Effects of Marine Bioactive Compounds on Gut Ecology Based on In Vitro Digestion and Colonic Fermentation Models
Abstract
:1. Introduction
2. In Vitro Gastrointestinal Digestion Models
2.1. Static Digestion Models
2.2. Dynamic Digestion Models
2.2.1. Mono-Compartment System
- Dynamic gastric model (DGM)
- Human gastric simulator (HGS)
2.2.2. Multi-Compartment System
- In vitro dynamic system (DIDGI)
- TNO gastrointestinal model (TIM)
- Simulator of the human intestinal microbial ecosystem (SHIME®)
- Engineered stomach and small intestinal system (ESIN)
3. In Vitro Colonic Fermentation
3.1. In Vitro Colonic Fermentation Models
3.1.1. In Vitro Static Batch Fermentation Models
3.1.2. In Vitro Dynamic Continuous Models
4. In Vitro Digestion and Fermentation of Marine Bioactive Compounds
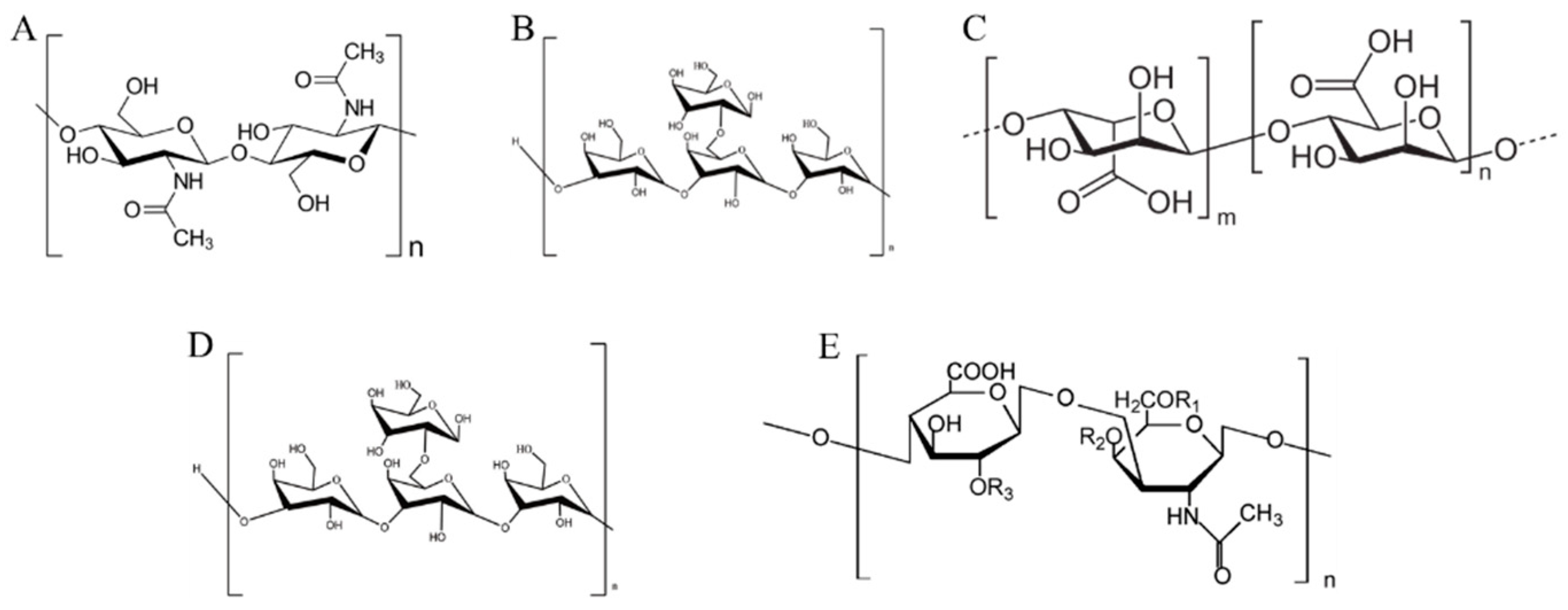
4.1. Polysaccharides
| Source | In Vitro Digestion Stages and Model | Colonic Fermentation | Results | Ref. |
|---|---|---|---|---|
| Gracilaria chouae sulfated polysaccharides | Static digestion model | Static batch fermentation | Polysaccharides are slightly degraded; different extraction methods have an effect on the enteric fermentation of polysaccharides. | [65] |
| Sea cucumber fucosylated glycosaminoglycan | Static digestion model | There is no release of free monosaccharides. | [66] | |
| Abalone sulfated polysaccharides | TIM model | Mice model/Static batch fermentation | Simulated digestive juices have no effect on polysaccharides, which regulate gut microbiota. | [68] |
| Gracilaria rubra sulfated polysaccharides | Static digestion model | Static batch fermentation | Polysaccharide is not digested by gastrointestinal tract;fermentation produces acetic acid, propionic acid, etc., and reduces the ratio of Firmicutes/Bacteroidetes. | [70] |
| Ascophyllum nodosum polysaccharides | Static digestion model | Static batch fermentation | Polysaccharide is not digested by saliva and gastrointestinal tract;colonic fermentation reduces the molecular weight of polysaccharides and reduces sugars;increases the relative abundance of Bacteroidetes and Firmicutes. | [71] |
| Coralline pilulifera polysaccharides | Static digestion model | Static batch fermentation | Polysaccharide is not digested by gastrointestinal tract;after 24 h of in vitro fermentation, polysaccharide content is reduced by 70%. | [72] |
| Nostoc commune Vauch polysaccharides (NCVPs) | Static digestion model | Static batch fermentation | Degradation of polysaccharides occurs during digestion process;NCVPs have the potential to promote intestinal metabolism. | [73] |
| κ-carrageenans | Static digestion model | Static batch fermentation | κ-carrageenan oligosaccharide was obtained after simulated gastric digestion;κ-carrageenan oligosaccharides with large degree of polymerization enhance the production of SCFAs and increase the abundance of Bifidobacteria and Lactobacillius. | [74] |
| Sea cucumber polysaccharides | Static batch fermentation | Fermentation contributes to the accumulation of beneficial microbial metabolites, including propionic acid, butyric acid, amino acid and derivatives. | [78] | |
| Oyster polysaccharides | Static digestion model | Static batch fermentation | A part of the polysaccharides is degraded during the digestion process;indigestible polysaccharides are utilized by the gut microbiota to contribute to SCFAs generation. | [79] |
| Gracilaria Lemaneiformis sulfated polysaccharide | Static digestion model | Static batch fermentation | Sulfated polysaccharide is degraded during fermentation;gut microbes are able to utilize sulfated polysaccharide and produce SCFAs. | [80] |
| Laminaria digitata polysaccharides | Static digestion model | Dynamic continuous models | Laminaria digitata polysaccharides resist degradation by digestive enzymes and are fermented by gut microbiota, changing the abundance of Streptococcus, Ruminococcus, etc. They also increase the concentration of SCFAs such as acetic acid and propionic acid. | [81] |
4.2. Protein
4.3. Lipids
4.4. Polyphenols and Other High-Value Components
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ueland, Ø.; Altintzoglou, T.; Kirkhus, B.; Lindberg, D.; Rognså, G.H.; Rosnes, J.T.; Rud, I.; Varela, P. Perspectives on personalised food. Trends Food Sci. Technol. 2020, 102, 169–177. [Google Scholar] [CrossRef]
- Verkempinck, S.; Pallares Pallares, A.; Hendrickx, M.; Grauwet, T. Processing as a tool to manage digestive barriers in plant-based foods: Recent advances. Curr. Opin. Food Sci. 2020, 35, 1–9. [Google Scholar] [CrossRef]
