 13, 1897212. [Google Scholar] [CrossRef]
- Salamone, D.; Rivellese, A.A.; Vetrani, C. The relationship between gut microbiota, short-chain fatty acids and type 2 diabetes mellitus: The possible role of dietary fibre. Acta Diabetol. 2021, 58, 1131–1138. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.K.; Abraham, B.; El-Serag, H. Dietary intake and risk of developing inflammatory bowel disease: A systematic review of the literature. Am. J. Gastroenterol. 2011, 106, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Vernia, P.; Annese, V.; Bresci, G.; d’Albasio, G.; D’Inca, R.; Giaccari, S.; Ingrosso, M.; Mansi, C.; Riegler, G.; Valpiani, D.; et al. Topical butyrate improves efficacy of 5-ASA in refractory distal ulcerative colitis: Results of a multicentre trial. Eur. J. Clin. Investig. 2003, 33, 244–248. [Google Scholar] [CrossRef]
- Roda, A.; Simoni, P.; Magliulo, M.; Nanni, P.; Baraldini, M.; Roda, G.; Roda, E. A new oral formulation for the release of sodium butyrate in the ileo-cecal region and colon. World J. Gastroenterol. 2007, 13, 1079–1084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaminski, M.; Skonieczna-Zydecka, K.; Nowak, J.K.; Stachowska, E. Global and local diet popularity rankings, their secular trends, and seasonal variation in Google Trends data. Nutrition 2020, 79–80, 110759. [Google Scholar] [CrossRef] [PubMed]
- Halmos, T.; Suba, I. Physiological patterns of intestinal microbiota. The role of dysbacteriosis in obesity, insulin resistance, diabetes and metabolic syndrome. Orv. Hetil. 2016, 157, 13–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boucher, J.; Kleinridders, A.; Kahn, C.R. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb. Perspect. Biol. 2014, 6, a009191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vital, M.; Karch, A.; Pieper, D.H. Colonic Butyrate-Producing Communities in Humans: An Overview Using Omics Data. mSystems 2017, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perraudeau, F.; McMurdie, P.; Bullard, J.; Cheng, A.; Cutcliffe, C.; Deo, A.; Eid, J.; Gines, J.; Iyer, M.; Justice, N.; et al. Improvements to postprandial glucose control in subjects with type 2 diabetes: A multicenter, double blind, randomized placebo-controlled trial of a novel probiotic formulation. BMJ Open Diabetes Res. Care 2020, 8. [Google Scholar] [CrossRef]
- Stoeva, M.K.; Garcia-So, J.; Justice, N.; Myers, J.; Tyagi, S.; Nemchek, M.; McMurdie, P.J.; Kolterman, O.; Eid, J. Butyrate-Producing human gut symbiont, Clostridium butyricum, and its role in health and disease. Gut Microbes 2021, 13, 1907272. [Google Scholar] [CrossRef] [PubMed]
- Boesmans, L.; Valles-Colomer, M.; Wang, J.; Eeckhaut, V.; Falony, G.; Ducatelle, R.; Van Immerseel, F.; Raes, J.; Verbeke, K. Butyrate Producers as Potential Next-Generation Probiotics: Safety Assessment of the Administration of Butyricicoccus pullicaecorum to Healthy Volunteers. mSystems 2018, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duysburgh, C.; Van den Abbeele, P.; Krishnan, K.; Bayne, T.F.; Marzorati, M. A synbiotic concept containing spore-forming Bacillus strains and a prebiotic fiber blend consistently enhanced metabolic activity by modulation of the gut microbiome in vitro. Int. J. Pharm. X 2019, 1, 100021. [Google Scholar] [CrossRef]
- Xu, Y.; Yu, Y.; Shen, Y.; Li, Q.; Lan, J.; Wu, Y.; Zhang, R.; Cao, G.; Yang, C. Effects of Bacillus subtilis and Bacillus licheniformis on growth performance, immunity, short chain fatty acid production, antioxidant capacity, and cecal microflora in broilers. Poult. Sci. 2021, 100, 101358. [Google Scholar] [CrossRef]
- Sokale, A.O.; Menconi, A.; Mathis, G.F.; Lumpkins, B.; Sims, M.D.; Whelan, R.A.; Doranalli, K. Effect of Bacillus subtilis DSM 32315 on the intestinal structural integrity and growth performance of broiler chickens under necrotic enteritis challenge. Poult. Sci. 2019, 98, 5392–5400. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, W.; Zhang, H.; Wang, J.; Zhang, W.; Gao, J.; Wu, S.; Qi, G. Supplemental Bacillus subtilis DSM 32315 manipulates intestinal structure and microbial composition in broiler chickens. Sci. Rep. 2018, 8, 15358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ladirat, S.E.; Schols, H.A.; Nauta, A.; Schoterman, M.H.; Keijser, B.J.; Montijn, R.C.; Gruppen, H.; Schuren, F.H. High-throughput analysis of the impact of antibiotics on the human intestinal microbiota composition. J. Microbiol. Methods 2013, 92, 387–397. [Google Scholar] [CrossRef]
- Eren, A.M.; Maignien, L.; Sul, W.J.; Murphy, L.G.; Grim, S.L.; Morrison, H.G.; Sogin, M.L. Oligotyping: Differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol. Evol. 2013, 4. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-Resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; R Version 3.6.3, accessed on 29 February 2020, Version 4.0.3, accessed on 10 October 2020. Available online: https://www.R-project.org/ (accessed on 10 October 2020).
