The Relationship between Blood Lipids and Risk of Atrial Fibrillation: Univariable and Multivariable Mendelian Randomization Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Resources and Study Design
2.2. Selection of Genetic Instrumental Variables
2.3. Statistical Analysis
2.4. Sensitivity Analysis
3. Results
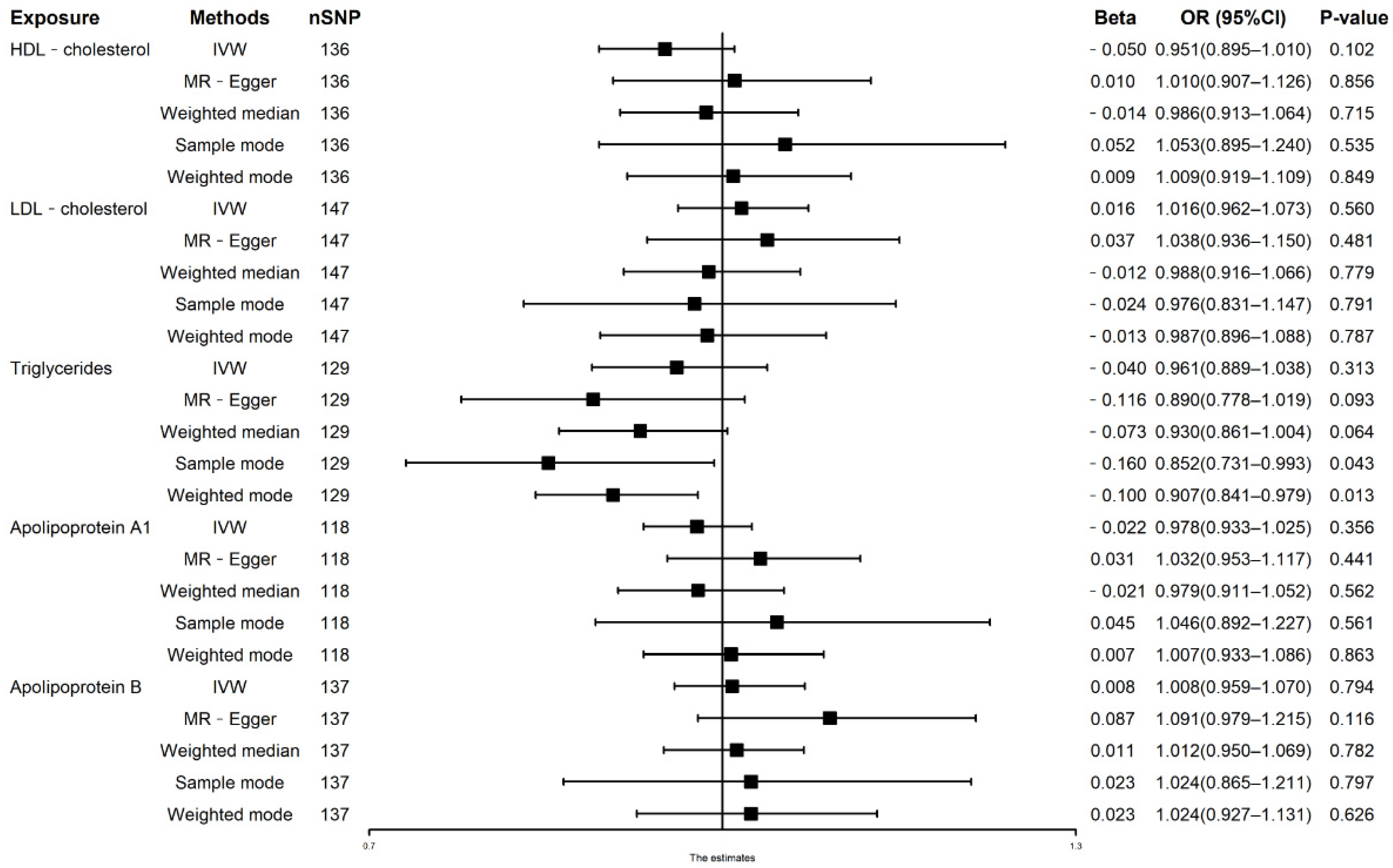
3.1. Univariable MR Analysis of Lipid Traits on AF Risks
3.2. Multivariable MR Analysis in Model 1
3.3. Multivariable MR Analysis in Model 2
3.4. Multivariable MR Analysis in Model 3
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Chung, M.K.; Refaat, M.; Shen, W.K.; Kutyifa, V.; Cha, Y.M.; Di Biase, L.; Baranchuk, A.; LampertC, R.; Natale, A.; Fisher, J.; et al. Atrial Fibrillation: JACC Council Perspectives. J. Am. Coll. Cardiol. 2020, 75, 1689–1713. [Google Scholar] [CrossRef]
- Benjamin, E.J.; Muntner, P.; Alonso, A.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Das, S.R.; et al. Heart Disease and Stroke Statistics-2019 Update: A Report From the American Heart Association. Circulation 2019, 13, e56–e528. [Google Scholar] [CrossRef]
- Georgiopoulos, G.; Ntritsos, G.; Stamatelopoulos, K.; Tsioufis, C.; Aimo, A.; Masi, S.; Evangelou, E. The relationship between blood pressure and risk of atrial fibrillation: A Mendelian randomization study. Eur. J. Prev. Cardiol. 2021, 28, 1617. [Google Scholar] [CrossRef] [PubMed]
- Wijesurendra, R.S.; Casadei, B. Mechanisms of atrial fibrillation. Heart 2019, 105, 1860–1867. [Google Scholar] [CrossRef]
- Lüscher, T.F. Frontiers in lipid research: Lipoprotein(a), apolipoprotein C-III and E, and PCSK9 and inflammation. Eur. Heart J. 2019, 40, 2741–2744. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Tanabe, N.; Yagihara, N.; Watanabe, T.; Aizawa, Y.; Kodama, M. Association between lipid profile and risk of atrial fibrillation. Circ. J. 2011, 75, 2767–2774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez, F.L.; Agarwal, S.K.; Maclehose, R.F.; Soliman, E.Z.; Sharrett, A.R.; Huxley, R.R.; Konety, S.; Ballantyne, C.M.; Alonso, A. Blood lipid levels, lipid-lowering medications, and the incidence of atrial fibrillation: The atherosclerosis risk in communities study. Circ. Arrhythm. Electrophysiol. 2012, 5, 155–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Psaty, B.M.; Manolio, T.A.; Kuller, L.H.; Kronmal, R.A.; Cushman, M.; Fried, L.P.; White, R.; Furberg, C.D.; Rautaharju, P.M. Incidence of and risk factors for atrial fibrillation in older adults. Circulation 1997, 96, 2455–2461. [Google Scholar] [CrossRef] [PubMed]
- Iguchi, Y.; Kimura, K.; Shibazaki, K.; Aoki, J.; Kobayashi, K.; Sakai, K.; Sakamoto, Y. Annual incidence of atrial fibrillation and related factors in adults. Am. J. Cardiol. 2010, 106, 1129–1133. [Google Scholar] [CrossRef]
- Mora, S.; Akinkuolie, A.O.; Sandhu, R.K.; Conen, D.; Albert, C.M. Paradoxical association of lipoprotein measures with incident atrial fibrillation. Circ. Arrhythm. Electrophysiol. 2014, 7, 612–619. [Google Scholar] [CrossRef] [Green Version]