- Dupont, D.; Le Feunteun, S.; Marze, S.; Souchon, I. Structuring food to control its disintegration in the gastrointestinal tract and optimize nutrient bioavailability. Innov. Food Sci. Emerg. Technol. 2018, 46, 83–90. [Google Scholar] [CrossRef]
- De Angelis, M.; Ferrocino, I.; Calabrese, F.M.; De Filippis, F.; Cavallo, N.; Siragusa, S.; Rampelli, S.; Di Cagno, R.; Rantsiou, K.; Vannini, L.; et al. Diet influences the functions of the human intestinal microbiome. Sci. Rep. 2020, 10, 4247. [Google Scholar] [CrossRef] [PubMed]
- Sensoy, I. A review on the food digestion in the digestive tract and the used in vitro models. Curr. Res. Food Sci. 2021, 4, 308–319. [Google Scholar] [CrossRef]
- Bescos, R.; Brookes, Z.L.S.; Belfield, L.A.; Fernandez-Sanjurjo, M.; Casas-Agustench, P. Modulation of oral microbiota: A new frontier in exercise supplementation. PharmaNutrition 2020, 14, 100230. [Google Scholar] [CrossRef]
- Helander, H.F.; Fändriks, L. Surface area of the digestive tract-revisited. Scand. J. Gastroenterol. 2014, 49, 681–689. [Google Scholar] [CrossRef]
- Wu, P.; Chen, X.D. On designing biomimic in vitro human and animal digestion track models: Ideas, current and future devices. Curr. Opin. Food Sci. 2020, 35, 10–19. [Google Scholar] [CrossRef]
- Hernalsteens, S.; Huang, S.; Cong, H.H.; Chen, X.D. The final fate of food: On the establishment of in vitro colon models. Food Res. Int. 2021, 150, 110743. [Google Scholar] [CrossRef]
- Shahidi, F.; Ambigaipalan, P. Novel functional food ingredients from marine sources. Curr. Opin. Food Sci. 2015, 2, 123–129. [Google Scholar] [CrossRef]
- Grienke, U.; Silke, J.; Tasdemir, D. Bioactive compounds from marine mussels and their effects on human health. Food Chem. 2014, 142, 48–60. [Google Scholar] [CrossRef]
- Atef, M.; Mahdi Ojagh, S. Health benefits and food applications of bioactive compounds from fish byproducts: A review. J. Funct. Foods 2017, 35, 673–681. [Google Scholar] [CrossRef]
- Ideia, P.; Pinto, J.; Ferreira, R.; Figueiredo, L.; Spínola, V.; Castilho, P.C. Fish processing industry residues: A review of valuable products extraction and characterization methods. Waste Biomass Valorization 2019, 11, 3223–3246. [Google Scholar] [CrossRef]
- Portela, L.C.P.N.; Cahú, T.B.; Bezerra, T.S.; do Nascimento Santos, D.K.D.; Sousa, G.F.; Portela, R.W.S.; Melo, C.M.L.; de Souza Bezerra, R. Biocompatibility and immunostimulatory properties of fish collagen and shrimp chitosan towards peripheral blood mononuclear cells (PBMCs). Int. J. Biol. Macromol. 2022, 210, 282–291. [Google Scholar] [CrossRef]
- Zhang, T.; Bai, X.; Dai, Y.; Dong, P.; Wang, J. Different n-6/n-3 PUFA diets with fish oil attenuated osteoarthritis in ovariectomized mice via targeting the NLRP3 inflammasome. Food Biosci. 2022, 45, 101220. [Google Scholar] [CrossRef]
- Xu, Q.; Ritzoulis, C.; Han, J.; Han, F.; Jin, W.; Liu, W. Particle degradation and nutrient bioavailability of soybean milk during in vitro digestion. Food Biophys. 2021, 16, 58–69. [Google Scholar] [CrossRef]
- Lucas-González, R.; Viuda-Martos, M.; Pérez-Alvarez, J.A.; Fernández-López, J. In vitro digestion models suitable for foods: Opportunities for new fields of application and challenges. Food Res. Int. 2018, 107, 423–436. [Google Scholar] [CrossRef]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised static in vitro digestion method suitable for food-an international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F.; et al. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef]
- Egger, L.; Ménard, O.; Delgado-Andrade, C.; Alvito, P.; Assunção, R.; Balance, S.; Barberá, R.; Brodkorb, A.; Cattenoz, T.; Clemente, A.; et al. The harmonized INFOGEST in vitro digestion method: From knowledge to action. Food Res. Int. 2016, 88, 217–225. [Google Scholar] [CrossRef]
- Bustos, M.C.; Vignola, M.B.; Pérez, G.T.; León, A.E. In vitro digestion kinetics and bioaccessibility of starch in cereal food products. J. Cereal Sci. 2017, 77, 243–250. [Google Scholar] [CrossRef]
- Calvo-Lerma, J.; Asensio-Grau, A.; Heredia, A.; Andrés, A. Lessons learnt from MyCyFAPP Project: Effect of cystic fibrosis factors and inherent-to-food properties on lipid digestion in foods. Food Res. Int. 2020, 133, 109198. [Google Scholar] [CrossRef] [PubMed]
- Shani-Levi, C.; Alvito, P.; Andrés, A.; Assunção, R.; Barberá, R.; Blanquet-Diot, S.; Bourlieu, C.; Brodkorb, A.; Cilla, A.; Deglaire, A.; et al. Extending in vitro digestion models to specific human populations: Perspectives, practical tools and bio-relevant information. Trends Food Sci. Technol. 2017, 60, 52–63. [Google Scholar] [CrossRef]
