The Relationship between Gut Microbiome and Cognition in Older Australians
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Faecal Sample Collection
2.3. 16S rRNA Sequencing and Data Processing
2.4. Functional Analysis
2.5. Cognition
2.6. Statistical Analysis
3. Results
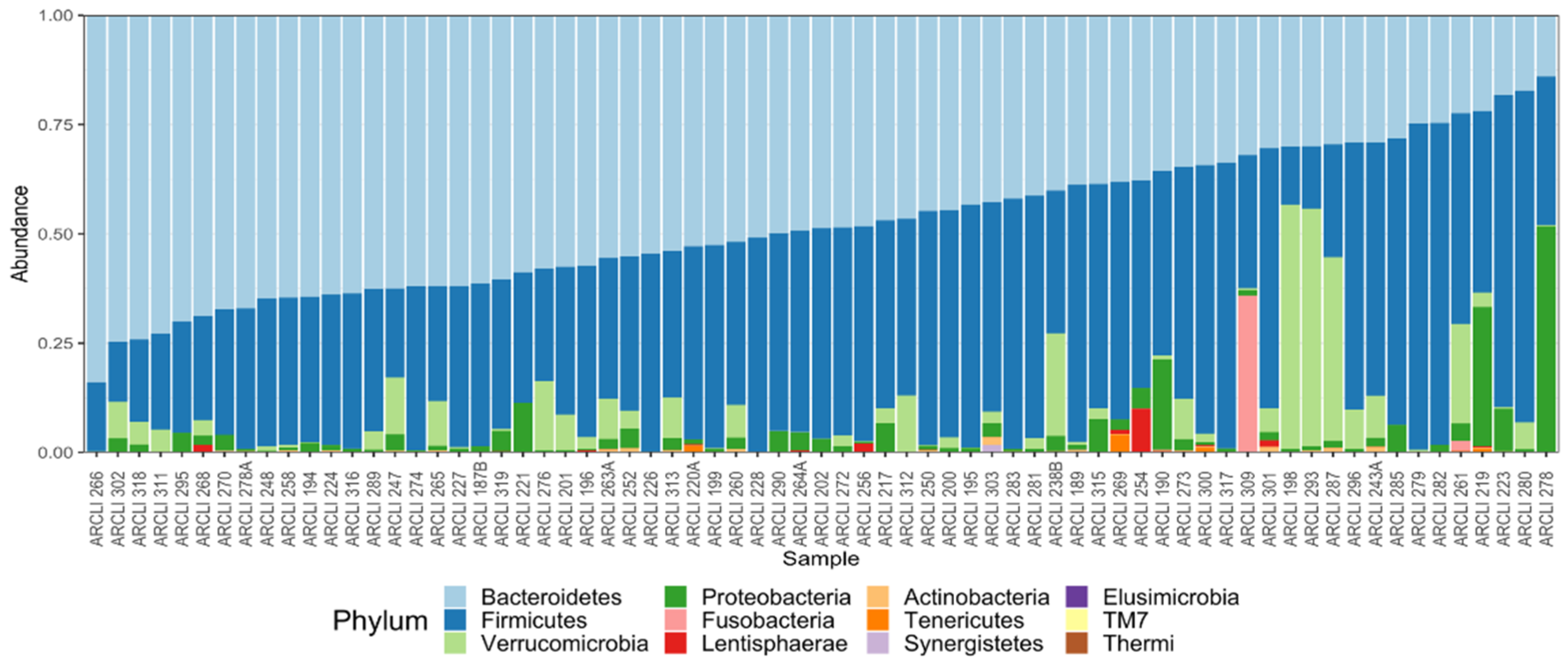
3.1. Gut Microbiome
3.2. Association between Microbial Diversity and Cognition
3.3. Association between Microbial Family and Cognition
3.4. Association between the Gut Microbiome and Demographic Measures
3.5. The Combined Effect of the Gut Microbiome on Cognition
3.6. Relation between Gut Microbial Function and Cognition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nagpal, R.; Mainali, R.; Ahmadi, S.; Wang, S.; Singh, R.; Kavanagh, K.; Kitzman, D.W.; Kushugulova, A.; Marotta, F.; Yadav, H. Gut microbiome and aging: Physiological and mechanistic insights. Nutr. Healthy Aging 2018, 4, 267–285. [Google Scholar] [CrossRef] [Green Version]
- Odamaki, T.; Kato, K.; Sugahara, H.; Hashikura, N.; Takahashi, S.; Xiao, J.Z.; Abe, F.; Osawa, R. Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbi. 2016, 16, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Claesson, M.J.; Cusack, S.; O’Sullivan, O.; Greene-Diniz, R.; de Weerd, H.; Flannery, E.; Marchesi, J.R.; Falush, D.; Dinan, T.; Fitzgerald, G.; et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. USA 2011, 108, 4586–4591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Toole, P.W.; Jeffery, I.B. Gut microbiota and aging. Science 2015, 350, 1214–1215. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef]
- Collino, S.; Montoliu, I.; Martin, F.-P.J.; Scherer, M.; Mari, D.; Salvioli, S.; Bucci, L.; Ostan, R.; Monti, D.; Biagi, E.; et al. Metabolic signatures of extreme longevity in northern Italian centenarians reveal a complex remodeling of lipids, amino acids, and gut microbiota metabolism. PLoS ONE 2013, 8, e56564. [Google Scholar] [CrossRef]
- Mitchell, E.L.; Davis, A.T.; Brass, K.; Dendinger, M.; Barner, R.; Gharaibeh, R.; Fodor, A.A.; Kavanagh, K. Reduced Intestinal Motility, Mucosal Barrier Function, and Inflammation in Aged Monkeys. J. Nutr. Health Aging 2017, 21, 354–361. [Google Scholar] [CrossRef]
- Braniste, V.; Al-Asmakh, M.; Kowal, C.; Anuar, F.; Abbaspour, A.; Tóth, M.; Korecka, A.; Bakocevic, N.; Ng, L.G.; Kundu, P.; et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 2014, 6, 263ra158. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.-J.; Zeng, B.-H.; Li, W.-W.; Zhou, C.-J.; Fan, S.-H.; Cheng, K.; Zeng, L.; Zheng, P.; Fang, L.; Wei, H.; et al. Effects of gut microbiota on the microRNA and mRNA expression in the hippocampus of mice. Behav. Brain Res. 2017, 322, 34–41. [Google Scholar] [CrossRef]
- Nishino, R.; Mikami, K.; Takahashi, H.; Tomonaga, S.; Furuse, M.; Hiramoto, T.; Aiba, Y.; Koga, Y.; Sudo, N. Commensal microbiota modulate murine behaviors in a strictly contamination-free environment confirmed by culture-based methods. Neurogastroenterol. Motil. 2013, 25, 521–528. [Google Scholar] [CrossRef]