- Alonso, A.; Yin, X.; Roetker, N.S.; Magnani, J.W.; Kronmal, R.A.; Ellinor, P.T.; Chen, L.Y.; Lubitz, S.A.; McClelland, R.L.; McManus, D.D.; et al. Blood lipids and the incidence of atrial fibrillation: The Multi-Ethnic Study of Atherosclerosis and the Framingham Heart Study. J. Am. Heart Assoc. 2014, 3, e001211. [Google Scholar] [CrossRef] [Green Version]
- Pol, T.; Held, C.; Westerbergh, J.; Lindbäck, J.; Alexander, J.H.; Alings, M.; Erol, C.; Goto, S.; Halvorsen, S.; Huber, K.; et al. Dyslipidemia and Risk of Cardiovascular Events in Patients With Atrial Fibrillation Treated With Oral Anticoagulation Therapy: Insights From the ARISTOTLE (Apixaban for Reduction in Stroke and Other Thromboembolic Events in Atrial Fibrillation) Trial. J. Am. Heart Assoc. 2018, 7, e007444. [Google Scholar] [CrossRef] [Green Version]
- Guan, B.; Li, X.; Xue, W.; Tse, G.; Waleed, K.B.; Liu, Y.; Zheng, M.; Wu, S.; Xia, Y.; Ding, Y. Blood lipid profiles and risk of atrial fibrillation: A systematic review and meta-analysis of cohort studies. J. Clin. Lipidol. 2020, 14, 133–142.e133. [Google Scholar] [CrossRef]
- Yao, Y.; Liu, F.; Wang, Y.; Liu, Z. Lipid levels and risk of new-onset atrial fibrillation: A systematic review and dose-response meta-analysis. Clin. Cardiol. 2020, 43, 935–943. [Google Scholar] [CrossRef]
- Emdin, C.A.; Khera, A.V.; Kathiresan, S. Mendelian Randomization. JAMA 2017, 318, 1925–1926. [Google Scholar] [CrossRef]
- Sheehan, N.A.; Didelez, V.; Burton, P.R.; Tobin, M.D. Mendelian randomisation and causal inference in observational epidemiology. PLoS Med. 2008, 5, e177. [Google Scholar] [CrossRef] [Green Version]
- Yuan, S.; Tang, B.; Zheng, J.; Larsson, S.C. Circulating Lipoprotein Lipids, Apolipoproteins and Ischemic Stroke. Ann. Neurol. 2020, 88, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Sinnott-Armstrong, N.; Tanigawa, Y.; Amar, D.; Mars, N.; Benner, C.; Aguirre, M.; Venkataraman, G.R.; Wainberg, M.; Ollila, H.M.; Kiiskinen, T.; et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet. 2021, 53, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Collins, R. What makes UK Biobank special? Lancet 2012, 379, 1173–1174. [Google Scholar] [CrossRef]
- Nielsen, J.B.; Thorolfsdottir, R.B.; Fritsche, L.G.; Zhou, W.; Skov, M.W.; Graham, S.E.; Herron, T.J.; McCarthy, S.; Schmidt, E.M.; Sveinbjornsson, G.; et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet. 2018, 50, 1234–1239. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, S.; Kim, Y.; Lee, Y.; Kang, M.W.; Kim, K.; Kim, Y.C.; Han, S.S.; Lee, H.H.; Lee, J.P.; et al. Atrial fibrillation and kidney function: A bidirectional Mendelian randomization study. Eur. Heart J. 2021, 42, 2816–2823. [Google Scholar] [CrossRef] [PubMed]
- Staley, J.R.; Blackshaw, J.; Kamat, M.A.; Ellis, S.; Surendran, P.; Sun, B.B.; Paul, D.S.; Freitag, D.; Burgess, S.; Danesh, J.; et al. PhenoScanner: A database of human genotype-phenotype associations. Bioinformatics 2016, 32, 3207–3209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Ferreira, T.; Morris, A.P.; Medland, S.E.; Madden, P.A.; Heath, A.C.; Martin, N.G.; Montgomery, G.W.; Weedon, M.N.; Loos, R.J.; et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012, 44, 361–363, 369–375. [Google Scholar] [CrossRef]