- Carbonell-Capella, J.M.; Buniowska, M.; Barba, F.J.; Esteve, M.J.; Frígola, A. Analytical Methods for Determining Bioavailability and Bioaccessibility of Bioactive Compounds from Fruits and Vegetables: A Review. Compr. Rev. Food Sci. Food Saf. 2014, 13, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Wickham, M.J.S.; Faulks, R.M.; Mann, J.; Mandalari, G. The design, operation, and application of a dynamic gastric model. Dissolution Technol. 2012, 19, 15–22. [Google Scholar] [CrossRef]
- Verhoeckx, K.; Cotter, P.; López-Expósito, I.; Kleiveland, C.; Lea, T.; Mackie, A.; Requena, T.; Swiatecka, D.; Wichers, H. The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Springer: Berlin/Heidelberg, Germany, 2015; ISBN 9783319161044. [Google Scholar]
- Kong, F.; Singh, R.P. A Human Gastric Simulator (HGS) to Study Food Digestion in Human Stomach. J. Food Sci. 2010, 75, E627–E635. [Google Scholar] [CrossRef] [PubMed]
- Bornhorst, G.M.; Rutherfurd, S.M.; Roman, M.J.; Burri, B.J.; Moughan, P.J.; Singh, R.P. Gastric pH distribution and mixing of soft and rigid food particles in the stomach using a Dual-Marker technique. Food Biophys. 2014, 9, 292–300. [Google Scholar] [CrossRef]
- Ménard, O.; Cattenoz, T.; Guillemin, H.; Souchon, I.; Deglaire, A.; Dupont, D.; Picque, D. Validation of a new in vitro dynamic system to simulate infant digestion. Food Chem. 2014, 145, 1039–1045. [Google Scholar] [CrossRef]
- De La Pomélie, D.; Santé-Lhoutellier, V.; Sayd, T.; Théron, L.; Gatellier, P. Using a dynamic artificial digestive system to investigate heme iron nitrosylation during gastro-intestinal transit. Food Chem. 2019, 281, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Reynaud, Y.; Couvent, A.; Manach, A.; Forest, D.; Lopez, M.; Picque, D.; Souchon, I.; Rémond, D.; Dupont, D. Food-dependent set-up of the DIDGI® dynamic in vitro system: Correlation with the porcine model for protein digestion of soya-based food. Food Chem. 2021, 341, 128276. [Google Scholar] [CrossRef]
- Bellmann, S.; Lelieveld, J.; Gorissen, T.; Minekus, M.; Havenaar, R. Development of an advanced in vitro model of the stomach and its evaluation versus human gastric physiology. Food Res. Int. 2016, 88, 191–198. [Google Scholar] [CrossRef]
- Aguirre, M.; Eck, A.; Koenen, M.E.; Savelkoul, P.H.M.; Budding, A.E.; Venema, K. Evaluation of an optimal preparation of human standardized fecal inocula for in vitro fermentation studies. J. Microbiol. Methods 2015, 117, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Verwei, M.; Minekus, M.; Zeijdner, E.; Schilderink, R.; Havenaar, R. Evaluation of two dynamic in vitro models simulating fasted and fed state conditions in the upper gastrointestinal tract (TIM-1 and tiny-TIM) for investigating the bioaccessibility of pharmaceutical compounds from oral dosage forms. Int. J. Pharm. 2016, 498, 178–186. [Google Scholar] [CrossRef]
- Blanquet, S.; Zeijdner, E.; Beyssac, E.; Meunier, J.P.; Denis, S.; Havenaar, R.; Alric, M. A dynamic artificial gastrointestinal system for studying the behavior of orally administered drug dosage forms under various physiological conditions. Pharm. Res. 2004, 21, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Molly, K.; Woestyne, M.V.; De Smet, I.; Verstraete, W. Validation of the simulator of the human intestinal microbial ecosystem (SHIME) reactor using microorganism-associated activities. Microb. Ecol. Health Dis. 1994, 7, 191–200. [Google Scholar] [CrossRef]
- Marzorati, M.; Vanhoecke, B.; De Ryck, T.; Sadaghian Sadabad, M.; Pinheiro, I.; Possemiers, S.; Van Den Abbeele, P.; Derycke, L.; Bracke, M.; Pieters, J.; et al. The HMI™ module: A new tool to study the host-microbiota interaction in the human gastrointestinal tract in vitro. BMC Microbiol. 2014, 14, 133. [Google Scholar] [CrossRef] [PubMed]
- Van Den Abbeele, P.; Belzer, C.; Goossens, M.; Kleerebezem, M.; De Vos, W.M.; Thas, O.; De Weirdt, R.; Kerckhof, F.M.; Van De Wiele, T. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013, 7, 949–961. [Google Scholar] [CrossRef]
- Voropaiev, M.; Nock, D. Onset of acid-neutralizing action of a calcium/magnesium carbonate-based antacid using an artificial stomach model: An in vitro evaluation. BMC Gastroenterol. 2021, 21, 1–6. [Google Scholar] [CrossRef]
- Guerra, A.; Etienne-Mesmin, L.; Livrelli, V.; Denis, S.; Blanquet-Diot, S.; Alric, M. Relevance and challenges in modeling human gastric and small intestinal digestion. Trends Biotechnol. 2012, 30, 591–600. [Google Scholar] [CrossRef]
