Vitamin K2 Improves Osteogenic Differentiation by Inhibiting STAT1 via the Bcl-6 and IL-6/JAK in C3H10 T1/2 Clone 8 Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Osteogenic Induction
2.2. Cell Proliferation Assay
2.3. Sample Preparation and RNA Sequencing (RNA-Seq)
2.4. Bioinformatics Analysis
2.5. Small Interfering RNA (siRNA) Assay
2.6. Construction of Recombinant Plasmids and Transfection Assay
2.7. Alkaline Phosphatase (ALP) Staining and Activity
2.8. Real-Time PCR Analysis
2.9. Western Blot Analysis
2.10. Statistical Analysis
3. Results
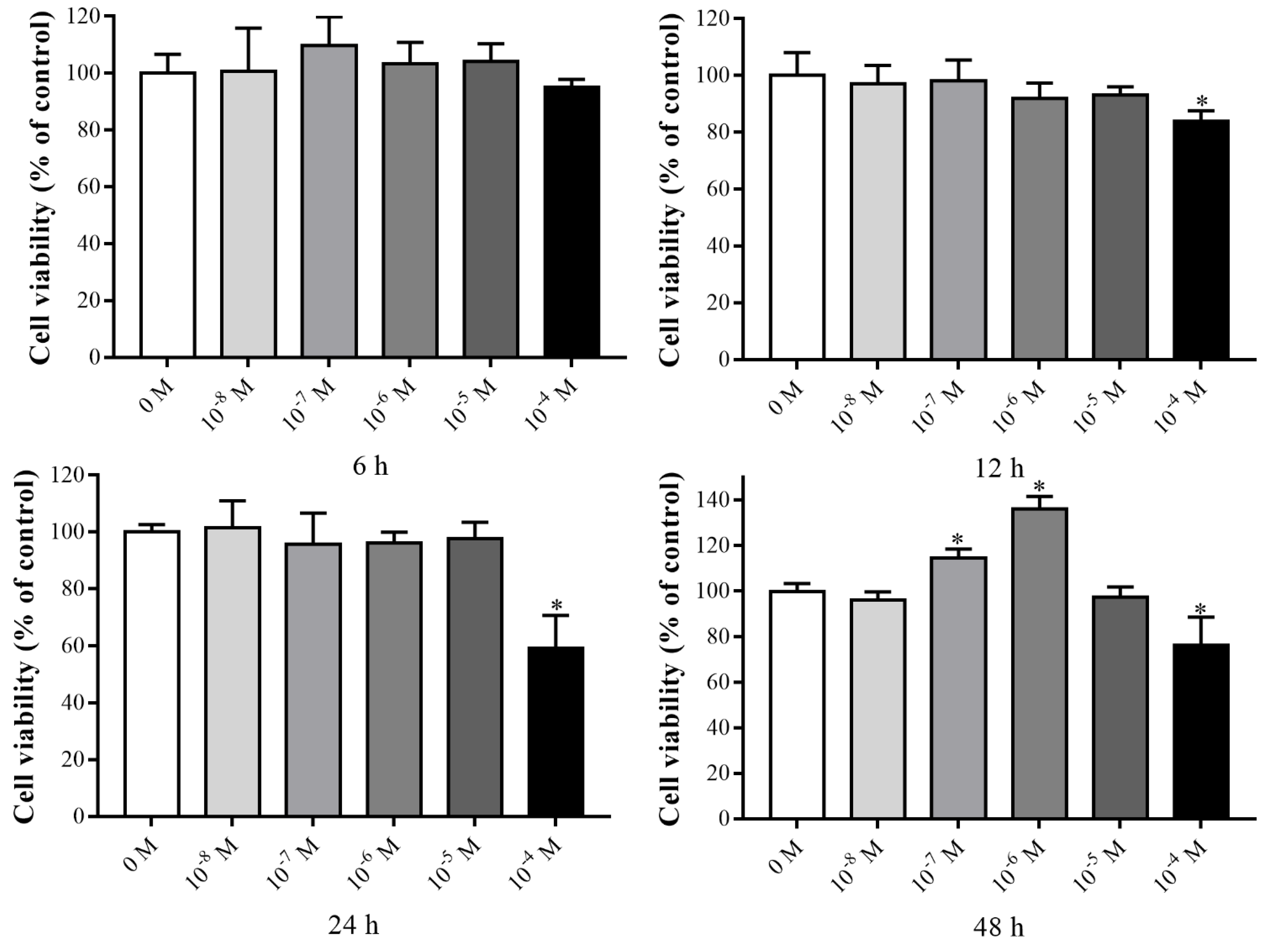
3.1. Effects of VK2 on the Proliferation of C3H10 T1/2 Clone 8 Cells
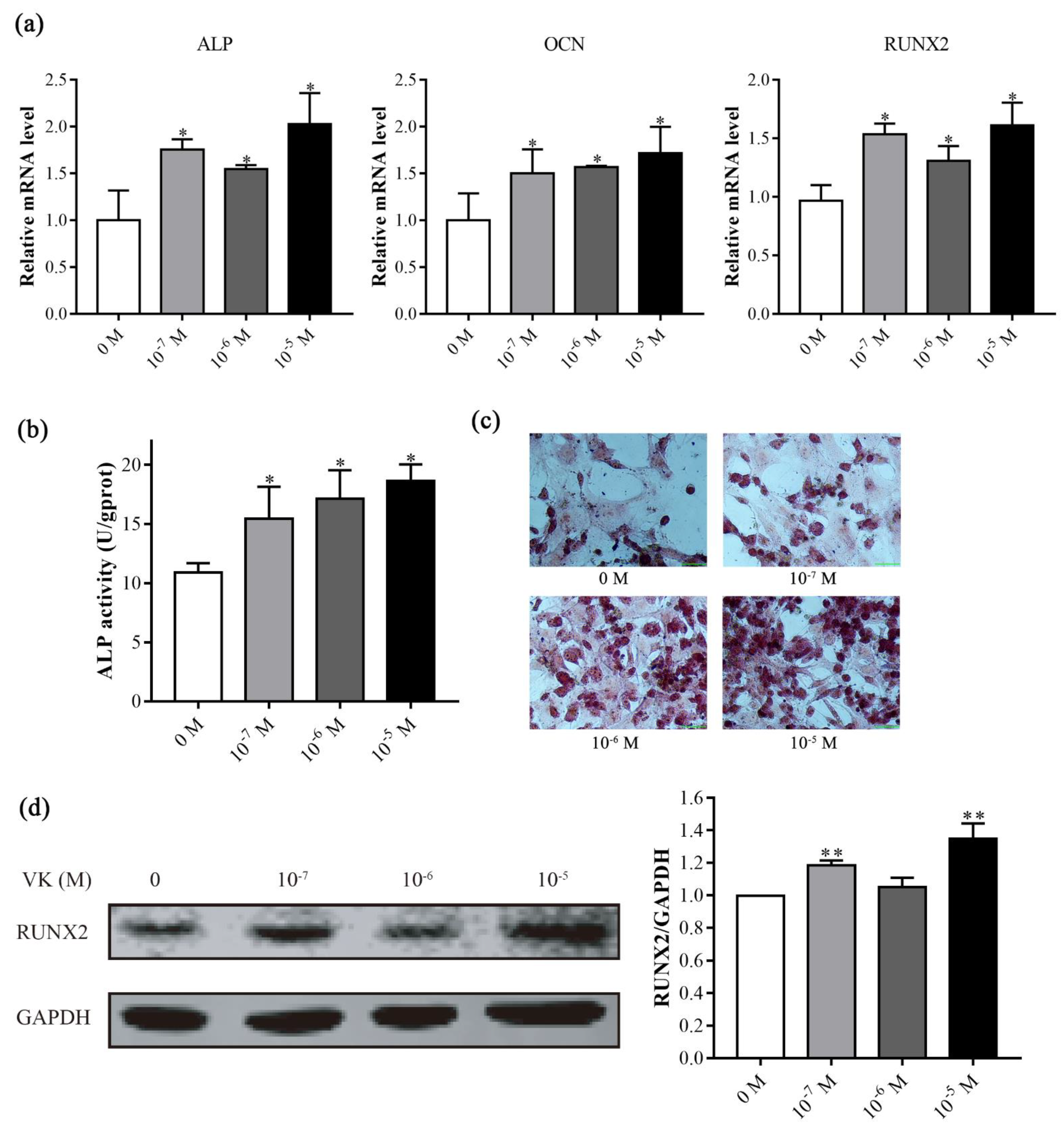
3.2. VK2 Promoted Osteogenic Differentiation in C3H10 T1/2 Clone 8 Cells
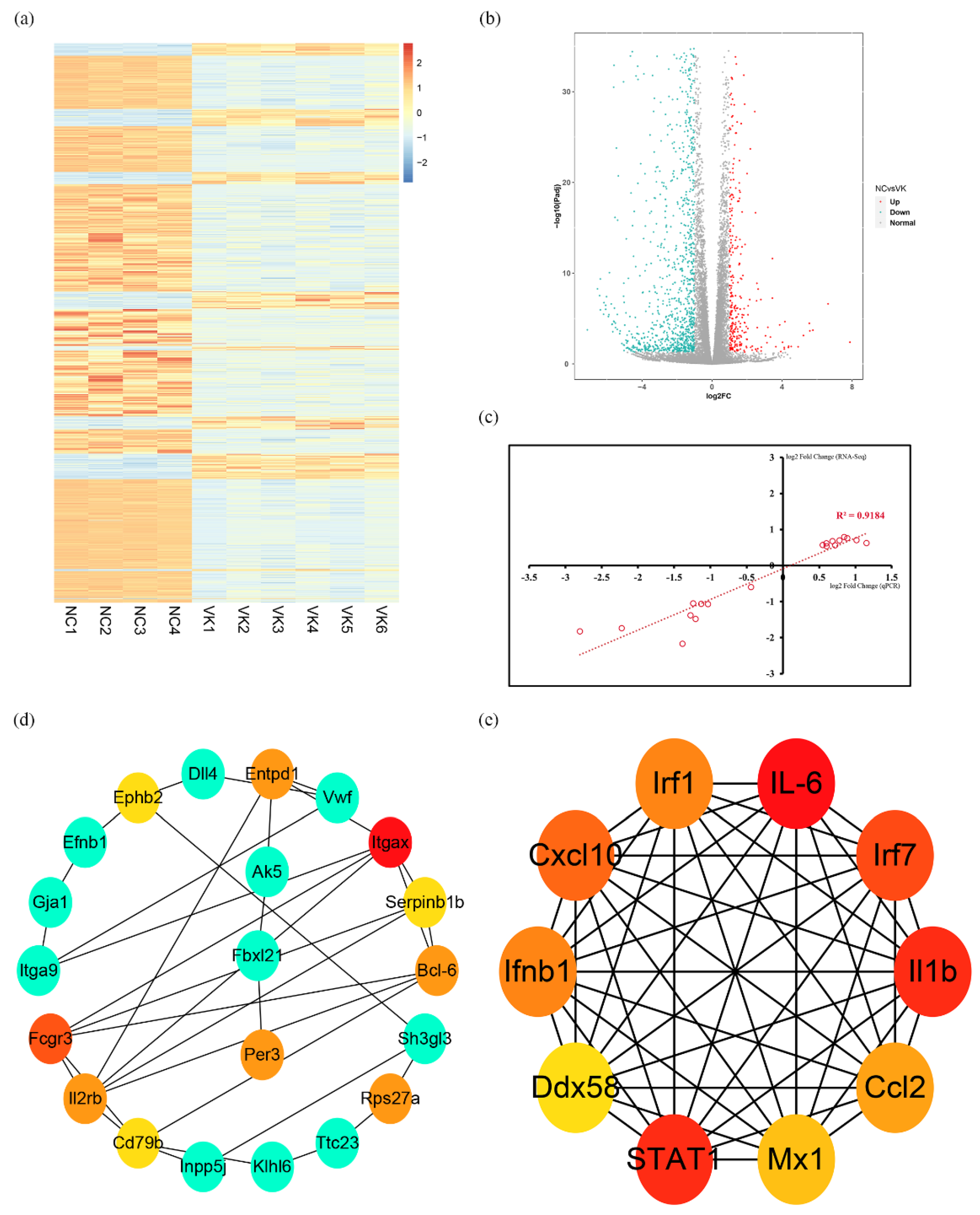
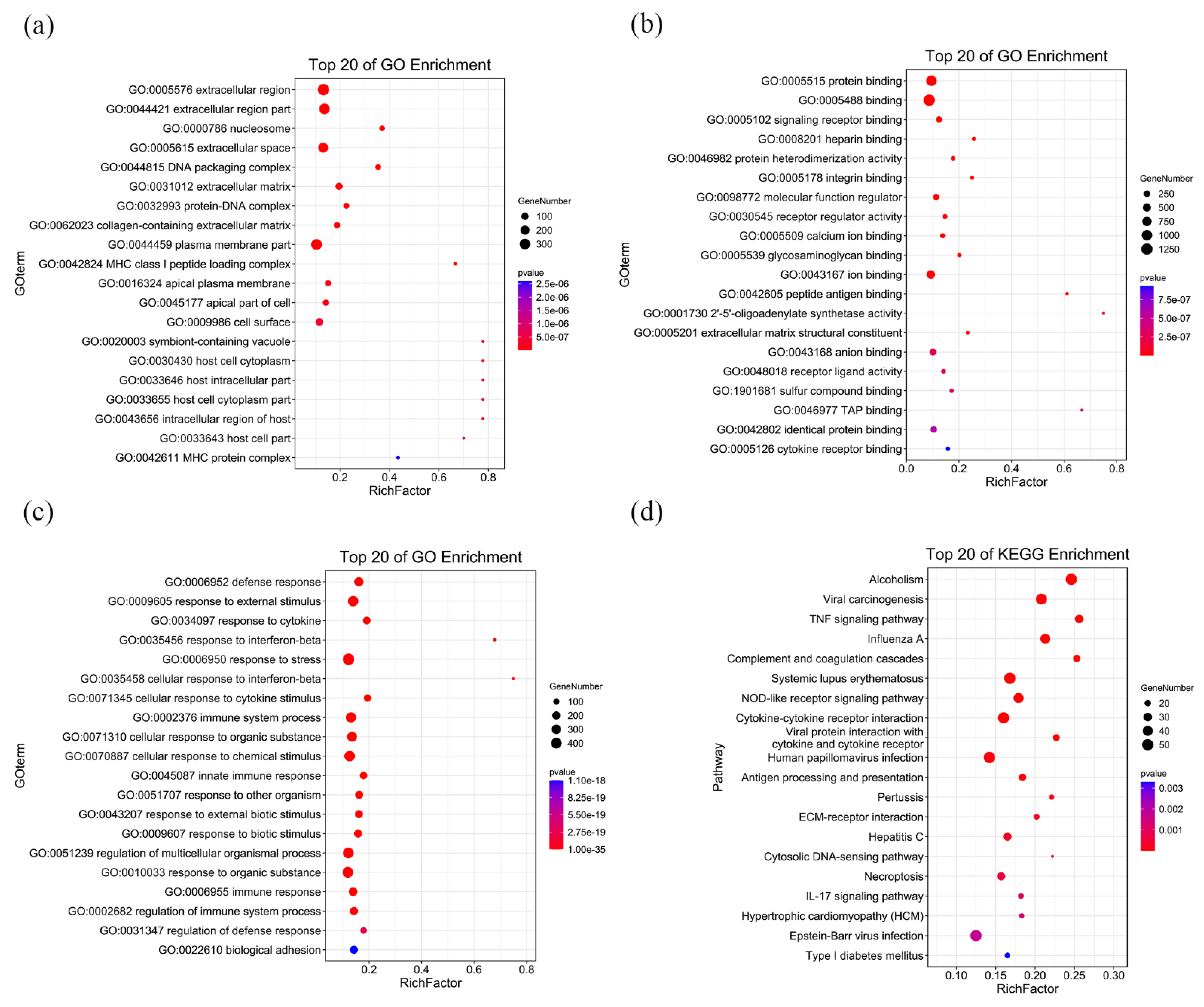
3.3. RNA-Seq Analysis of C3H10 T1/2 Clone 8 Cells upon VK2 Treatment
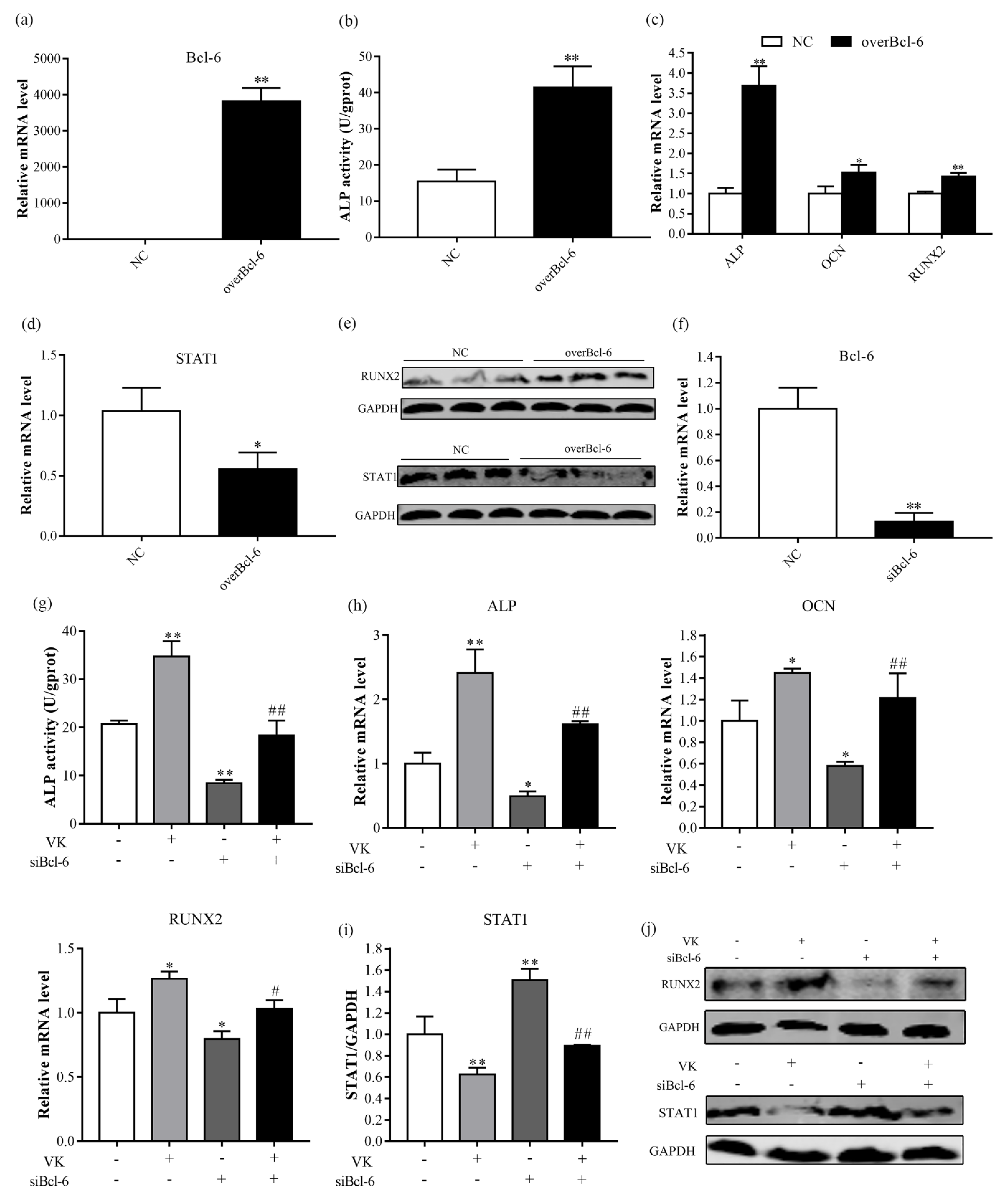
3.4. VK2 Promotes Osteogenic Differentiation by Upregulating Bcl-6
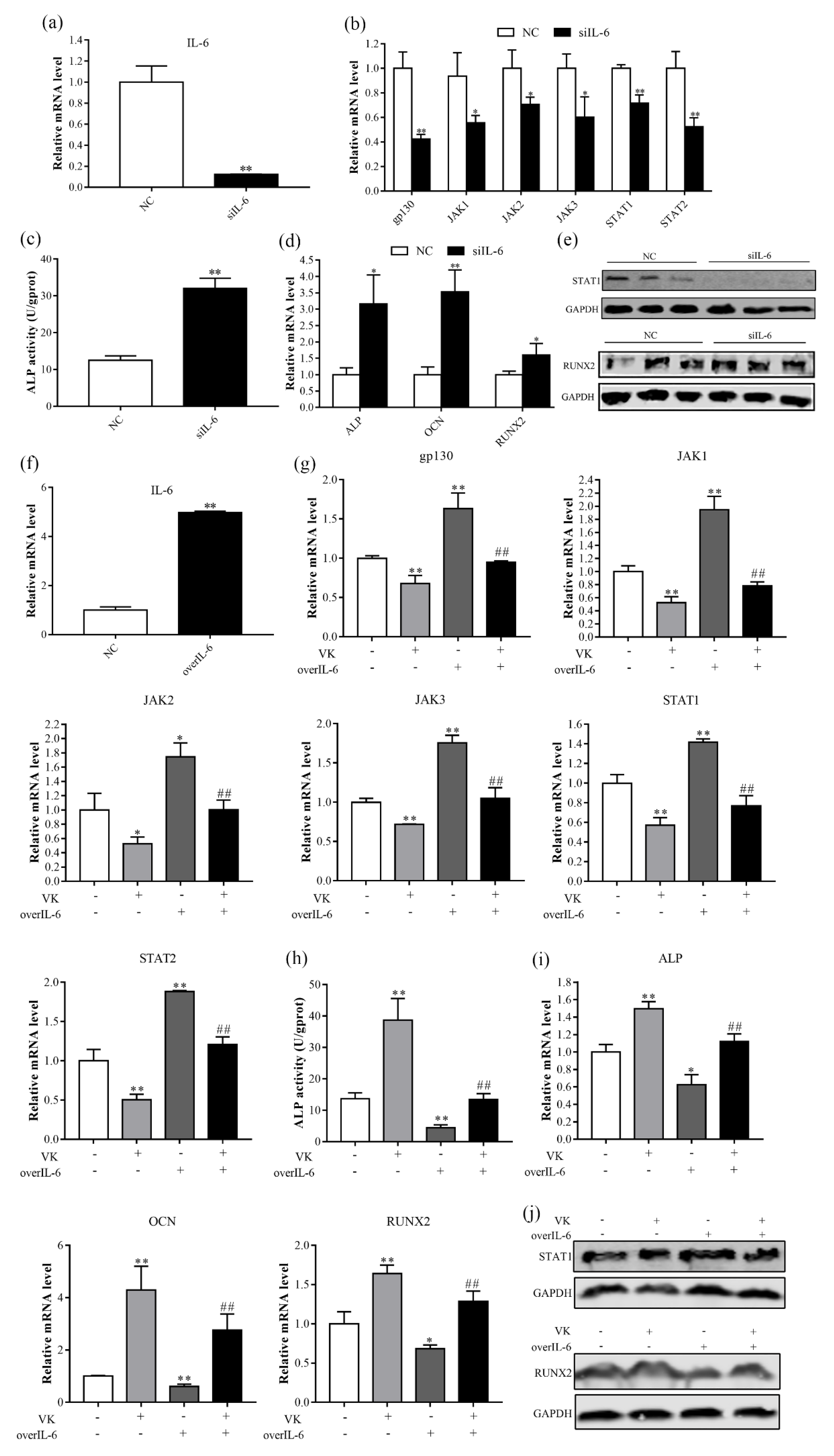
3.5. VK2 Promotes Osteogenic Differentiation by Downregulating IL-6