- Minter, M.R.; Hinterleitner, R.; Meisel, M.; Zhang, C.; Leone, V.; Zhang, X.; Oyler-Castrillo, P.; Zhang, X.; Musch, M.W.; Shen, X.; et al. Antibiotic-induced perturbations in microbial diversity during post-natal development alters amyloid pathology in an aged APPSWE/PS1ΔE9 murine model of Alzheimer’s disease. Sci. Rep. 2017, 7, 10411. [Google Scholar] [CrossRef]
- Komanduri, M.; Gondalia, S.; Scholey, A.; Stough, C. The microbiome and cognitive aging: A review of mechanisms. Psychopharmacology 2019, 236, 1559–1571. [Google Scholar] [CrossRef]
- Sochocka, M.; Donskow-Łysoniewska, K.; Diniz, B.S.; Kurpas, D.; Brzozowska, E.; Leszek, J. The Gut Microbiome Alterations and Inflammation-Driven Pathogenesis of Alzheimer’s Disease—A Critical Review. Mol. Neurobiol. 2019, 56, 1841–1851. [Google Scholar] [CrossRef] [Green Version]
- Dinan, T.G.; Stanton, C.; Cryan, J.F. Psychobiotics: A novel class of psychotropic. Biol. Psychiatry 2013, 74, 720–726. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Baptista, L.C.; Roberts, L.M.; Jumbo-Lucioni, P.; McMahon, L.L.; Buford, T.W.; Carter, C.S. The Gut Microbiome as a Therapeutic Target for Cognitive Impairment. J. Gerontol. Ser. A 2019, 75, 1242–1250. [Google Scholar] [CrossRef] [PubMed]
- Benton, D.; Williams, C.; Brown, A. Impact of consuming a milk drink containing a probiotic on mood and cognition. Eur. J. Clin. Nutr. 2007, 61, 355–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, Y.-C.; Jin, H.-M.; Cui, Y.; Kim, D.; Jung, J.; Park, J.-I.; Jung, E.-S.; Choi, E.-K.; Chae, S.-W. Fermented milk of Lactobacillus helveticus IDCC3801 improves cognitive functioning during cognitive fatigue tests in healthy older adults. J. Funct. Foods 2014, 10, 465–474. [Google Scholar] [CrossRef]
- Akbari, E.; Asemi, Z.; Daneshvar Kakhaki, R.; Bahmani, F.; Kouchaki, E.; Tamtaji, O.R.; Hamidi, G.A.; Salami, M. Effect of Probiotic Supplementation on Cognitive Function and Metabolic Status in Alzheimer’s Disease: A Randomized, Double-Blind and Controlled Trial. Front. Aging Neurosci. 2016, 8, 256. [Google Scholar] [CrossRef] [Green Version]
- Gill, P.A.; van Zelm, M.C.; Muir, J.G.; Gibson, P.R. Review article: Short chain fatty acids as potential therapeutic agents in human gastrointestinal and inflammatory disorders. Aliment. Pharmacol. Ther. 2018, 48, 15–34. [Google Scholar] [CrossRef] [Green Version]
- Pluznick, J.L. Microbial Short-Chain Fatty Acids and Blood Pressure Regulation. Curr. Hypertens. Rep. 2017, 19, 25. [Google Scholar] [CrossRef] [Green Version]
- Tahara, Y.; Yamazaki, M.; Sukigara, H.; Motohashi, H.; Sasaki, H.; Miyakawa, H.; Haraguchi, A.; Ikeda, Y.; Fukuda, S.; Shibata, S. Gut Microbiota-Derived Short Chain Fatty Acids Induce Circadian Clock Entrainment in Mouse Peripheral Tissue. Sci. Rep. 2018, 8, 1395. [Google Scholar] [CrossRef]
- Erny, D.; Hrabě de Angelis, A.L.; Prinz, M. Communicating systems in the body: How microbiota and microglia cooperate. Immunology 2017, 150, 7–15. [Google Scholar] [CrossRef]