- Guerra, A.; Denis, S.; le Goff, O.; Sicardi, V.; François, O.; Yao, A.F.; Garrait, G.; Manzi, A.P.; Beyssac, E.; Alric, M.; et al. Development and validation of a new dynamic computer-controlled model of the human stomach and small intestine. Biotechnol. Bioeng. 2016, 113, 1325–1335. [Google Scholar] [CrossRef]
- Barroso, E.; Cueva, C.; Peláez, C.; Martínez-Cuesta, M.C.; Requena, T. Development of human colonic microbiota in the computer-controlled dynamic SIMulator of the GastroIntestinal tract SIMGI. LWT-Food Sci. Technol. 2015, 61, 283–289. [Google Scholar] [CrossRef]
- Chen, J.; Gaikwad, V.; Holmes, M.; Murray, B.; Povey, M.; Wang, Y.; Zhang, Y. Development of a simple model device for in vitro gastric digestion investigation. Food Funct. 2011, 2, 174–182. [Google Scholar] [CrossRef]
- Barros, L.; Retamal, C.; Torres, H.; Zúñiga, R.N.; Troncoso, E. Development of an in vitro mechanical gastric system (IMGS) with realistic peristalsis to assess lipid digestibility. Food Res. Int. 2016, 90, 216–225. [Google Scholar] [CrossRef]
- Wang, J.; Wu, P.; Liu, M.; Liao, Z.; Wang, Y.; Dong, Z.; Chen, X.D. An advanced near real dynamic: In vitro human stomach system to study gastric digestion and emptying of beef stew and cooked rice. Food Funct. 2019, 10, 2914–2925. [Google Scholar] [CrossRef]
- Passannanti, F.; Nigro, F.; Gallo, M.; Tornatore, F.; Frasso, A.; Saccone, G.; Budelli, A.; Barone, M.V.; Nigro, R. In vitro dynamic model simulating the digestive tract of 6-month-old infants. PLoS ONE 2017, 12, e0189807. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Ye, A.; Han, F.; Han, J. Advances and challenges in liposome digestion: Surface interaction, biological fate, and GIT modeling. Adv. Colloid Interface Sci. 2019, 263, 52–67. [Google Scholar] [CrossRef]
- Lazar, V.; Ditu, L.M.; Pircalabioru, G.G.; Gheorghe, I.; Curutiu, C.; Holban, A.M.; Picu, A.; Petcu, L.; Chifiriuc, M.C. Aspects of gut microbiota and immune system interactions in infectious diseases, immunopathology, and cancer. Front. Immunol. 2018, 9, 01830. [Google Scholar] [CrossRef] [PubMed]
- Hollister, E.B.; Gao, C.; Versalovic, J. Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology 2014, 146, 1449–1458. [Google Scholar] [CrossRef]
- Zhang, H.; Jiang, F.; Zhang, J.; Wang, W.; Li, L.; Yan, J. Modulatory effects of polysaccharides from plants, marine algae and edible mushrooms on gut microbiota and related health benefits: A review. Int. J. Biol. Macromol. 2022, 204, 169–192. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Mukherjee, S.S.; Chakraborty, R.; Roy, R.R.; Pandey, A.; Patra, S.; Dey, S. The emerging role of gut microbiota in cardiovascular diseases. Indian Heart J. 2021, 73, 264–272. [Google Scholar] [CrossRef]
- Tremlett, H.; Bauer, K.C.; Appel-Cresswell, S.; Finlay, B.B.; Waubant, E. The gut microbiome in human neurological disease: A review. Ann. Neurol. 2017, 81, 369–382. [Google Scholar] [CrossRef]
- Mithul Aravind, S.; Wichienchot, S.; Tsao, R.; Ramakrishnan, S.; Chakkaravarthi, S. Role of dietary polyphenols on gut microbiota, their metabolites and health benefits. Food Res. Int. 2021, 142, 110189. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wang, Y.; Yang, G.; Zhang, Q.; Meng, L.; Xin, Y.; Jiang, X. The role of short-chain fatty acids in intestinal barrier function, inflammation, oxidative stress, and colonic carcinogenesis. Pharmacol. Res. 2021, 165, 105420. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Mikami, D.; Kimura, H.; Kamiyama, K.; Morikawa, Y.; Yokoi, S.; Kasuno, K.; Takahashi, N.; Taniguchi, T.; Iwano, M. Short-chain fatty acids, GPR41 and GPR43 ligands, inhibit TNF-α-induced MCP-1 expression by modulating p38 and JNK signaling pathways in human renal cortical epithelial cells. Biochem. Biophys. Res. Commun. 2017, 486, 499–505. [Google Scholar] [CrossRef]
- Barry, J.-L.; Hoebler, C.; MacFarlane, G.T.; MacFarlane, S.; Mathers, J.C.; Reed, K.A.; Mortensen, P.B.; Nordgaard, I.; Rowland, I.R.; Rumney, C.J. Estimation of the fermentability of dietary fibre in vitro: A European interlaboratory study. Br. J. Nutr. 1995, 74, 303–322. [Google Scholar] [CrossRef]
- Li, Z.T.; Hu, G.A.; Zhu, L.; Zhao, Z.C.; Jiang, Y.; Gao, M.J.; Zhan, X.B. In vitro digestion and fecal fermentation of highly resistant starch rice and its effect on the gut microbiota. Food Chem. 2021, 361, 130095. [Google Scholar] [CrossRef]
- Yadav, V.; Gaisford, S.; Merchant, H.A.; Basit, A.W. Colonic bacterial metabolism of corticosteroids. Int. J. Pharm. 2013, 457, 268–274. [Google Scholar] [CrossRef]