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Pankoke, H.; Maus, I.; Loh, G.; Huser, A.; Seifert, J.; Tilker, A.; Hark, S.; Sczyrba, A.; Pelzer, S.; Kleinbolting, J. F5Evaluation of commercially available DNA extraction kits for the analysis of the broiler chicken cecal microbiota. FEMS Microbiol. Lett. 2019. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Cohen, R. Weighted false discovery rate controlling procedures for clinical trials. Biostatistics 2017, 18, 91–104. [Google Scholar] [CrossRef] [Green Version]
- Rios-Covian, D.; Gonzalez, S.; Nogacka, A.M.; Arboleya, S.; Salazar, N.; Gueimonde, M.; de Los Reyes-Gavilan, C.G. An Overview on Fecal Branched Short-Chain Fatty Acids Along Human Life and as Related With Body Mass Index: Associated Dietary and Anthropometric Factors. Front. Microbiol. 2020, 11, 973. [Google Scholar] [CrossRef]
- Rios-Covian, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; de Los Reyes-Gavilan, C.G.; Salazar, N. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef] [Green Version]
- Riviere, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arora, T.; Tremaroli, V. Therapeutic Potential of Butyrate for Treatment of Type 2 Diabetes. Front. Endocrinol. 2021, 12, 761834. [Google Scholar] [CrossRef] [PubMed]
- Elshaghabee, F.M.F.; Rokana, N.; Gulhane, R.D.; Sharma, C.; Panwar, H. Bacillus As Potential Probiotics: Status, Concerns, and Future Perspectives. Front. Microbiol. 2017, 8, 1490. [Google Scholar] [CrossRef] [Green Version]
- Tsukuda, N.; Yahagi, K.; Hara, T.; Watanabe, Y.; Matsumoto, H.; Mori, H.; Higashi, K.; Tsuji, H.; Matsumoto, S.; Kurokawa, K.; et al. Key bacterial taxa and metabolic pathways affecting gut short-chain fatty acid profiles in early life. ISME J. 2021, 15, 2574–2590. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Falcone, G.; Salvatore, G.; Covelli, I. Mechanism of induction of spore germination in Bacillus subtilis by L-alanine and hydrogen peroxide. Biochim. Biophys. Acta 1959, 36, 390–397. [Google Scholar] [CrossRef]
- Pan, Y.Q.; Zheng, Q.X.; Jiang, X.M.; Chen, X.Q.; Zhang, X.Y.; Wu, J.L. Probiotic Supplements Improve Blood Glucose and Insulin Resistance/Sensitivity among Healthy and GDM Pregnant Women: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Evid. Based Complement. Alternat. Med. 2021, 2021, 9830200. [Google Scholar] [CrossRef]
- Wu, H.; Tremaroli, V.; Schmidt, C.; Lundqvist, A.; Olsson, L.M.; Kramer, M.; Gummesson, A.; Perkins, R.; Bergstrom, G.; Backhed, F. The Gut Microbiota in Prediabetes and Diabetes: A Population-Based Cross-Sectional Study. Cell Metab. 2020, 32, 379–390.e3. [Google Scholar] [CrossRef]
- Zhang, X.; Shen, D.; Fang, Z.; Jie, Z.; Qiu, X.; Zhang, C.; Chen, Y.; Ji, L. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS ONE 2013, 8, e71108. [Google Scholar] [CrossRef]
- Ojo, O.; Feng, Q.Q.; Ojo, O.O.; Wang, X.H. The Role of Dietary Fibre in Modulating Gut Microbiota Dysbiosis in Patients with Type 2 Diabetes: A Systematic Review and Meta-Analysis of Randomised Controlled Trials. Nutrients 2020, 12, 3239. [Google Scholar] [CrossRef] [PubMed]