- Mitchell, R.W.; On, N.H.; Del Bigio, M.R.; Miller, D.W.; Hatch, G.M. Fatty acid transport protein expression in human brain and potential role in fatty acid transport across human brain microvessel endothelial cells. J. Neurochem. 2011, 117, 735–746. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.; Xiayu, X.; Shi, C.; Chen, W.; Song, N.; Fu, X.; Zhou, R.; Xu, Y.F.; Huang, L.; et al. Altered Gut Microbiota in a Mouse Model of Alzheimer’s Disease. J. Alzheimers Dis. 2017, 60, 1241–1257. [Google Scholar] [CrossRef] [PubMed]
- Unger, M.M.; Spiegel, J.; Dillmann, K.U.; Grundmann, D.; Philippeit, H.; Bürmann, J.; Faßbender, K.; Schwiertz, A.; Schäfer, K.H. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Parkinsonism Relat. Disord. 2016, 32, 66–72. [Google Scholar] [CrossRef]
- Frost, G.; Sleeth, M.L.; Sahuri-Arisoylu, M.; Lizarbe, B.; Cerdan, S.; Brody, L.; Anastasovska, J.; Ghourab, S.; Hankir, M.; Zhang, S.; et al. The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat. Commun. 2014, 5, 3611. [Google Scholar] [CrossRef] [Green Version]
- Huuskonen, J.; Suuronen, T.; Nuutinen, T.; Kyrylenko, S.; Salminen, A. Regulation of microglial inflammatory response by sodium butyrate and short-chain fatty acids. Br. J. Pharmacol. 2004, 141, 874–880. [Google Scholar] [CrossRef]
- Stough, C.K.; Pase, M.P.; Cropley, V.; Myers, S.; Nolidin, K.; King, R.; Camfield, D.; Wesnes, K.; Pipingas, A.; Croft, K.; et al. A randomized controlled trial investigating the effect of Pycnogenol and BacopaCDRI08 herbal medicines on cognitive, cardiovascular, and biochemical functioning in cognitively healthy elderly people: The Australian Research Council Longevity Intervention (ARCLI) study protocol (ANZCTR12611000487910). Nutr. J. 2012, 11, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, T.; Deleuil, S.; Echeverria, N.; Komanduri, M.; Macpherson, H.; Suo, C.; Gondalia, S.; Fard, M.T.; Pipingas, A.; Scholey, A.; et al. The Australian Research Council Longevity Intervention (ARCLI) study protocol (ANZCTR12611000487910) addendum: Neuroimaging and gut microbiota protocol. Nutr. J. 2019, 18, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folstein, M.F.; Folstein, S.E.; McHugh, P.R. “Mini-mental state”: A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 1975, 12, 189–198. [Google Scholar] [CrossRef]
- Yesavage, J.A.; Brink, T.L.; Rose, T.L.; Lum, O.; Huang, V.; Adey, M.; Leirer, V.O. Development and validation of a geriatric depression screening scale: A preliminary report. J. Psychiatr. Res. 1982, 17, 37–49. [Google Scholar] [CrossRef]
- Goldberg, D.P.; Gater, R.; Sartorius, N.; Ustun, T.B.; Piccinelli, M.; Gureje, O.; Rutter, C. The validity of two versions of the GHQ in the WHO study of mental illness in general health care. Psychol. Med. 1997, 27, 191–197. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069. [Google Scholar] [CrossRef] [Green Version]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Darzi, Y.; Falony, G.; Vieira-Silva, S.; Raes, J. Towards biome-specific analysis of meta-omics data. ISME J. 2016, 10, 1025–1028. [Google Scholar] [CrossRef] [Green Version]
