The Gut Microbial Metabolite Pyrogallol Is a More Potent Inducer of Nrf2-Associated Gene Expression Than Its Parent Compound Green Tea (-)-Epigallocatechin Gallate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Cell Lines
2.3. Reporter Gene Assay
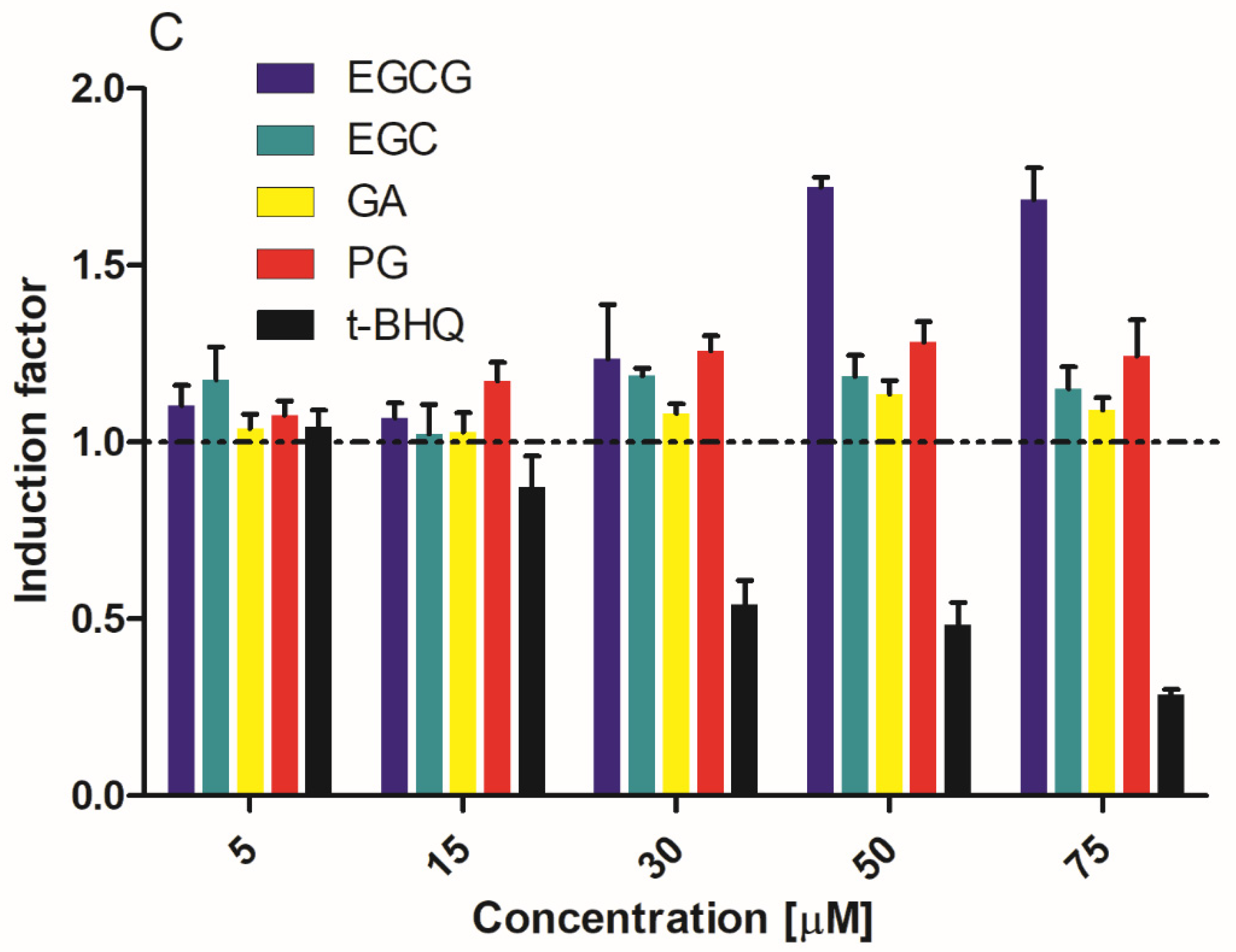
2.4. Cell Viability Assay
2.5. Sample Preparation and Liquid Chromatography–Mass Spectrometry (LC-MS) Analysis for Label-Free Quantitative Proteomics
2.6. Data Analysis and Bioinformatics
2.7. RT-qPCR
2.8. Benchmark Dose Analysis
2.9. Statistical Analysis
3. Results
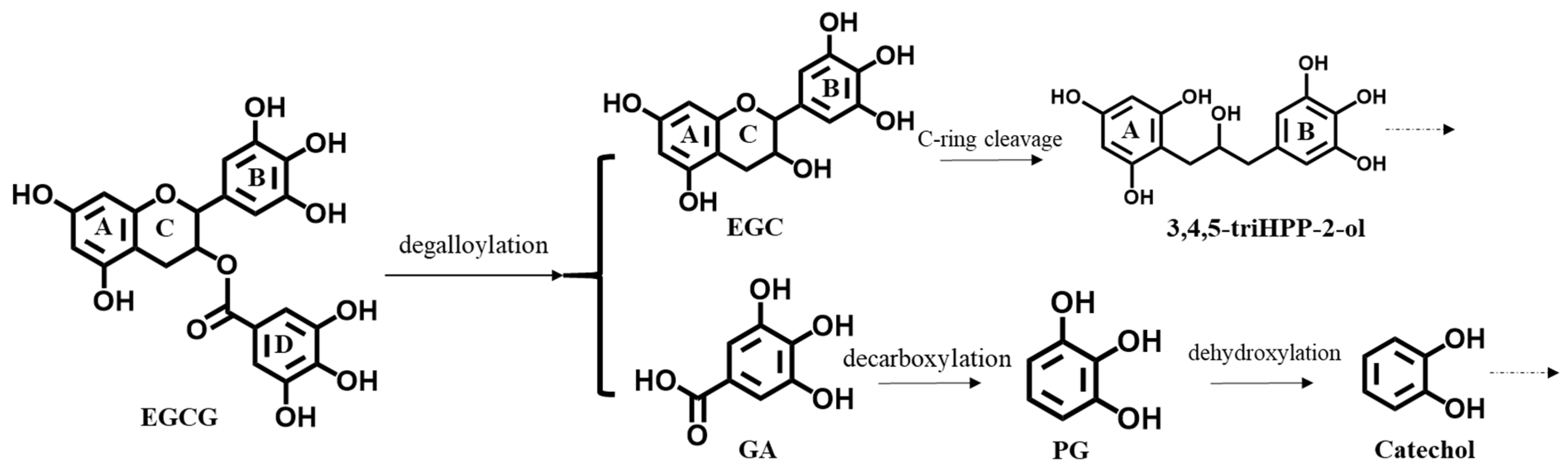
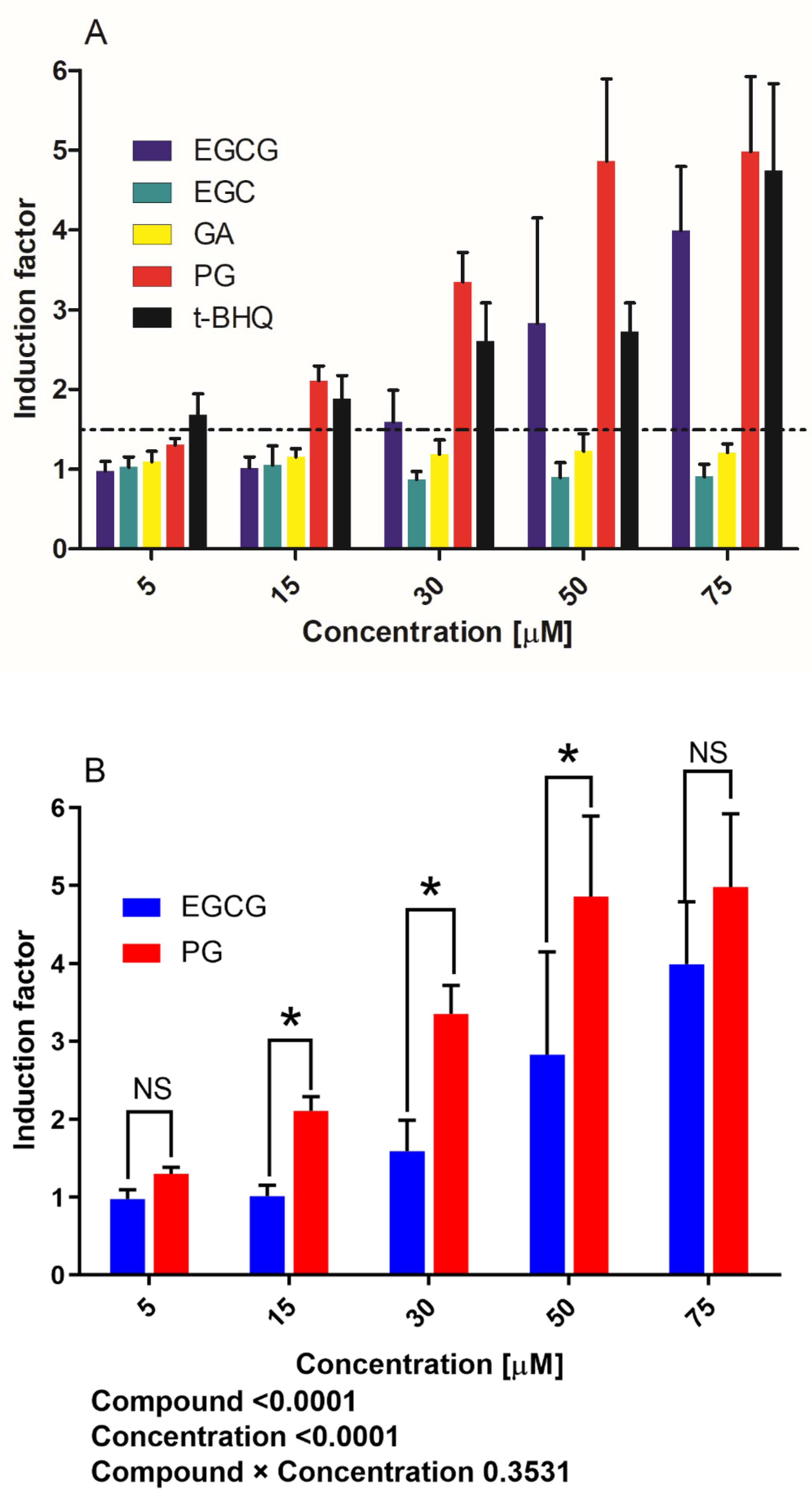
3.1. Activation of Nrf2-Mediated Luciferase Expression by EGCG and Its Metabolites
3.2. Nrf2-Related Protein Expression in Hepa1c1c7 Cells
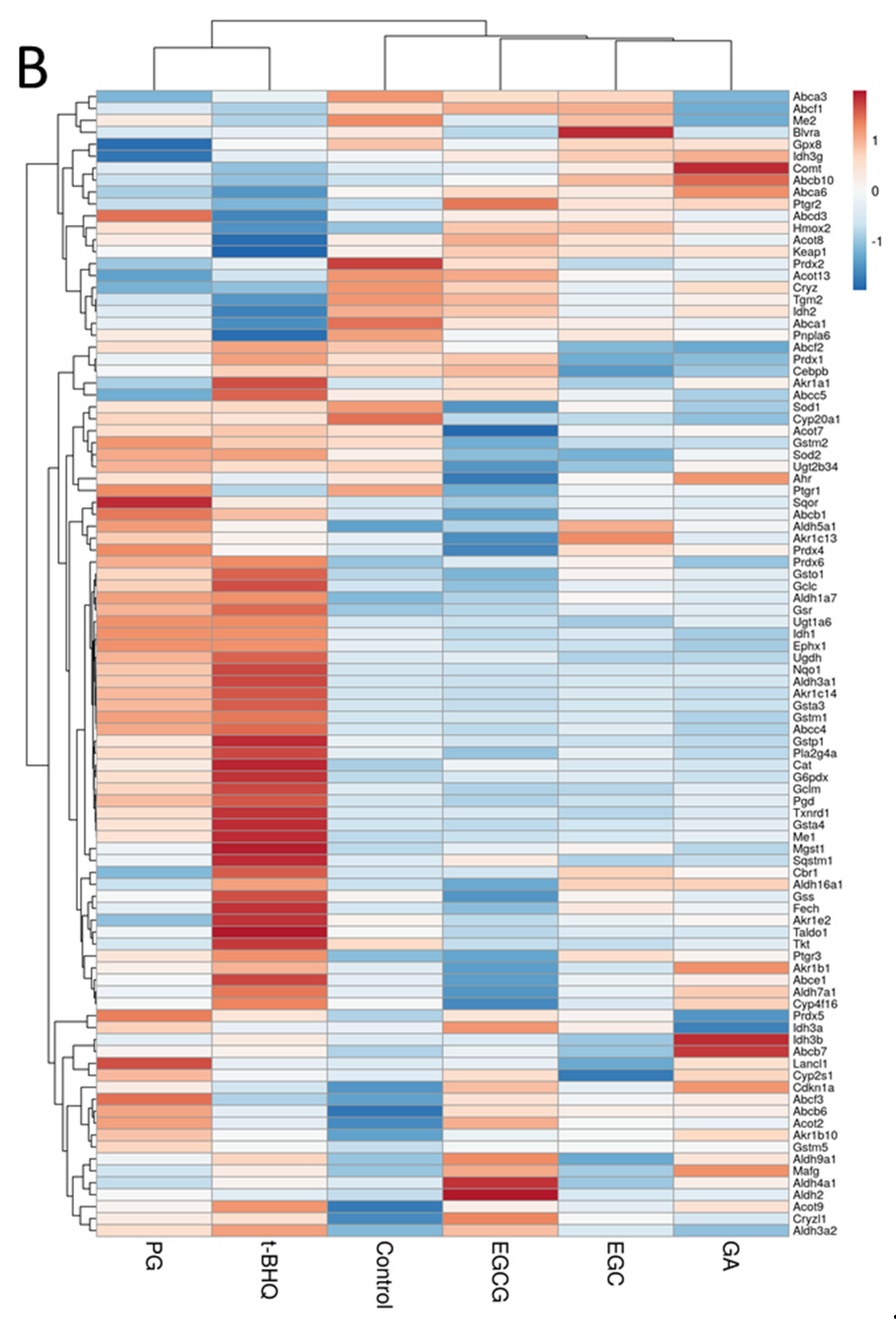
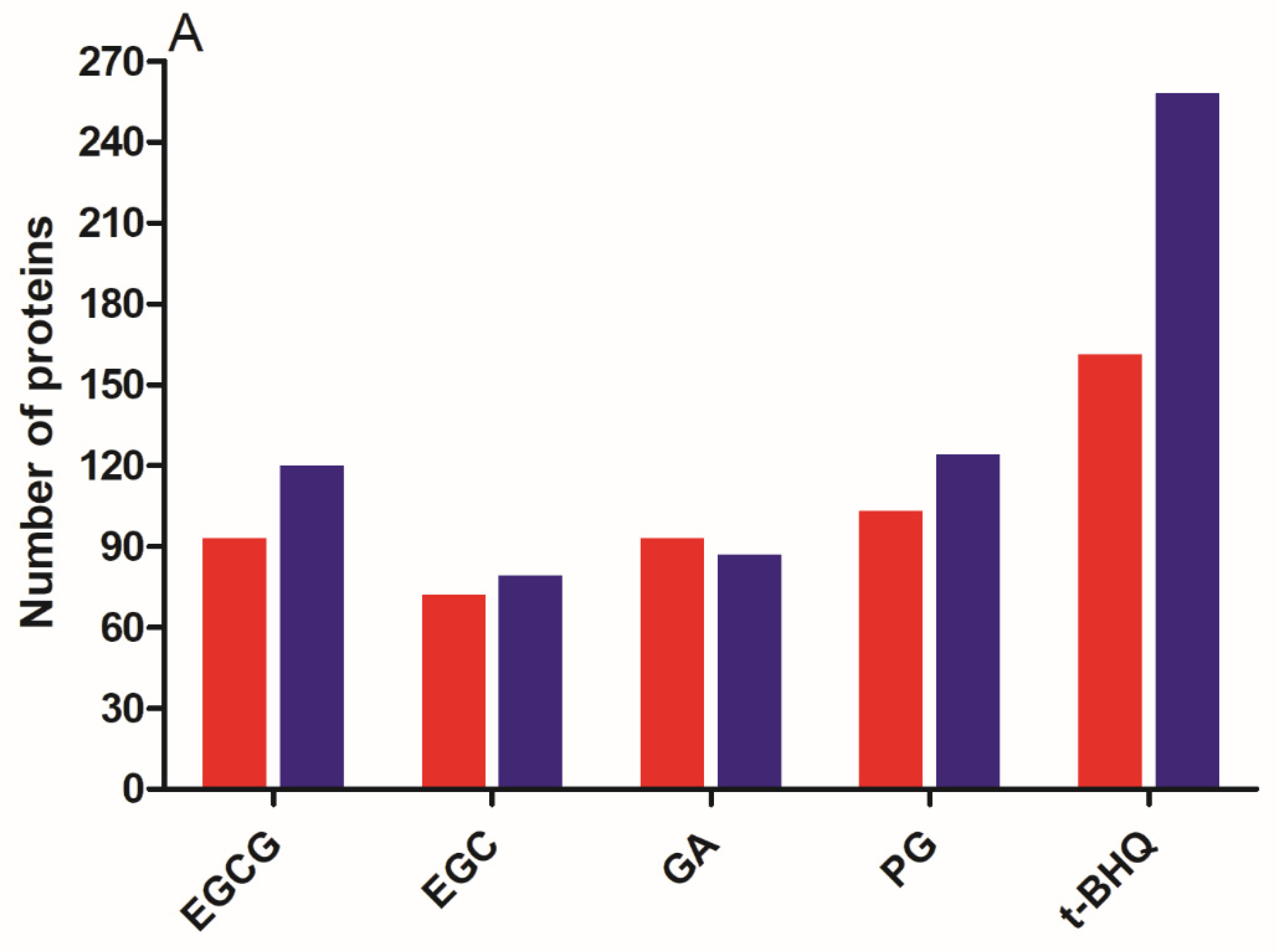