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lane, J.M.; Russell, L.; Khan, S.N. Osteoporosis. Clin. Orthop. Relat. Res. 2000, 372, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Long, F. Building strong bones: Molecular regulation of the osteoblast lineage. Nat. Rev. Mol. Cell Biol. 2011, 13, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Odén, A.; McCloskey, E.V.; Kanis, J.A.; Harvey, N.C.; Johansson, H. Burden of high fracture probability worldwide: Secular increases 2010–2040. Osteoporos. Int. 2015, 26, 2243–2248. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Sterbenz, J.M.; Malay, S.; Zhong, L.; MacEachern, M.P.; Chung, K.C. Effectiveness of anti-osteoporotic drugs to prevent secondary fragility fractures: Systematic review and meta-analysis. Osteoporos Int 2017, 28, 3289–3300. [Google Scholar] [CrossRef]
- Duque, G.; Vidal, C.; Li, W.; Al Saedi, A.; Khalil, M.; Lim, C.K.; Myers, D.E.; Guillemin, G.J. Picolinic Acid, a Catabolite of Tryptophan, Has an Anabolic Effect on Bone in vivo. J. Bone Miner. Res. 2010, 35, 2275–2288. [Google Scholar] [CrossRef]
- Palermo, A.; Tuccinardi, D.; D’Onofrio, L.; Watanabe, M.; Maggi, D.; Maurizi, A.R.; Greto, V.; Buzzetti, R.; Napoli, N.; Pozzilli, P.; et al. Vitamin K and osteoporosis: Myth or reality? Metabolism 2017, 70, 57–71. [Google Scholar] [CrossRef]