- Wesnes, K.A.; McNamara, C.; Annas, P. Norms for healthy adults aged 18–87 years for the Cognitive Drug Research System: An automated set of tests of attention, information processing and memory for use in clinical trials. J. Psychopharmacol. 2016, 30, 263–272. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S.A. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Lahti, L.; Shetty, S. Microbiome R Package. 2012–2019. Available online: http://microbiome.github.io (accessed on 18 November 2021).
- Wickham, H.; François, R.; Henry; Kirill, L.a.M. dplyr: A Grammar of Data Manipulation; R Package Version 1.0.7.; R Foundation Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Jari Oksanen, F.G.; Blanchet, M.F.; Roeland Kindt, P.L.; Dan McGlinn, P.R.M.; O’Hara, G.L.S.; Peter Solymos, M.H.H.; Stevens, E.S.a.H.; Wagner Mackenzie, B. Vegan: Community. Ecology Package; R Package Version 2.5-6; R Foundation Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Wesnes, K.A. The value of assessing cognitive function in drug development. Dialogues Clin. Neurosci. 2000, 2, 183–202. [Google Scholar]
- Wesnes, K.A.; Ward, T.; McGinty, A.; Petrini, O. The memory enhancing effects of a Ginkgo biloba/Panax ginseng combination in healthy middle-aged volunteers. Psychopharmacology 2000, 152, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Bennett, S.; Magnus, P.; Gibson, D. Obesity Trends in Older Australians; Australian Institute of Health and Welfare: Darlinghurst, Australia, 2004. [Google Scholar]
- Gao, R.; Zhu, C.; Li, H.; Yin, M.; Pan, C.; Huang, L.; Kong, C.; Wang, X.; Zhang, Y.; Qu, S.; et al. Dysbiosis Signatures of Gut Microbiota Along the Sequence from Healthy, Young Patients to Those with Overweight and Obesity. Obesity 2018, 26, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Takagi, T.; Naito, Y.; Inoue, R.; Kashiwagi, S.; Uchiyama, K.; Mizushima, K.; Tsuchiya, S.; Dohi, O.; Yoshida, N.; Kamada, K.; et al. Differences in gut microbiota associated with age, sex, and stool consistency in healthy Japanese subjects. J. Gastroenterol. 2019, 54, 53–63. [Google Scholar] [CrossRef]
- Verdi, S.; Jackson, M.A.; Beaumont, M.; Bowyer, R.C.E.; Bell, J.T.; Spector, T.D.; Steves, C.J. An Investigation Into Physical Frailty as a Link Between the Gut Microbiome and Cognitive Health. Front. Aging Neurosci. 2018, 10. [Google Scholar] [CrossRef] [Green Version]
- Wasser, C.I.; Mercieca, E.-C.; Kong, G.; Hannan, A.J.; McKeown, S.J.; Glikmann-Johnston, Y.; Stout, J.C. Gut dysbiosis in Huntington’s disease: Associations among gut microbiota, cognitive performance and clinical outcomes. Brain Commun. 2020, 2, fcaa110. [Google Scholar] [CrossRef]
- Badal, V.D.; Vaccariello, E.D.; Murray, E.R.; Yu, K.E.; Knight, R.; Jeste, D.V.; Nguyen, T.T. The Gut Microbiome, Aging, and Longevity: A Systematic Review. Nutrients 2020, 12, 3759. [Google Scholar] [CrossRef]