- Holmes, M.V.; Ala-Korpela, M.; Smith, G.D. Mendelian randomization in cardiometabolic disease: Challenges in evaluating causality. Nat. Rev. Cardiol. 2017, 14, 577–590. [Google Scholar] [CrossRef] [Green Version]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef] [Green Version]
- Hartwig, F.P.; Davey Smith, G.; Bowden, J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 2017, 46, 1985–1998. [Google Scholar] [CrossRef] [Green Version]
- Burgess, S.; Bowden, J.; Fall, T.; Ingelsson, E.; Thompson, S.G. Sensitivity Analyses for Robust Causal Inference from Mendelian Randomization Analyses with Multiple Genetic Variants. Epidemiology 2017, 28, 30–42. [Google Scholar] [CrossRef] [Green Version]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Davey Smith, G.; Sheehan, N.; Thompson, J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 2017, 36, 1783–1802. [Google Scholar] [CrossRef] [Green Version]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef] [Green Version]
- Verbanck, M.; Chen, C.Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Thompson, S.G. Multivariable Mendelian randomization: The use of pleiotropic genetic variants to estimate causal effects. Am. J. Epidemiol. 2015, 181, 251–260. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Gao, L.; Wang, Z.; Guan, B.; Guan, X.; Wang, B.; Han, X.; Xiao, X.; Waleed, K.B.; Chandran, C.; et al. Lipid profile and incidence of atrial fibrillation: A prospective cohort study in China. Clin. Cardiol. 2018, 41, 314–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balse, E.; El-Haou, S.; Dillanian, G.; Dauphin, A.; Eldstrom, J.; Fedida, D.; Coulombe, C.; Hatem, S.N.; Jan, L.Y. Cholesterol modulates the recruitment of Kv1.5 channels from Rab11-associated recycling endosome in native atrial myocytes. Proc. Natl. Acad. Sci. USA 2009, 106, 14681–14686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epshtein, Y.; Chopra, A.P.; Rosenhouse-Dantsker, A.; Kowalsky, G.B.; Logothetis, D.E.; Levitan, I. Identification of a C-terminus domain critical for the sensitivity of Kir2.1 to cholesterol. Proc. Natl. Acad. Sci. USA 2009, 106, 8055–8060. [Google Scholar] [CrossRef] [Green Version]
- Baartscheer, A.; Schumacher, C.A.; Wekker, V.; Verkerk, A.O.; Veldkamp, M.W.; van Oort, R.J.; Elzenaar, L.; Ottenhoff, R.; van Roomen, C.; Aerts, H.; et al. Dyscholesterolemia Protects Against Ischemia-Induced Ventricular Arrhythmias. Circ. Arrhythm. Electrophysiol. 2015, 8, 1481–1490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundbaek, J.A.; Birn, P.; Hansen, A.J.; Søgaard, R.; Nielsen, C.; Girshman, J.; Bruno, M.J.; Tape, S.E.; Egebjerg, J.; Greathouse, D.V.; et al. Regulation of sodium channel function by bilayer elasticity: The importance of hydrophobic coupling. Effects of Micelle-forming amphiphiles and cholesterol. J. Gen. Physiol. 2004, 123, 599–621. [Google Scholar] [CrossRef] [Green Version]
- Duntas, L.H.; Brenta, G. The effect of thyroid disorders on l.l.lipid levels and metabolism. Med. Clin. N. Am. 2012, 96, 269–281. [Google Scholar] [CrossRef]