- Akbari, S.; Rasouli-Ghahroudi, A.A. Vitamin K and Bone Metabolism: A Review of the Latest Evidence in Preclinical Studies. Biomed. Res. Int. 2018, 2018, 4629383. [Google Scholar] [CrossRef]
- Li, W.; Zhang, S.; Liu, J.; Liu, Y.; Liang, Q. Vitamin K2 stimulates MC3T3-E1 osteoblast differentiation and mineralization through autophagy induction. Mol. Med. Rep. 2019, 19, 3676–3684. [Google Scholar] [CrossRef]
- Wu, W.J.; Gao, H.; Jin, J.S.; Ahn, B.Y. A comparatively study of menaquinone-7 isolated from Cheonggukjang with vitamin K1 and menaquinone-4 on osteoblastic cells differentiation and mineralization. Food Chem. Toxicol. 2019, 131, 110540. [Google Scholar] [CrossRef]
- Kaneki, M.; Hodges, S.J.; Hosoi, T.; Fujiwara, S.; Lyons, A.; Crean, S.J.; Ishida, N.; Nakagawa, M.; Takechi, M.; Sano, Y.; et al. Japanese fermented soybean food as the major determinant of the large geographic difference in circulating levels of vitamin K2: Possible implications for hip-fracture risk. Nutrition 2001, 17, 315–321. [Google Scholar] [CrossRef]