- Saji, N.; Niida, S.; Murotani, K.; Hisada, T.; Tsuduki, T.; Sugimoto, T.; Kimura, A.; Toba, K.; Sakurai, T. Analysis of the relationship between the gut microbiome and dementia: A cross-sectional study conducted in Japan. Sci. Rep. 2019, 9, 1008. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wu, L.; Peng, G.; Han, Y.; Tang, R.; Ge, J.; Zhang, L.; Jia, L.; Yue, S.; Zhou, K.; et al. Altered microbiomes distinguish Alzheimer’s disease from amnestic mild cognitive impairment and health in a Chinese cohort. Brain Behav. Immun. 2019, 80, 633–643. [Google Scholar] [CrossRef]
- Vogt, N.M.; Kerby, R.L.; Dill-McFarland, K.A.; Harding, S.J.; Merluzzi, A.P.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Zetterberg, H.; Blennow, K.; et al. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 2017, 7, 13537. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, M.J.; Sharp, R.; Macfarlane, G.T. Variation in human intestinal microbiota with age. Dig. Liver Dis. 2002, 34, S12–S18. [Google Scholar] [CrossRef]
- Tuikhar, N.; Keisam, S.; Labala, R.K.; Imrat; Ramakrishnan, P.; Arunkumar, M.C.; Ahmed, G.; Biagi, E.; Jeyaram, K. Comparative analysis of the gut microbiota in centenarians and young adults shows a common signature across genotypically non-related populations. Mech. Ageing Dev. 2019, 179, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Biagi, E.; Franceschi, C.; Rampelli, S.; Severgnini, M.; Ostan, R.; Turroni, S.; Consolandi, C.; Quercia, S.; Scurti, M.; Monti, D.; et al. Gut Microbiota and Extreme Longevity. Curr. Biol. 2016, 26, 1480–1485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biagi, E.; Nylund, L.; Candela, M.; Ostan, R.; Bucci, L.; Pini, E.; Nikkïla, J.; Monti, D.; Satokari, R.; Franceschi, C.; et al. Through Ageing, and Beyond: Gut Microbiota and Inflammatory Status in Seniors and Centenarians. PLoS ONE 2010, 5, 10667. [Google Scholar] [CrossRef]
- Zhuang, Z.-Q.; Shen, L.-L.; Li, W.-W.; Fu, X.; Zeng, F.; Gui, L.; Lü, Y.; Cai, M.; Zhu, C.; Tan, Y.-L.; et al. Gut Microbiota is Altered in Patients with Alzheimer’s Disease. J. Alzheimer’s Dis. 2018, 63, 1337–1346. [Google Scholar] [CrossRef] [Green Version]
- Park, M.J.; Sohrabji, F. The histone deacetylase inhibitor, sodium butyrate, exhibits neuroprotective effects for ischemic stroke in middle-aged female rats. J. Neuroinflamm. 2016, 13, 300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Cai, Y.-Y.; Yan, Z.-X. Brain-derived neurotrophic factor preserves intestinal mucosal barrier function and alters gut microbiota in mice. Kaohsiung J. Med. Sci. 2018, 34, 134–141. [Google Scholar] [CrossRef]
- Distrutti, E.; O’Reilly, J.A.; McDonald, C.; Cipriani, S.; Renga, B.; Lynch, M.A.; Fiorucci, S. Modulation of intestinal microbiota by the probiotic VSL#3 resets brain gene expression and ameliorates the age-related deficit in LTP. PLoS ONE 2014, 9, 106503. [Google Scholar] [CrossRef]