- Cárdenas-Castro, A.P.; Zamora-Gasga, V.M.; Alvarez-Parrilla, E.; Ruíz-Valdiviezo, V.M.; Venema, K.; Sáyago-Ayerdi, S.G. In vitro gastrointestinal digestion and colonic fermentation of tomato (Solanum lycopersicum L.) and husk tomato (Physalis ixocarpa Brot.): Phenolic compounds released and bioconverted by gut microbiota. Food Chem. 2021, 360, 130051. [Google Scholar] [CrossRef] [PubMed]
- Geirnaert, A.; Calatayud, M.; Grootaert, C.; Laukens, D.; Devriese, S.; Smagghe, G.; De Vos, M.; Boon, N.; Van De Wiele, T. Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci. Rep. 2017, 7, 11450. [Google Scholar] [CrossRef]
- Ferreira-Lazarte, A.; Moreno, F.J.; Cueva, C.; Gil-Sánchez, I.; Villamiel, M. Behaviour of citrus pectin during its gastrointestinal digestion and fermentation in a dynamic simulator (simgi®). Carbohydr. Polym. 2019, 207, 382–390. [Google Scholar] [CrossRef]
- Cordonnier, C.; Thévenot, J.; Etienne-Mesmin, L.; Denis, S.; Alric, M.; Livrelli, V.; Blanquet-Diot, S. Dynamic in vitro models of the human gastrointestinal tract as relevant tools to assess the survival of probiotic strains and their interactions with gut microbiota. Microorganisms 2015, 3, 725–745. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Cummings, J.H.; Macfarlane, G.T. Use of a three-stage continuous culture system to study the effect of mucin on dissimilatory sulfate reduction and methanogenesis by mixed populations of human gut bacteria. Appl. Environ. Microbiol. 1988, 54, 2750–2755. [Google Scholar] [CrossRef]
- De Medeiros, V.P.B.; de Souza, E.L.; de Albuquerque, T.M.R.; da Costa Sassi, C.F.; dos Santos Lima, M.; Sivieri, K.; Pimentel, T.C.; Magnani, M. Freshwater microalgae biomasses exert a prebiotic effect on human colonic microbiota. Algal Res. 2021, 60, 102547. [Google Scholar] [CrossRef]
- Li, X.; Xie, Q.; Huang, S.; Shao, P.; You, L.; Pedisić, S. Digestion & fermentation characteristics of sulfated polysaccharides from Gracilaria chouae using two extraction methods in vitro and in vivo. Food Res. Int. 2021, 145, 110406. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Qin, Y.; Guan, R.; Zheng, W.; Liu, J.; Zhao, J. Digestibility of fucosylated glycosaminoglycan from sea cucumber and its effects on digestive enzymes under simulated salivary and gastrointestinal conditions. Carbohydr. Polym. 2018, 186, 217–225. [Google Scholar] [CrossRef]
- Seong, H.; Bae, J.H.; Seo, J.S.; Kim, S.A.; Kim, T.J.; Han, N.S. Comparative analysis of prebiotic effects of seaweed polysaccharides laminaran, porphyran, and ulvan using in vitro human fecal fermentation. J. Funct. Foods 2019, 57, 408–416. [Google Scholar] [CrossRef]
- Ai, C.; Ma, N.; Sun, X.; Duan, M.; Wu, S.; Yang, J.; Wen, C.; Song, S. Absorption and degradation of sulfated polysaccharide from pacific abalone in in vitro and in vivo models. J. Funct. Foods 2017, 35, 127–133. [Google Scholar] [CrossRef]
- Nie, Q.; Chen, H.; Hu, J.; Fan, S.; Nie, S. Dietary compounds and traditional Chinese medicine ameliorate type 2 diabetes by modulating gut microbiota. Crit. Rev. Food Sci. Nutr. 2019, 59, 848–863. [Google Scholar] [CrossRef]
- Zheng, L.X.; Chen, X.Q.; Cheong, K.L. Current trends in marine algae polysaccharides: The digestive tract, microbial catabolism, and prebiotic potential. Int. J. Biol. Macromol. 2020, 151, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xu, W.; Chen, D.; Chen, G.; Liu, J.; Zeng, X.; Shao, R.; Zhu, H. Digestibility of sulfated polysaccharide from the brown seaweed Ascophyllum nodosum and its effect on the human gut microbiota in vitro. Int. J. Biol. Macromol. 2018, 112, 1055–1061. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, G.; Peng, Y.; Rui, Y.; Zeng, X.; Ye, H. Simulated digestion and fermentation in vitro with human gut microbiota of polysaccharides from Coralline pilulifera. LWT-Food Sci. Technol. 2019, 100, 167–174. [Google Scholar] [CrossRef]
- Li, H.; Liu, S.; Liu, Y.; Li, W.; Niu, A.; Ren, P.; Liu, Y.; Jiang, C.; Inam, M.; Guan, L. Effects of in vitro digestion and fermentation of Nostoc commune Vauch. polysaccharides on properties and gut microbiota. Carbohydr. Polym. 2022, 281, 119055. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Cui, X.; Duan, M.; Ai, C.; Song, S.; Chen, X. In vitro fermentation of κ-carrageenan oligosaccharides by human gut microbiota and its inflammatory effect on HT29 cells. J. Funct. Foods 2019, 59, 80–91. [Google Scholar] [CrossRef]
- Vázquez-Rodríguez, B.; Santos-Zea, L.; Heredia-Olea, E.; Acevedo-Pacheco, L.; Santacruz, A.; Gutiérrez-Uribe, J.A.; Cruz-Suárez, L.E. Effects of phlorotannin and polysaccharide fractions of brown seaweed Silvetia compressa on human gut microbiota composition using an in vitro colonic model. J. Funct. Foods 2021, 84, 104596. [Google Scholar] [CrossRef]