- Binkley, N.; Harke, J.; Krueger, D.; Engelke, J.; Vallarta-Ast, N.; Gemar, D.; Checovich, M.; Chappell, R.; Suttie, J. Vitamin K treatment reduces undercarboxylated osteocalcin but does not alter bone turnover, density, or geometry in healthy postmenopausal North American women. J. Bone Miner. Res. 2009, 24, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Cockayne, S.; Adamson, J.; Lanham-New, S.; Shearer, M.J.; Gilbody, S.; Torgerson, D.J. Vitamin K and the prevention of fractures: Systematic review and meta-analysis of randomized controlled trials. Arch. Intern. Med. 2006, 166, 1256–1261. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, N.; Li, L.; Yang, P.; Ma, Y. Menaquinone 4 Reduces Bone Loss in Ovariectomized Mice through Dual Regulation of Bone Remodeling. Nutrients 2021, 13, 2570. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.L.; Weintraub, H.; Lassar, A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987, 51, 987–1000. [Google Scholar] [CrossRef]
- Konieczny, S.F.; Emerson, C.P., Jr. 5-Azacytidine induction of stable mesodermal stem cell lineages from 10T1/2 cells: Evidence for regulatory genes controlling determination. Cell 1984, 38, 791–800. [Google Scholar] [CrossRef]