- Ren, T.; Gao, Y.; Qiu, Y.; Jiang, S.; Zhang, Q.; Zhang, J.; Wang, L.; Zhang, Y.; Wang, L.; Nie, K. Gut Microbiota Altered in Mild Cognitive Impairment Compared With Normal Cognition in Sporadic Parkinson’s Disease. Front. Neurol. 2020, 11, 137. [Google Scholar] [CrossRef]
- Sethi, K.D.; Ray, R.; Roesel, R.A.; Carter, A.L.; Gallagher, B.B.; Loring, D.W.; Hommes, F.A. Adult-onset chorea and dementia with propionic acidemia. Neurology 1989, 39, 1343–1345. [Google Scholar] [CrossRef]
- Morland, C.; Frøland, A.S.; Pettersen, M.N.; Storm-Mathisen, J.; Gundersen, V.; Rise, F.; Hassel, B. Propionate enters GABAergic neurons, inhibits GABA transaminase, causes GABA accumulation and lethargy in a model of propionic acidemia. Biochem. J. 2018, 475, 749–758. [Google Scholar] [CrossRef]
- Killingsworth, J.; Sawmiller, D.; Shytle, R.D. Propionate and Alzheimer’s Disease. Front. Aging Neurosci. 2021, 12, 501. [Google Scholar] [CrossRef]
- Hou, Y.-F.; Shan, C.; Zhuang, S.-Y.; Zhuang, Q.-Q.; Ghosh, A.; Zhu, K.-C.; Kong, X.-K.; Wang, S.-M.; Gong, Y.-L.; Yang, Y.-Y.; et al. Gut microbiota-derived propionate mediates the neuroprotective effect of osteocalcin in a mouse model of Parkinson’s disease. Microbiome 2021, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Toczylowska, B.; Jamrozik, Z.; Liebert, A.; Kwiecinski, H. NMR-based Metabonomics of Cerebrospinal Fluid Applied to Amyotrophic Lateral Sclerosis. Biocybern. Biomed. Eng. 2013, 33, 21–32. [Google Scholar] [CrossRef]
- Schneider, W.J.; McGrew, K.S. The Cattell-Horn-Carroll model of intelligence. In Contemporary Intellectual Assessment: Theories, Tests, and Issues; Flanagan, D.P., Harrison, P.L., Eds.; Guilford Press: New York, NY, USA, 2012; pp. 99–144. [Google Scholar]
- Pase, M.P.; Stough, C. Describing a taxonomy of cognitive processes for clinical trials assessing cognition. Am. J. Clin. Nutr. 2013, 98, 509–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Characteristic | Mean | SD |
|---|---|---|
| Sample Size | 69 | |
| Gender | Male (34), Female (35) | |
| Age | 65.06 | 4.01 |
| BMI | 26.57 | 4.76 |
| MMSE | 28.78 | 1.29 |
| GDS | 3.91 | 3.34 |
| General Health Questionnaire (GHQ-12) | 8.66 | 2.74 |
| Cognition | ||
| Word Recall Original Accuracy | 70.69 | 16.30 |
| Word Recall Novel Accuracy | 86.96 | 12.10 |
| Picture Recall Original Accuracy | 92.57 | 8.92 |
| Picture Recall Novel Accuracy | 87.21 | 10.42 |
| Immediate Word Recall Accuracy | 38.53 | 11.91 |
| Immediate Word Recall Error | 0.32 | 0.63 |
| Delayed Word Recall Accuracy | 22.26 | 11.89 |
| Delayed Word Recall Error | 0.74 | 1.05 |
| Spatial Working Memory Sensitivity Index | 0.83 | 0.30 |
| Numeric Working Memory Sensitivity Index | 0.91 | 0.10 |
| Simple Reaction Time | 299.60 | 39.76 |
| Digit Vigilance | 441.29 | 49.42 |
| Choice Reaction Time | 510.88 | 49.32 |
| Spatial Working Memory Reaction Time | 1099.83 | 346.92 |
| Numeric Working Memory Reaction Time | 848.11 | 163.61 |
| Word Recall Reaction Time | 1006.38 | 194.81 |