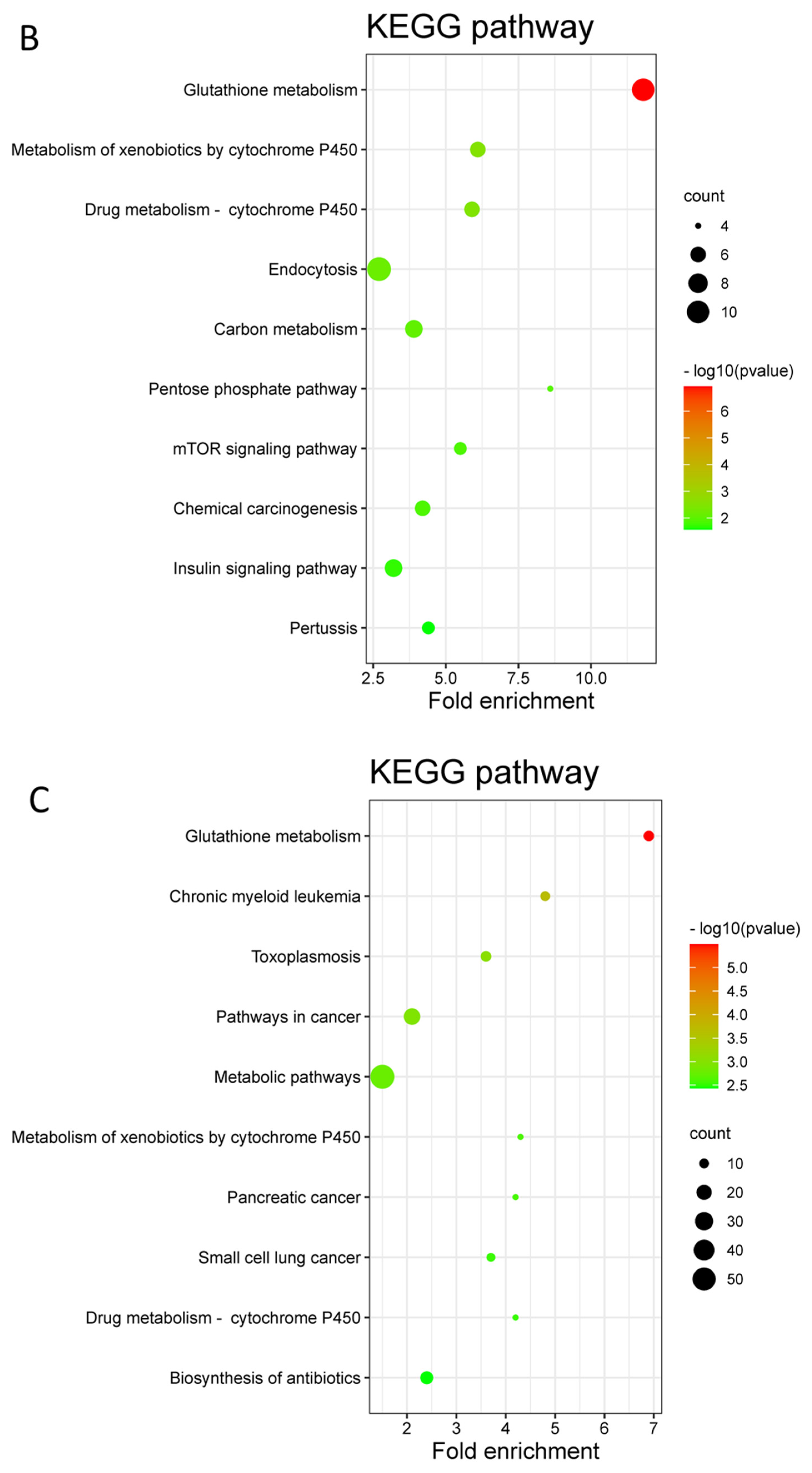
3.3. Differentially Expressed Proteins (DEPs) and Functional Enrichment Analysis
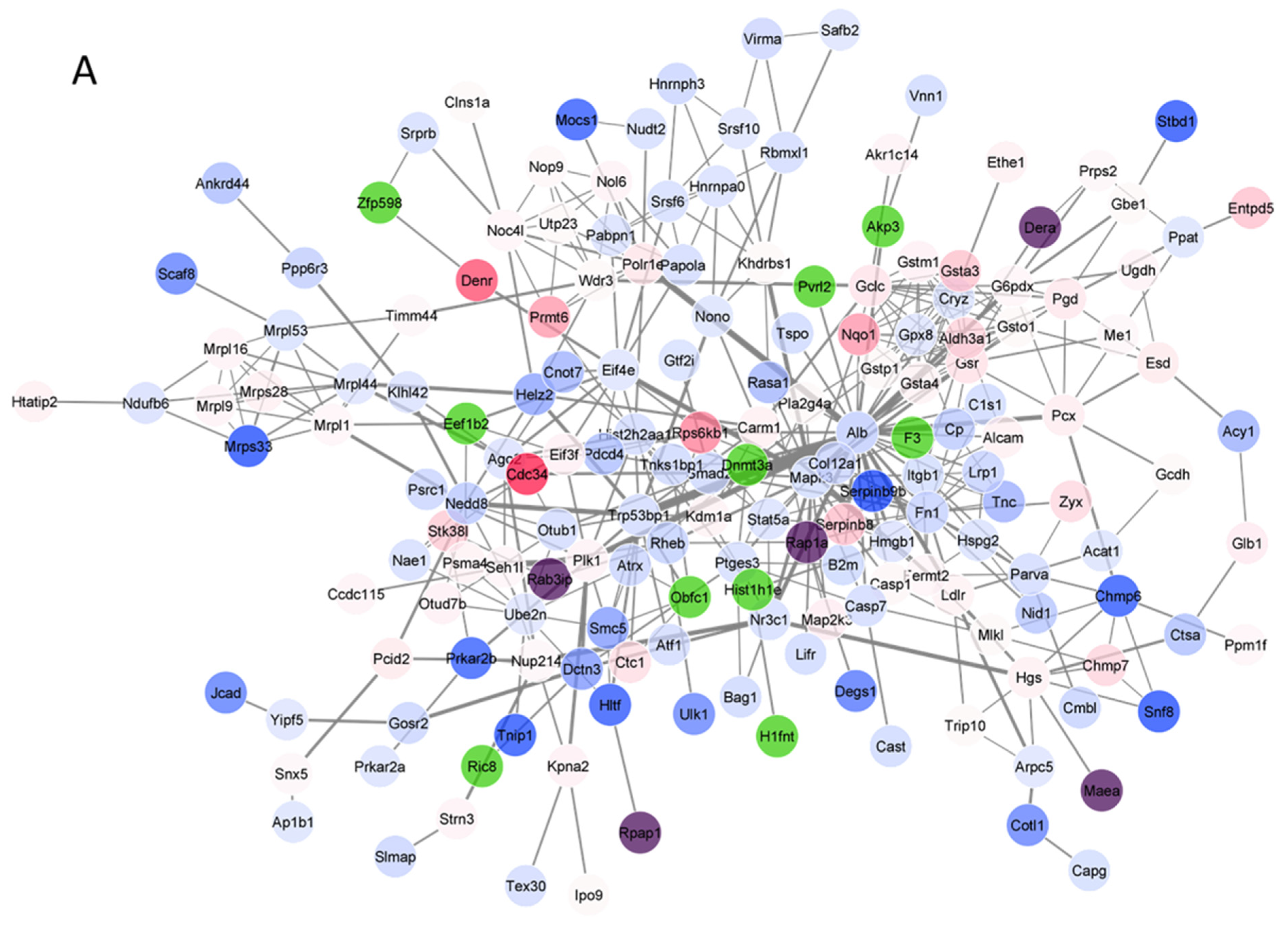
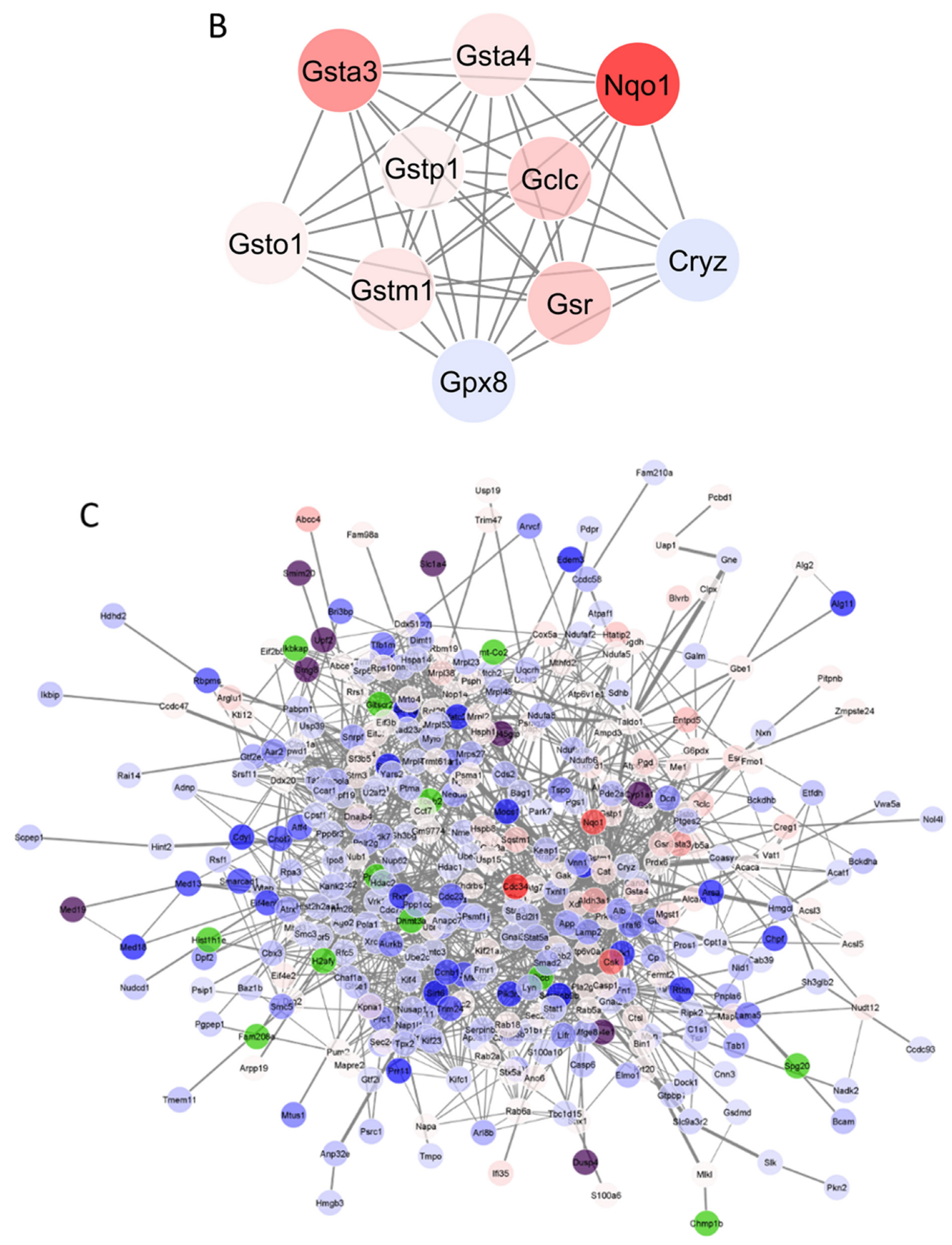
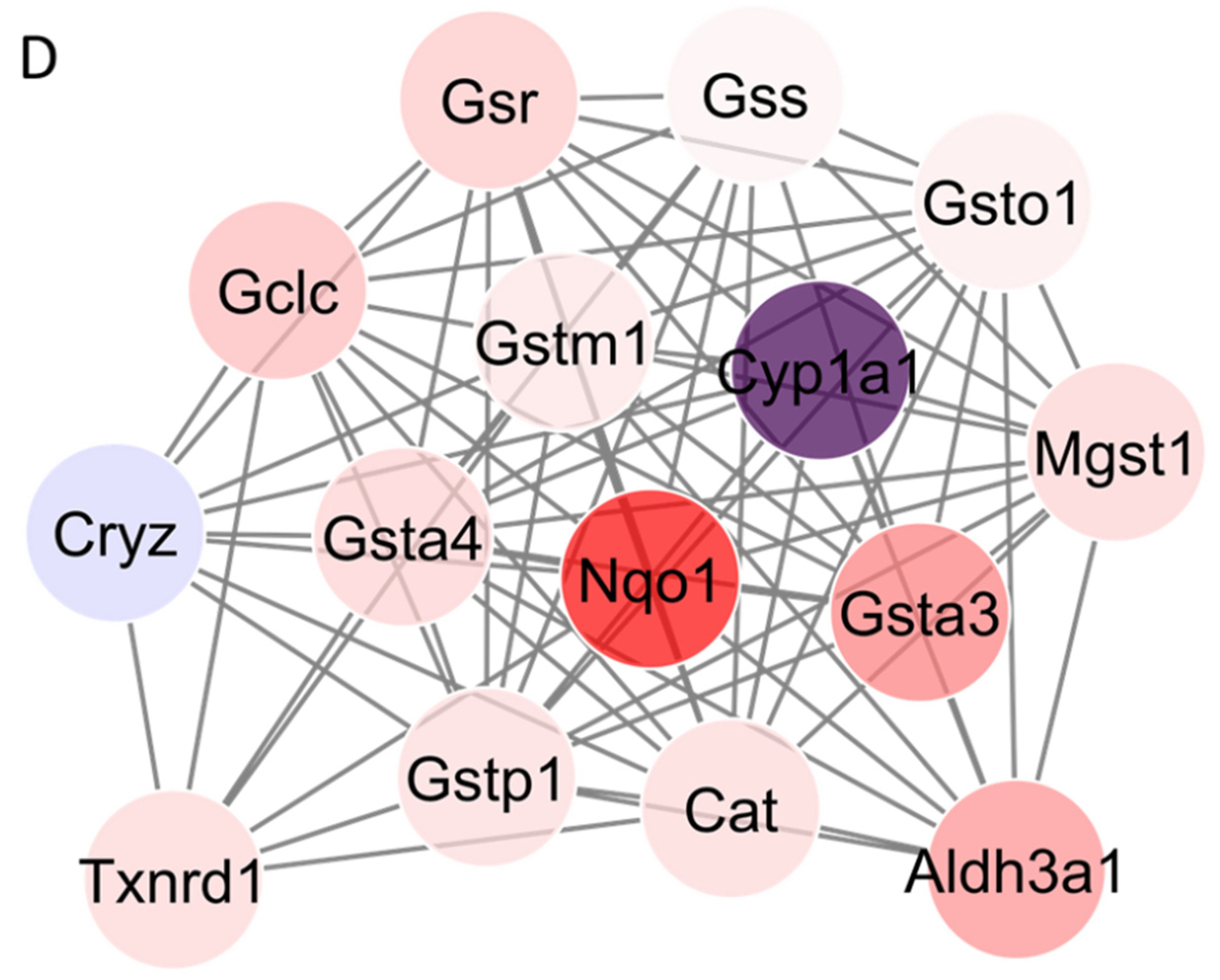
3.4. Protein–Protein Interaction (PPI) Network Analysis
3.5. A Further Analysis of “Target” Proteins from PG and t-BHQ Treatments
3.6. RT-qPCR Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kakutani, S.; Watanabe, H.; Murayama, N. Green tea intake and risks for dementia, Alzheimer’s disease, mild cognitive impairment, and cognitive impairment: A systematic review. Nutrients 2019, 11, 1165. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, C.; Artacho, R.; Giménez, R. Beneficial effects of green tea—A review. J. Am. Coll. Nutr. 2006, 25, 79–99. [Google Scholar] [CrossRef] [PubMed]