- Deehan, E.C.; Yang, C.; Perez-Munoz, M.E.; Nguyen, N.K.; Cheng, C.C.; Triador, L.; Zhang, Z.; Bakal, J.A.; Walter, J. Precision Microbiome Modulation with Discrete Dietary Fiber Structures Directs Short-Chain Fatty Acid Production. Cell Host Microbe 2020, 27, 389–404.e6. [Google Scholar] [CrossRef] [PubMed]
- Palacios, T.; Vitetta, L.; Coulson, S.; Madigan, C.D.; Lam, Y.Y.; Manuel, R.; Briskey, D.; Hendy, C.; Kim, J.N.; Ishoey, T.; et al. Targeting the Intestinal Microbiota to Prevent Type 2 Diabetes and Enhance the Effect of Metformin on Glycaemia: A Randomised Controlled Pilot Study. Nutrients 2020, 12, 2041. [Google Scholar] [CrossRef]
- Faerch, K.; Torekov, S.S.; Vistisen, D.; Johansen, N.B.; Witte, D.R.; Jonsson, A.; Pedersen, O.; Hansen, T.; Lauritzen, T.; Sandbaek, A.; et al. GLP-1 Response to Oral Glucose Is Reduced in Prediabetes, Screen-Detected Type 2 Diabetes, and Obesity and Influenced by Sex: The ADDITION-PRO Study. Diabetes 2015, 64, 2513–2525. [Google Scholar] [CrossRef] [Green Version]
- Seon, M.J.; Hwang, S.Y.; Son, Y.; Song, J.; Kim, O.Y. Circulating GLP-1 Levels as a Potential Indicator of Metabolic Syndrome Risk in Adult Women. Nutrients 2021, 13, 865. [Google Scholar] [CrossRef]
- Adam, T.C.; Jocken, J.; Westerterp-Plantenga, M.S. Decreased glucagon-like peptide 1 release after weight loss in overweight/obese subjects. Obes. Res. 2005, 13, 710–716. [Google Scholar] [CrossRef] [Green Version]
- De Luis, D.A.; Gonzalez Sagrado, M.; Conde, R.; Aller, R.; Izaola, O. Decreased basal levels of glucagon-like peptide-1 after weight loss in obese subjects. Ann. Nutr. Metab. 2007, 51, 134–138. [Google Scholar] [CrossRef]
- He, J.; Zhang, F.; Han, Y. Effect of probiotics on lipid profiles and blood pressure in patients with type 2 diabetes: A meta-analysis of RCTs. Medicine 2017, 96, e9166. [Google Scholar] [CrossRef] [PubMed]
- Mo, R.; Zhang, X.; Yang, Y. Effect of probiotics on lipid profiles in hypercholesterolaemic adults: A meta-analysis of randomized controlled trials. Med. Clin. 2019, 152, 473–481. [Google Scholar] [CrossRef]
- Costabile, A.; Buttarazzi, I.; Kolida, S.; Quercia, S.; Baldini, J.; Swann, J.R.; Brigidi, P.; Gibson, G.R. An in vivo assessment of the cholesterol-lowering efficacy of Lactobacillus plantarum ECGC 13110402 in normal to mildly hypercholesterolaemic adults. PLoS ONE 2017, 12, e0187964. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Nagpal, R.; Kumar, R.; Hemalatha, R.; Verma, V.; Kumar, A.; Chakraborty, C.; Singh, B.; Marotta, F.; Jain, S.; et al. Cholesterol-lowering probiotics as potential biotherapeutics for metabolic diseases. Exp. Diabetes Res. 2012, 2012, 902917. [Google Scholar] [CrossRef] [Green Version]
| Variable | Study Population (n = 18) |
|---|---|
| Mean (95% CI) | |
| Age [years] | 31.7 (29.5–33.9) |
| BMI [kg/m²] | 24.8 (23.5–26.1) |
| Cholesterol [mg/dL] | 180.2 (166.9–193.4) |
| LDL-cholesterol [mg/dL] | 123.9 (106.1–141.7) |
| GPT [U/L] | 27.7 (21.8–33.6) |
| GOT [U/L] | 19.3 (16.9–21.8) |