| Picture Recall Reaction Time | 1166.98 | 237.06 |
| Digit Vigilance Accuracy | 96.64 | 6.79 |
| Choice Reaction Time Accuracy | 98.15 | 1.74 |
| Digit Vigilance False Alarms | 3.50 | 14.03 |
| CDR factors | ||
| Quality of Episodic Secondary Memory (QESM) | 191.15 | 39.38 |
| Quality of Working Memory (QWM) | 1.77 | 0.26 |
| Power of Concentration (PoC) | 1251.77 | 99.64 |
| Continuity of Attention (CoA) | 90.88 | 6.76 |
| Speed of Memory (SoM) | 4115.16 | 718.37 |
| Alpha Diversity Index | Observed | Shannon | Chao1 | Fisher | Simpson | Invsimpson | ACE | B/F Ratio |
|---|---|---|---|---|---|---|---|---|
| Age | 0.157 | 0.066 | 0.085 | 0.084 | 0.103 | 0.103 | 0.081 | 0.006 |
| Sex | −0.01 | −0.082 | 0.032 | −0.14 | −0.01 | −0.01 | −0.006 | −0.042 |
| BMI | −0.097 | −0.111 | −0.025 | −0.042 | −0.136 | −0.136 | −0.039 | 0.023 |
| QESM | −0.113 | −0.011 | −0.067 | −0.072 | 0.003 | 0.003 | −0.057 | −0.035 |
| QWM | −0.034 | −0.193 | −0.094 | −0.042 | −0.202 | −0.202 | −0.083 | −0.171 |
| PoC | 0.011 | 0.079 | −0.008 | −0.001 | 0.131 | 0.131 | −0.054 | −0.144 |
| CoA | −0.021 | 0.131 | −0.02 | −0.003 | 0.186 | 0.186 | −0.075 | −0.182 |
| SoM | 0.061 | 0.21 | 0.021 | 0.103 | 0.218 | 0.218 | −0.014 | −0.171 |
| Bacterial Family | QESM | QWM | PoC | CoA | SoM | Age | Sex | BMI |
|---|---|---|---|---|---|---|---|---|
| Alcaligenaceae | −0.031 | −0.294 * | 0.103 | 0.119 | 0.198 | −0.026 | 0.013 | 0.095 |
| Bacteroidaceae | −0.006 | 0.064 | −0.247 * | 0.03 | −0.265 * | 0.015 | 0.173 | −0.033 |
| Barnesiellaceae | 0.043 | 0.221 | −0.413 ** | 0.109 | −0.328 ** | −0.283 * | 0.058 | 0.041 |
| Carnobacteriaceae | 0.273 * | 0.217 | −0.062 | −0.057 | −0.238 | −0.077 | 0.105 | 0.03 |
| Clostridiaceae | 0.229 | 0.265 * | −0.017 | 0.261 * | −0.015 | 0.025 | 0.119 | −0.018 |
| Desulfovibrionaceae | −0.019 | −0.148 | −0.098 | −0.054 | −0.03 | 0.247 * | 0.007 | 0.1 |
| Gemellaceae | −0.05 | −0.029 | −0.252 * | −0.111 | −0.245 * | −0.051 | 0.127 | 0.156 |
| Lactobacillaceae | 0.121 | −0.165 | 0.184 | −0.033 | 0.152 | −0.317 ** | −0.015 | −0.129 |
| Micrococcaceae | 0.087 | 0.07 | −0.057 | −0.093 | −0.255 * | −0.102 | 0.016 | 0.054 |
| Odoribacteraceae | 0.073 | 0.123 | −0.172 | 0.149 | −0.075 | −0.051 | 0.320 ** | −0.027 |
| Porphyromonadaceae | −0.026 | −0.159 | −0.146 | −0.011 | −0.183 | 0.240 * | −0.084 | 0.055 |
| Prevotellaceae | −0.107 | −0.168 | 0.163 | 0.032 | 0.126 | 0.13 | −0.269 * | 0.03 |
| Rikenellaceae | 0.167 | 0.027 | −0.248 * | 0.288 * | −0.075 | −0.121 | 0.355 ** | −0.174 |
| Tissierellaceae | 0.223 | 0.001 | 0.163 | −0.132 | 0.014 | −0.057 | 0.272 * | −0.096 |
| Verrucomicrobiaceae | −0.051 | 0.008 | −0.052 | −0.247 * | 0.139 | 0.08 | 0.025 | −0.048 |
| Bacterial Family | Cognitive Domain | Unadjusted | Adjusted + | ||||||
|---|---|---|---|---|---|---|---|---|---|
| β | CI (2.5, 97.5) | p Value | β | CI (2.5, 97.5) | p Value | ||||
| Carnobacteriaceae | QESM | 10.27 | 3.14 | 17.40 | 0.006 | 9.25 | 2.12 | 16.39 | 0.014 |