- Gan, R.-Y.; Li, H.-B.; Sui, Z.-Q.; Corke, H. Absorption, metabolism, anti-cancer effect and molecular targets of epigallocatechin gallate (EGCG): An updated review. Crit. Rev. Food Sci. Nutr. 2018, 58, 924–941. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-J.; Maliakal, P.; Chen, L.; Meng, X.; Bondoc, F.Y.; Prabhu, S.; Lambert, G.; Mohr, S.; Yang, C.S. Pharmacokinetics of tea catechins after ingestion of green tea and (−)-epigallocatechin-3-gallate by humans: Formation of different metabolites and individual variability. Cancer Epidemiol. Prev. Biomark. 2002, 11, 1025–1032. [Google Scholar]
- Monagas, M.; Urpi-Sarda, M.; Sánchez-Patán, F.; Llorach, R.; Garrido, I.; Gómez-Cordovés, C.; Andres-Lacueva, C.; Bartolomé, B. Insights into the metabolism and microbial biotransformation of dietary flavan-3-ols and the bioactivity of their metabolites. Food Funct. 2010, 1, 233–253. [Google Scholar] [CrossRef]
- Liu, C.; Vervoort, J.; van den Elzen, J.; Beekmann, K.; Baccaro, M.; de Haan, L.; Rietjens, I.M. Interindividual Differences in Human In Vitro Intestinal Microbial Conversion of Green Tea (-)-Epigallocatechin-3-O-Gallate and Consequences for Activation of Nrf2 Mediated Gene Expression. Mol. Nutr. Food Res. 2021, 65, 2000934. [Google Scholar] [CrossRef]
- Gross, G.; Jacobs, D.M.; Peters, S.; Possemiers, S.; van Duynhoven, J.; Vaughan, E.E.; Van de Wiele, T. In vitro bioconversion of polyphenols from black tea and red wine/grape juice by human intestinal microbiota displays strong interindividual variability. J. Agric. Food Chem. 2010, 58, 10236–10246. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, M.; Wang, T.; Liu, T.; Guo, Y.; Granato, D. Green tea polyphenols mitigate the plant lectins-induced liver inflammation and immunological reaction in C57BL/6 mice via NLRP3 and Nrf2 signaling pathways. Food Chem. Toxicol. 