- Wongcharoen, W.; Lin, Y.J.; Chang, S.L.; Lo, L.W.; Hu, Y.F.; Chung, F.P.; Chong, E.; Chao, T.-F.; Tuan, T.-C.; Chang, Y.-T.; et al. History of hyperthyroidism and long-term outcome of catheter ablation of drug-refractory atrial fibrillation. Heart Rhythm. 2015, 12, 1956–1962. [Google Scholar] [CrossRef]
- Ellervik, C.; Roselli, C.; Christophersen, I.E.; Alonso, A.; Pietzner, M.; Sitlani, C.M.; Trompet, S.; Arking, D.E.; Geelhoed, B.; Guo, X.; et al. Assessment of the Relationship Between Genetic Determinants of Thyroid Function and Atrial Fibrillation: A Mendelian Randomization Study. JAMA Cardiol. 2019, 4, 144–152. [Google Scholar] [CrossRef]
- Sanchez, O.A.; Duprez, D.A.; Bahrami, H.; Daniels, L.B.; Folsom, A.R.; Lima, J.A.; Maisel, A.; Peraltag, C.M.; Jacobs, D.R., Jr. The associations between metabolic variables and NT-proBNP are blunted at pathological ranges: The Multi-Ethnic Study of Atherosclerosis. Metabolism 2014, 63, 475–483. [Google Scholar] [CrossRef] [Green Version]
- Patton, K.K.; Heckbert, S.R.; Alonso, A.; Bahrami, H.; Lima, J.A.; Burke, G.; Kronmal, R.A. N-terminal pro-B-type natriuretic peptide as a predictor of incident atrial fibrillation in the Multi-Ethnic Study of Atherosclerosis: The effects of age, sex and ethnicity. Heart 2013, 99, 1832–1836. [Google Scholar] [CrossRef]
- Schnabel, R.B.; Larson, M.G.; Yamamoto, J.F.; Sullivan, L.M.; Pencina, M.J.; Meigs, J.B.; Tofler, G.H.; Selhub, J.; Jacques, P.F.; Wolf, P.A.; et al. Relations of biomarkers of distinct pathophysiological pathways and atrial fibrillation incidence in the community. Circulation 2010, 121, 200–207. [Google Scholar] [CrossRef] [Green Version]
- Sinner, M.F.; Stepas, K.A.; Moser, C.B.; Krijthe, B.P.; Aspelund, T.; Sotoodehnia, N.; Fontes, J.D.; Janssens, C.J.W.; Kronmal, R.A.; Magnani, J.W.; et al. B-type natriuretic peptide and C-reactive protein in the prediction of atrial fibrillation risk: The CHARGE-AF Consortium of community-based cohort studies. Europace 2014, 16, 1426–1433. [Google Scholar] [CrossRef]
- Folsom, A.R.; Pankow, J.S.; Tracy, R.P.; Arnett, D.K.; Peacock, J.M.; Hong, Y.; Djoussé, L.; Eckfeldt, J.H. Association of C-reactive protein with markers of prevalent atherosclerotic disease. Am. J. Cardiol. 2001, 88, 112–117. [Google Scholar] [CrossRef]
- Richardson, T.G.; Sanderson, E.; Palmer, T.M.; Ala-Korpela, M.; Ference, B.A.; Davey Smith, G.; Holmes, M.V. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: A multivariable Mendelian randomisation analysis. PLoS Med. 2020, 17, e1003062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sniderman, A.D.; Thanassoulis, G.; Glavinovic, T.; Navar, A.M.; Pencina, M.; Catapano, A.; Ference, B.A. Apolipoprotein B Particles and Cardiovascular Disease: A Narrative Review. JAMA Cardiol. 2019, 4, 1287–1295. [Google Scholar] [CrossRef] [PubMed]
- Avogaro, P.; Bon, G.B.; Cazzolato, G.; Quinci, G.B. Are apolipoproteins better discriminators than lipids for atherosclerosis? Lancet 1979, 1, 901–903. [Google Scholar] [CrossRef]