| Alcaligenaceae | QWM | −0.08 | −0.12 | −0.03 | 0.002 | −0.08 | −0.13 | −0.04 | 0.001 |
| Clostridiaceae | 0.05 | −0.01 | 0.11 | 0.13 | 0.05 | −0.01 | 0.11 | 0.12 | |
| Bacteroidaceae | PoC | −21.8 | −44.57 | 0.98 | 0.07 | −23.16 | −46.68 | 0.36 | 0.06 |
| Barnesiellaceae | −19.74 | −34.78 | −4.69 | 0.01 | −21.15 | −37.12 | −5.13 | 0.01 | |
| Gemellaceae | −30.46 | −57.30 | −3.62 | 0.03 | −31.05 | −58.97 | −3.14 | 0.03 | |
| Rikenellaceae | −22.85 | −44.71 | −0.99 | 0.05 | −27.24 | −50.47 | −4.00 | 0.03 | |
| Clostridiaceae | CoA | 1.32 | 0.06 | 2.57 | 0.0 | 1.23 | −0.06 | 2.52 | 0.07 |
| Rikenellaceae | 1.26 | −0.24 | 2.77 | 0.10 | 1.28 | −0.31 | 2.87 | 0.12 | |
| Verrucomicrobiaceae | 0.27 | −0.28 | 0.83 | 0.34 | 0.25 | −0.33 | 0.80 | 0.42 | |
| Bacteroidaceae | SoM | −181.87 | −347.11 | −16.62 | 0.04 | −191.14 | −362.41 | −19.87 | 0.03 |
| Barnesiellaceae | −91.79 | −203.62 | 20.03 | 0.11 | −115.65 | −234.15 | 2.88 | 0.06 | |
| Gemellaceae | −224.97 | −420.95 | −28.98 | 0.03 | −238.58 | −442.16 | −35.00 | 0.023 | |
| Micrococcaceae | −259.40 | −475.02 | −43.76 | 0.02 | −277.84 | −498.70 | −56.97 | 0.012 | |
| Bacterial Family | Cognition | F-Statistic | R2 | Adjusted R2 | p Value |
|---|---|---|---|---|---|
| Carnobacteriaceae | QESM | 7.966 | 0.108 | 0.094 | 0.006 |
| Alcaligenaceae + Clostridiaceae | QWM | 6.973 | 0.177 | 0.151 | 0.002 |
| Bacteroidaceae + Barnesiellaceae + Rikenellaceae + Gemellaceae | PoC | 3.031 | 0.161 | 0.108 | 0.024 |
| Rikenellaceae + Clostridiaceae + Verrucomicrobiaceae | CoA | 2.039 | 0.088 | 0.045 | 0.118 |
| Bacteroidaceae + Barnesiellaceae + Gemellaceae + Micrococcaceae | SoM | 2.475 | 0.138 | 0.082 | 0.053 |
| GBM | Cognition | Rho | p Value |
|---|---|---|---|
| Propionate Production III | CoA | −0.311 | 0.011 |
| Tyrosine Degradation I | PoC | 0.274 | 0.024 |
| Phenylalanine Degradation | 0.274 | 0.024 | |
| Tyrosine Degradation I | QWM | −0.246 | 0.045 |
| Phenylalanine Degradation | −0.246 | 0.045 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komanduri, M.; Savage, K.; Lea, A.; McPhee, G.; Nolidin, K.; Deleuil, S.; Stough, C.; Gondalia, S. The Relationship between Gut Microbiome and Cognition in Older Australians. Nutrients 2022, 14, 64. https://doi.org/10.3390/nu14010064
Komanduri M, Savage K, Lea A, McPhee G, Nolidin K, Deleuil S, Stough C, Gondalia S. The Relationship between Gut Microbiome and Cognition in Older Australians. Nutrients. 2022; 14(1):64. https://doi.org/10.3390/nu14010064
Chicago/Turabian StyleKomanduri, Mrudhula, Karen Savage, Ana Lea, Grace McPhee, Karen Nolidin, Saurenne Deleuil, Con Stough, and Shakuntla Gondalia. 2022. "The Relationship between Gut Microbiome and Cognition in Older Australians" Nutrients 14, no. 1: 64. https://doi.org/10.3390/nu14010064
APA StyleKomanduri, M., Savage, K., Lea, A., McPhee, G., Nolidin, K., Deleuil, S., Stough, C., & Gondalia, S. (2022). The Relationship between Gut Microbiome and Cognition in Older Australians. Nutrients, 14(1), 64. https://doi.org/10.3390/nu14010064