| Fasting blood glucose [mg/dL] | 92.9 (90.4–95.4) |
| Calorie intake | 3005 (1738–4272) |
| Fiber intake [g/d] | 18.5 (14.6–22.38) |
| Stool frequency [stools/week] | 7.2 (5.3–9.0) |
| Stool consistency [Bristol] | 3.7 (3.3–4.2) |
| Systolic blood pressure | 130 (125–136) |
| Diastolic blood pressure | 77 (74–81) |
| Variables | Baseline | 2 Weeks | 4 Weeks |
|---|---|---|---|
| Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | |
| Total SCFA [µmol/g] | 34.72 (28.55–40.88) | 37.55 (31.22–43.88) | 35.06 (29.23–40.88) |
| Butyrate [µmol/g] | 3.88 (2.68–5.09) | 4.70 (3.50–5.90) p = 0.0278 * | 4.70 (3.59–5.81) p = 0.0728 * |
| Propionate [µmol/g] | 6.16 (4.51–7.81) | 6.45 (4.55–8.35) | 6.24 (4.71–7.78) |
| Acetate [µmol/g] | 24.64 (20.70–28.58) | 26.36 (22.21–30.51) | 24.12 (20.09–28.15) |
| Variables | Baseline | 2 Weeks | 4 Weeks |
|---|---|---|---|
| Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | |
| Total GLP-1 [pmol/L] | 23.11 (19.33–26.88) | 14.09 (11.29–16.89) p < 0.0001 | 14.89 (11.37–18.42) p < 0.0001 |
| PYY [pmol/L] | 96.44 (79.53–113.40) | 78.04 (61.43–94.65) p = 0.0139 | 57.52 (44.60–70.44) p < 0.0001 |
| Variables | Baseline | 2 Weeks | 4 Weeks |
|---|---|---|---|
| Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | |
| Total cholesterol [mg/dL] | 179.3 (164.5–194.2) | 172.4 (157.5–187.4) | 169.10 (155.5–182.7) p = 0.0037 * |
| LDL cholesterol [mg/dL] | 120.6 (103.4–137.7) | 117.80 (100.3–135.3) | 113.10 (95.0–131.2) p = 0.0313 * |
| HDL cholesterol [mg/dL] | 46.1 (41.9–50.2) | 46.9 (42.7–51.2) | 47.3 (42.6–52.1) |
| Triglycerides [mg/dL] | 116.3 (95.0–137.5) | 119.4 (98.7–140.2) | 124.2 (89.1–159.4) |
| Fasting blood glucose [mg/dL) | 92.1 (88.7–95.4) | 90.9 (88.7–93.2) | 89.5 (86.8–92.2) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
tom Dieck, H.; Schön, C.; Wagner, T.; Pankoke, H.C.; Fluegel, M.; Speckmann, B. A Synbiotic Formulation Comprising Bacillus subtilis DSM 32315 and L-Alanyl-L-Glutamine Improves Intestinal Butyrate Levels and Lipid Metabolism in Healthy Humans. Nutrients 2022, 14, 143. https://doi.org/10.3390/nu14010143
tom Dieck H, Schön C, Wagner T, Pankoke HC, Fluegel M, Speckmann B. A Synbiotic Formulation Comprising Bacillus subtilis DSM 32315 and L-Alanyl-L-Glutamine Improves Intestinal Butyrate Levels and Lipid Metabolism in Healthy Humans. Nutrients. 2022; 14(1):143. https://doi.org/10.3390/nu14010143
Chicago/Turabian Styletom Dieck, Heike, Christiane Schön, Tanja Wagner, Helga Carola Pankoke, Monika Fluegel, and Bodo Speckmann. 2022. "A Synbiotic Formulation Comprising Bacillus subtilis DSM 32315 and L-Alanyl-L-Glutamine Improves Intestinal Butyrate Levels and Lipid Metabolism in Healthy Humans" Nutrients 14, no. 1: 143. https://doi.org/10.3390/nu14010143
APA Styletom Dieck, H., Schön, C., Wagner, T., Pankoke, H. C., Fluegel, M., & Speckmann, B. (2022). A Synbiotic Formulation Comprising Bacillus subtilis DSM 32315 and L-Alanyl-L-Glutamine Improves Intestinal Butyrate Levels and Lipid Metabolism in Healthy Humans. Nutrients, 14(1), 143. https://doi.org/10.3390/nu14010143