- Pakizeh, M.; Moradi, A.; Ghassemi, T. Chemical extraction and modification of chitin and chitosan from shrimp shells. Eur. Polym. J. 2021, 159, 110709. [Google Scholar] [CrossRef]
- Zhou, C.; Mi, S.; Li, J.; Gao, J.; Wang, X.; Sang, Y. Purification, characterisation and antioxidant activities of chondroitin sulphate extracted from Raja porosa cartilage. Carbohydr. Polym. 2020, 241, 1–8. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, Y.; Ai, C.; Wen, C.; Dong, X.; Sun, X.; Cao, C.; Zhang, X.; Zhu, B.; Song, S. Gut microbiota response to sulfated sea cucumber polysaccharides in a differential manner using an in vitro fermentation model. Food Res. Int. 2021, 148, 110562. [Google Scholar] [CrossRef]
- Ma, Y.; Jiang, S.; Zeng, M. In vitro simulated digestion and fermentation characteristics of polysaccharide from oyster (Crassostrea gigas), and its effects on the gut microbiota. Food Res. Int. 2021, 149, 110646. [Google Scholar] [CrossRef]
- Han, R.; Pang, D.; Wen, L.; You, L.; Huang, R.; Kulikouskaya, V. In vitro digestibility and prebiotic activities of a sulfated polysaccharide from Gracilaria lemaneiformis. J. Funct. Foods 2020, 64, 103652. [Google Scholar] [CrossRef]
- Strain, C.R.; Collins, K.C.; Naughton, V.; McSorley, E.M.; Stanton, C.; Smyth, T.J.; Soler-Vila, A.; Rea, M.C.; Ross, P.R.; Cherry, P.; et al. Effects of a polysaccharide-rich extract derived from Irish-sourced Laminaria digitata on the composition and metabolic activity of the human gut microbiota using an in vitro colonic model. Eur. J. Nutr. 2020, 59, 309–325. [Google Scholar] [CrossRef]
- Wang, M.; Zhou, J.; Selma-Royo, M.; Simal-Gandara, J.; Collado, M.C.; Barba, F.J. Potential benefits of high-added-value compounds from aquaculture and fish side streams on human gut microbiota. Trends Food Sci. Technol. 2021, 112, 484–494. [Google Scholar] [CrossRef]
- Chen, H.; Wierenga, P.A.; Hendriks, W.H.; Jansman, A.J.M. In vitro protein digestion kinetics of protein sources for pigs. Animal 2019, 13, 1154–1164. [Google Scholar] [CrossRef] [PubMed]
- Peretti, N.; Emmanuel, M.A.S. Congenital disorders of intestinal digestion and absorption (sugars, proteins, lipids, ions). Best Pract. Res. Clin. Gastroenterol. 2022, 56–57, 101785. [Google Scholar] [CrossRef]
- Yao, C.K.; Muir, J.G.; Gibson, P.R. Review article: Insights into colonic protein fermentation, its modulation and potential health implications. Aliment. Pharmacol. Ther. 2016, 43, 181–196. [Google Scholar] [CrossRef]
- Wu, L.; Tang, Z.; Chen, H.; Ren, Z.; Ding, Q.; Liang, K.; Sun, Z. Mutual interaction between gut microbiota and protein/amino acid metabolism for host mucosal immunity and health. Anim. Nutr. 2021, 7, 11–16. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, H.; Dong, S.; Jin, W.; Han, K.; Ren, Y.; Zeng, M. Glycation of fish protein impacts its fermentation metabolites and gut microbiota during in vitro human colonic fermentation. Food Res. Int. 2018, 113, 189–196. [Google Scholar] [CrossRef]
- Han, K.; Yao, Y.; Dong, S.; Jin, S.; Xiao, H.; Wu, H.; Zeng, M. Chemical characterization of the glycated myofibrillar proteins from grass carp (Ctenopharyngodon idella) and their impacts on the human gut microbiota in vitro fermentation. Food Funct. 2017, 8, 1184–1194. [Google Scholar] [CrossRef]
- Lima, K.O.; da Costa de Quadros, C.; da Rocha, M.; Gomes de Lacerda, J.T.J.; Juliano, M.A.; Dias, M.; Mendes, M.A.; Prentice, C. Bioactivity and bioaccessibility of protein hydrolyzates from industrial byproducts of Stripped weakfish (Cynoscion guatucupa). LWT-Food Sci. Technol. 2019, 111, 408–413. [Google Scholar] [CrossRef]
- Niccolai, A.; Chini Zittelli, G.; Rodolfi, L.; Biondi, N.; Tredici, M.R. Microalgae of interest as food source: Biochemical composition and digestibility. Algal Res. 2019, 42, 101617. [Google Scholar] [CrossRef]
- Tibbetts, S.M.; Milley, J.E.; Lall, S.P. Chemical composition and nutritional properties of freshwater and marine microalgal biomass cultured in photobioreactors. J. Appl. Phycol. 2015, 27, 1109–1119. [Google Scholar] [CrossRef]
- Pateiro, M.; Domínguez, R.; Munekata, P.E.S.; Barba, F.J.; Lorenzo, J.M. Lipids and Fatty Acids; Elsevier Inc.: Amsterdam, The Netherlands, 2019; ISBN 9780128141748. [Google Scholar]