- Taylor, S.M.; Jones, P.A. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell 1979, 17, 771–779. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Chen, Q.; Yu, D.; Zhao, Y.; Qiu, J.; Xie, Y.; Tao, M. Screening and identification of hub genes in pancreatic cancer by integrated bioinformatics analysis. J. Cell Biochem. 2019, 120, 19496–19508. [Google Scholar] [CrossRef]
- Fujie, A.; Funayama, A.; Miyauchi, Y.; Sato, Y.; Kobayashi, T.; Kanagawa, H.; Katsuyama, E.; Hao, W.; Tando, T.; Watanabe, R.; et al. Bcl6 promotes osteoblastogenesis through Stat1 inhibition. Biochem. Biophys. Res. Commun. 2015, 457, 451–456. [Google Scholar] [CrossRef]
- Hirano, T.; Ishihara, K.; Hibi, M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene 2000, 19, 2548–2556. [Google Scholar] [CrossRef] [PubMed]
- Eastell, R.; Szulc, P. Use of bone turnover markers in postmenopausal osteoporosis. Lancet Diabetes Endocrinol. 2017, 5, 908–923. [Google Scholar] [CrossRef]
- Cui, Q.; Li, N.; Nie, F.; Yang, F.; Li, H.; Zhang, J. Vitamin K2 promotes the osteogenic differentiation of periodontal ligament stem cells via the Wnt/β-catenin signaling pathway. Arch. Oral Biol. 2021, 124, 105057. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Weng, S.; Yin, J.; Ding, H.; Zhang, C.; Gao, Y. Vitamin K2 promotes mesenchymal stem cell differentiation by inhibiting miR-133a expression. Mol. Med. Rep. 2017, 15, 2473–2480. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nizet, A.; Cavalier, E.; Stenvinkel, P.; Haarhaus, M.; Magnusson, P. Bone alkaline phosphatase: An important biomarker in chronic kidney disease-mineral and bone disorder. Clin. Chim. Acta 2020, 501, 198–206. [Google Scholar] [CrossRef]
- Lian, J.B.; Stein, G.S. Development of the osteoblast phenotype: Molecular mechanisms mediating osteoblast growth and differentiation. Iowa Orthop. J. 1995, 15, 118–140. [Google Scholar] [PubMed]
- Vimalraj, S. Alkaline phosphatase: Structure, expression and its function in bone mineralization. Gene 2000, 754, 144855. [Google Scholar] [CrossRef]
- Komori, T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 2010, 339, 189–195. [Google Scholar] [CrossRef]
- Komori, T. Functions of Osteocalcin in Bone, Pancreas, Testis, and Muscle. Int. J. Mol. Sci. 2020, 21, 7513. [Google Scholar] [CrossRef]
- Komori, T. Runx2, an inducer of osteoblast and chondrocyte differentiation. Histochem. Cell Biol. 2018, 149, 313–323. [Google Scholar] [CrossRef]