- Sniderman, A.; Shapiro, S.; Marpole, D.; Skinner, B.; Teng, B.; Kwiterovich, P.O., Jr. Association of coronary atherosclerosis with hyperapobetalipoproteinemia [increased protein but normal cholesterol levels in human plasma low density (beta) lipoproteins]. Proc. Natl. Acad. Sci. USA 1980, 77, 604–608. [Google Scholar] [CrossRef] [Green Version]
- Sniderman, A.D.; Robinson, J.G. ApoB in clinical care: Pro and Con. Atherosclerosis 2019, 282, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Budoff, M. Triglycerides and Triglyceride-Rich Lipoproteins in the Causal Pathway of Cardiovascular Disease. Am. J. Cardiol. 2016, 118, 138–145. [Google Scholar] [CrossRef] [Green Version]
- Lawlor, D.A. Commentary: Two-sample Mendelian randomization: Opportunities and challenges. Int. J. Epidemiol. 2016, 45, 908–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Xu, L. Circulating Vitamin E Levels and Risk of Coronary Artery Disease and Myocardial Infarction: A Mendelian Randomization Study. Nutrients. 2019, 11, 2153. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Exposures/Outcomes | Consortium | Ethnicity | Sample Sizes | R-Squared % (of Variance in AF) | F-Statistic (Total) |
|---|---|---|---|---|---|
| HDL-C | UKB | European | 291,830 | 1.582 | 34.375 |
| LDL-C | UKB | European | 318,340 | 1.296 | 28.422 |
| TG | UKB | European | 318,674 | 1.543 | 38.704 |
| Apolipoprotein A1 | UKB | European | 290,198 | 1.976 | 49.559 |
| Apolipoprotein B | UKB | European | 317,412 | 1.354 | 31.788 |
| Atrial fibrillation | HUNT, DECODE, DiscovEHR, MGI, UKB, and AF Gen Consortium | European | 1,030,836 | NA | NA |
| Exposure | Raw Estimates | Outlier Corrected Estimates | Distortion Test | ||||||
|---|---|---|---|---|---|---|---|---|---|
| nSNP | Beta | OR (95%CI) | p-Value | nSNP | Beta | OR (95%CI) | p-Value | p-Value | |
| HDL-C | 138 | −0.025 | 0.975(0.920,1.030) | 0.379 | 137 | −0.018 | 0.982(0.928,1.036) | 0.517 | 0.695 |
| LDL-C | 147 | −0.001 | 0.999(0.947,1.051) | 0.966 | 145 | −0.004 | 0.996(0.946,1.046) | 0.882 | 0.942 |
| TG | 129 | −0.041 | 0.960(0.885,1.035) | 0.291 | 125 | −0.047 | 0.954(0.897,1.011) | 0.114 | 0.847 |
| Apolipoprotein A1 | 118 | −0.002 | 0.998(0.994,1.002) | 0.921 | NA | NA | NA | NA | NA |
| Apolipoprotein B | 137 | 0.008 | 1.008(0.959,1.070) | 0.786 | NA | NA | NA | NA | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Pudasaini, R.; Zhi, H.; Wang, L. The Relationship between Blood Lipids and Risk of Atrial Fibrillation: Univariable and Multivariable Mendelian Randomization Analysis. Nutrients 2022, 14, 181. https://doi.org/10.3390/nu14010181
Yang S, Pudasaini R, Zhi H, Wang L. The Relationship between Blood Lipids and Risk of Atrial Fibrillation: Univariable and Multivariable Mendelian Randomization Analysis. Nutrients. 2022; 14(1):181. https://doi.org/10.3390/nu14010181
Chicago/Turabian StyleYang, Shengyi, Rupak Pudasaini, Hong Zhi, and Lina Wang. 2022. "The Relationship between Blood Lipids and Risk of Atrial Fibrillation: Univariable and Multivariable Mendelian Randomization Analysis" Nutrients 14, no. 1: 181. https://doi.org/10.3390/nu14010181