- Xu, E.; Chen, C.; Fu, J.; Zhu, L.; Shu, J.; Jin, M.; Wang, Y.; Zong, X. Dietary fatty acids in gut health: Absorption, metabolism and function. Anim. Nutr. 2021, 7, 1337–1344. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Xu, Y.J.; Liu, Y. Influences of dietary oils and fats, and the accompanied minor content of components on the gut microbiota and gut inflammation: A review. Trends Food Sci. Technol. 2021, 113, 255–276. [Google Scholar] [CrossRef]
- Liu, W.; Luo, X.; Liu, T.; Feng, F. Study on the digestive characteristics of short-and medium-chain fatty acid structural lipid and its rapid intervention on gut microbes: In vivo and in vitro studies. Food Chem. 2022, 380, 131792. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Lu, T.; Liu, X.Y.; Zhao, M.T.; Yin, F.W.; Rakariyatham, K.; Zhou, D.Y. Improving the oxidative stability and lengthening the shelf life of DHA algae oil with composite antioxidants. Food Chem. 2020, 313, 126139. [Google Scholar] [CrossRef]
- Dasilva, G.; Boller, M.; Medina, I.; Storch, J. Relative levels of dietary EPA and DHA impact gastric oxidation and essential fatty acid uptake. J. Nutr. Biochem. 2018, 55, 68–75. [Google Scholar] [CrossRef]
- Francisco, J.; Horta, A.; Pedrosa, R.; Afonso, C.; Cardoso, C.; Bandarra, N.M.; Gil, M.M. Bioaccessibility of antioxidants and fatty acids from Fucus spiralis. Foods 2020, 9, 440. [Google Scholar] [CrossRef]
- Canelli, G.; Neutsch, L.; Carpine, R.; Tevere, S.; Giuffrida, F.; Rohfritsch, Z.; Dionisi, F.; Bolten, C.J.; Mathys, A. Chlorella vulgaris in a heterotrophic bioprocess: Study of the lipid bioaccessibility and oxidative stability. Algal Res. 2020, 45, 101754. [Google Scholar] [CrossRef]
- Besednova, N.N.; Andryukov, B.G.; Zaporozhets, T.S.; Kryzhanovsky, S.P.; Kuznetsova, T.A.; Fedyanina, L.N.; Makarenkova, I.D.; Zvyagintseva, T.N. Algae polyphenolic compounds and modern antibacterial strategies: Current achievements and immediate prospects. Biomedicines 2020, 8, 342. [Google Scholar] [CrossRef]
- Liu, M.; Hansen, P.E.; Lin, X. Bromophenols in marine algae and their bioactivities. Mar. Drugs 2011, 9, 1273–1292. [Google Scholar] [CrossRef]
- Williamson, G.; Clifford, M.N. Role of the small intestine, colon and microbiota in determining the metabolic fate of polyphenols. Biochem. Pharmacol. 2017, 139, 24–39. [Google Scholar] [CrossRef]
- Tarko, T.; Duda-Chodak, A.; Zajac, N. Digestion and absorption of phenolic compounds assessed by in vitro simulation methods. A review. Rocz. Państwowego Zakładu Hig. 2013, 64, 79–84. [Google Scholar]
- Corona, G.; Coman, M.M.; Guo, Y.; Hotchkiss, S.; Gill, C.; Yaqoob, P.; Spencer, J.P.E.; Rowland, I. Effect of simulated gastrointestinal digestion and fermentation on polyphenolic content and bioactivity of brown seaweed phlorotannin-rich extracts. Mol. Nutr. Food Res. 2017, 61, 1–10. [Google Scholar] [CrossRef]
- Gu, C.; Suleria, H.A.R.; Dunshea, F.R.; Howell, K. Dietary lipids influence bioaccessibility of polyphenols from black carrots and affect microbial diversity under simulated gastrointestinal digestion. Antioxidants 2020, 9, 762. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Chan, Y.H.; Rathinasabapathy, T.; Grace, M.H.; Komarnytsky, S.; Lila, M.A. Enhanced stability of berry pomace polyphenols delivered in protein-polyphenol aggregate particles to an in vitro gastrointestinal digestion model. Food Chem. 2020, 331, 127279. [Google Scholar] [CrossRef]
- Moreda-Piñeiro, J.; Moreda-Piñeiro, A.; Romarís-Hortas, V.; Domínguez-González, R.; Alonso-Rodríguez, E.; López-Mahía, P.; Muniategui-Lorenzo, S.; Prada-Rodríguez, D.; Bermejo-Barrera, P. Trace metals in marine foodstuff: Bioavailability estimation and effect of major food constituents. Food Chem. 2012, 134, 339–345. [Google Scholar] [CrossRef]
- Chen, K.; Roca, M. Cooking effects on bioaccessibility of chlorophyll pigments of the main edible seaweeds. Food Chem. 2019, 295, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Alves, R.N.; Rambla-Alegre, M.; Braga, A.C.; Maulvault, A.L.; Barbosa, V.; Campàs, M.; Reverté, L.; Flores, C.; Caixach, J.; Kilcoyne, J.; et al. Bioaccessibility of lipophilic and hydrophilic marine biotoxins in seafood: An in vitro digestion approach. Food Chem. Toxicol. 2019, 129, 153–161. [Google Scholar] [CrossRef]
- Cruz, R.; Mendes, E.; Maulvault, A.L.; Marques, A.; Casal, S.; Cunha, S.C. Bioaccessibility of polybrominated diphenyl ethers and their methoxylated metabolites in cooked seafood after using a multi-compartment in vitro digestion model. Chemosphere 2020, 252, 126462. [Google Scholar] [CrossRef]
- Fu, Y.; Yin, N.; Cai, X.; Du, H.; Wang, P.; Sultana, M.S.; Sun, G.; Cui, Y. Arsenic speciation and bioaccessibility in raw and cooked seafood: Influence of seafood species and gut microbiota. Environ. Pollut. 2021, 280, 116958. [Google Scholar] [CrossRef]