- Datta, H.K.; Ng, W.F.; Walker, J.A.; Tuck, S.P.; Varanasi, S.S. The cell biology of bone metabolism. J. Clin. Pathol. 2008, 61, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.C.W.; Tan, Z.; To, M.K.T.; Chan, D. Regulation and Role of Transcription Factors in Osteogenesis. Int. J. Mol. Sci. 2021, 22, 5445. [Google Scholar] [CrossRef] [PubMed]
- Ayturk, U. RNA-seq in Skeletal Biology. Curr. Osteoporos. Rep. 2019, 17, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Basso, K.; Dalla-Favera, R. BCL6: Master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv. Immunol. 2010, 105, 193–210. [Google Scholar]
- Miyauchi, Y.; Ninomiya, K.; Miyamoto, H. The Blimp1-Bcl6 axis is critical to regulate osteoclast differentiation and bone homeostasis. J. Exp. Med. 2010, 207, 751–762. [Google Scholar] [CrossRef]
- Damerau, A.; Gaber, T.; Ohrndorf, S. JAK/STAT Activation: A General Mechanism for Bone Development, Homeostasis, and Regeneration. Int. J. Mol. Sci. 2020, 21, 9004. [Google Scholar] [CrossRef]
- Morris, R.; Kershaw, N.J.; Babon, J.J. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 2018, 27, 1984–2009. [Google Scholar] [CrossRef]
- Wu, Q.; Zhou, X.; Huang, D. IL-6 Enhances Osteocyte-Mediated Osteoclastogenesis by Promoting JAK2 and RANKL Activity In Vitro. Cell. Physiol. Biochem. 2017, 41, 1360–1369. [Google Scholar] [CrossRef]