| Model | Stage | Advantage | Disadvantage |
|---|---|---|---|
| INFOGEST | Oral-gastric-small intestine | Simple operation, short time consuming, suitable for single component digestion. Several samples simultaneously | The physiological environment is simplified and cannot simulate dynamic processes (digestive fluids secretion flow rate, removal of the products of digestion). |
| DGM | Gastric (Fundus/main body and antrum) | Digestion and emptying of food in the stomach can be monitored in real-time. | The position of the main body and the gastric antrum is different from the real; it is necessary to combine the duodenum model to track the further form changes of food after DGM |
| HGS | Gastric (Antrum) | The mechanical force is more reasonable, and the digestion parameters can be changed. | Only simulates stomach digestion, with limitations; compartments fail to model the true shape of the stomach. |
| DIDGI | Gastric-small intestine | The device is transparent and the morphological changes of food during digestion can be monitored. | Absorption in the small intestine phase has not been simulated and needs to be combined with other models. |
| TIM | Gastric and small intestine (duodenum, jejunum and ileum) | Simulates the complete digestive system and can be used to explore the digestive process of various foods. | Inaccurate simulation on shear/grinding of the gastrointestinal tract. |
| SHIME | Oral-gastric-small and large intestine | Simultaneous sampling and automatic parameter definition: adding fluid flow rate, emptying time and flow rate, with better stability and reproducibility. | Experiments take at least 4–5 weeks and the equipment is fed 3 times a day with the study compounds for at least 2 weeks. More suitable for studies of extracts or pure substances. In addition, the absorption of metabolites was not considered. |
| ESIN | Oral, stomach, duodenum, jejunum and ileum | Ability to simulate digestion of foods of a similar size to normal meals. | Simulates digestion from oral to small intestine only, needs to be combined with colonic fermentation models. |
| SIMGI | Stomach, small intestine, colon | Digestion parameters can be controlled, including digester flow rate, digestion volume and time, pH, temperature and pressure, etc. | Absorption of metabolites and interactions between gut microbiota and host cannot be simulated. |
| ARCOL | Colon | The anaerobic environment of the colon and the passive absorption of metabolites can be simulated. | The different physiological conditions of the three parts of the colon cannot be distinguished. |
| Bioactive Compounds | Oral | Gastric | Small Intestinal | Colon |
|---|---|---|---|---|
| Polysaccharides | It is not degraded by digestive enzymes | Participate in the fermentation of gut microbiota and increase the abundance of beneficial bacteria such as Bifidobacterium and Lactobacillus. | ||
| Protein | Structural degeneration | It is degraded into oligopeptides and amino acids, which enter the body fluid circulation. | The undigested protein enters the distal colon and supplies nitrogen to the gut microbiota. | |
| Lipids | The structure starts to fall apart | Partial lipid hydrolysis | The lipids are hydrolyzed into smaller molecules of fatty acids that are absorbed by the intestine. | |
| Phenolic | Digestive enzymes hydrolyze food matrix and release polyphenols | Polyphenols undergo glycosidic hydrolysis and methylation, and some of them are absorbed by small intestinal. | Undigested polyphenols are degraded into phenolic acids, which participate in colonic fermentation. | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Zhou, J.; Calvo-Lerma, J.; Liu, Y.; Collado, M.C.; Barba, F.J. Effects of Marine Bioactive Compounds on Gut Ecology Based on In Vitro Digestion and Colonic Fermentation Models. Nutrients 2022, 14, 3307. https://doi.org/10.3390/nu14163307
Wang M, Zhou J, Calvo-Lerma J, Liu Y, Collado MC, Barba FJ. Effects of Marine Bioactive Compounds on Gut Ecology Based on In Vitro Digestion and Colonic Fermentation Models. Nutrients. 2022; 14(16):3307. https://doi.org/10.3390/nu14163307
Chicago/Turabian StyleWang, Min, Jianjun Zhou, Joaquim Calvo-Lerma, Yixuan Liu, María Carmen Collado, and Francisco J. Barba. 2022. "Effects of Marine Bioactive Compounds on Gut Ecology Based on In Vitro Digestion and Colonic Fermentation Models" Nutrients 14, no. 16: 3307. https://doi.org/10.3390/nu14163307