- Kim, S.; Koga, T.; Isobe, M. Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation. Genes Dev. 2003, 17, 1979–1991. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Li, L.; Zhang, N.; Ma, Y. Vitamin K2 Improves Osteogenic Differentiation by Inhibiting STAT1 via the Bcl-6 and IL-6/JAK in C3H10 T1/2 Clone 8 Cells. Nutrients 2022, 14, 2934. https://doi.org/10.3390/nu14142934
Wang H, Li L, Zhang N, Ma Y. Vitamin K2 Improves Osteogenic Differentiation by Inhibiting STAT1 via the Bcl-6 and IL-6/JAK in C3H10 T1/2 Clone 8 Cells. Nutrients. 2022; 14(14):2934. https://doi.org/10.3390/nu14142934
Chicago/Turabian StyleWang, Huakai, Longxian Li, Nan Zhang, and Yongxi Ma. 2022. "Vitamin K2 Improves Osteogenic Differentiation by Inhibiting STAT1 via the Bcl-6 and IL-6/JAK in C3H10 T1/2 Clone 8 Cells" Nutrients 14, no. 14: 2934. https://doi.org/10.3390/nu14142934
APA StyleWang, H., Li, L., Zhang, N., & Ma, Y. (2022). Vitamin K2 Improves Osteogenic Differentiation by Inhibiting STAT1 via the Bcl-6 and IL-6/JAK in C3H10 T1/2 Clone 8 Cells. Nutrients, 14(14), 2934. https://doi.org/10.3390/nu14142934