2020, 144, 111576. [Google Scholar] [CrossRef]
- van Duynhoven, J.; van der Hooft, J.J.; van Dorsten, F.A.; Peters, S.; Foltz, M.; Gomez-Roldan, V.; Vervoort, J.; de Vos, R.C.; Jacobs, D.M. Rapid and sustained systemic circulation of conjugated gut microbial catabolites after single-dose black tea extract consumption. J. Proteome Res. 2014, 13, 2668–2678. [Google Scholar] [CrossRef]
- Kohri, T.; Matsumoto, N.; Yamakawa, M.; Suzuki, M.; Nanjo, F.; Hara, Y.; Oku, N. Metabolic fate of (−)-[4-3H] epigallocatechin gallate in rats after oral administration. J. Agric. Food Chem. 2001, 49, 4102–4112. [Google Scholar] [CrossRef]
- Liu, Z.; de Bruijn, W.J.; Bruins, M.E.; Vincken, J.-P. Reciprocal interactions between epigallocatechin-3-gallate (EGCG) and human gut microbiota in vitro. J. Agric. Food Chem. 2020, 68, 9804–9815. [Google Scholar] [CrossRef] [PubMed]
- Baranowska, M.; Suliborska, K.; Chrzanowski, W.; Kusznierewicz, B.; Namieśnik, J.; Bartoszek, A. The relationship between standard reduction potentials of catechins and biological activities involved in redox control. Redox Biol. 2018, 17, 355–366. [Google Scholar] [CrossRef]
- Sirota, R.; Gibson, D.; Kohen, R. The role of the catecholic and the electrophilic moieties of caffeic acid in Nrf2/Keap1 pathway activation in ovarian carcinoma cell lines. Redox Biol. 2015, 4, 48–59. [Google Scholar] [CrossRef]
- Wu, R.P.; Hayashi, T.; Cottam, H.B.; Jin, G.; Yao, S.; Wu, C.C.; Rosenbach, M.D.; Corr, M.; Schwab, R.B.; Carson, D.A. Nrf2 responses and the therapeutic selectivity of electrophilic compounds in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2010, 107, 7479–7484. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Kensler, T.W.; Motohashi, H. The KEAP1-NRF2 system: A thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 2018, 98, 1169–1203. [Google Scholar] [CrossRef] [PubMed]
- Muzolf-Panek, M.; Gliszczyńska-Świgło, A.; de Haan, L.; Aarts, J.M.; Szymusiak, H.; Vervoort, J.M.; Tyrakowska, B.; Rietjens, I.M. Role of catechin quinones in the induction of EpRE-mediated gene expression. Chem. Res. Toxicol. 2008, 21, 2352–2360. [Google Scholar] [CrossRef] [PubMed]
- Talebi, M.; Talebi, M.; Farkhondeh, T.; Mishra, G.; İlgün, S.; Samarghandian, S. New insights into the role of the Nrf2 signaling pathway in green tea catechin applications. Phytother. Res. 2021, 35, 3078–3112. [Google Scholar] [CrossRef]
- He, F.; Ru, X.; Wen, T. NRF2, a transcription factor for stress response and beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef]
- Park, H.J.; DiNatale, D.A.; Chung, M.-Y.; Park, Y.-K.; Lee, J.-Y.; Koo, S.I.; O’Connor, M.; Manautou, J.E.; Bruno, R.S. Green tea extract attenuates hepatic steatosis by decreasing adipose lipogenesis and enhancing hepatic antioxidant defenses in ob/ob mice. J. Nutr. Biochem. 2011, 22, 393–400. [Google Scholar] [CrossRef]
- Park, H.J.; Lee, J.-Y.; Chung, M.-Y.; Park, Y.-K.; Bower, A.M.; Koo, S.I.; Giardina, C.; Bruno, R.S. Green tea extract suppresses NFκB activation and inflammatory responses in diet-induced obese rats with nonalcoholic steatohepatitis. J. Nutr. 2012, 142, 57–63. [Google Scholar] [CrossRef]
- Thavasi, V.; Leong, L.P.; Bettens, R.P.A. Investigation of the influence of hydroxy groups on the radical scavenging ability of polyphenols. J. Phys. Chem. A 2006, 110, 4918–4923. [Google Scholar] [CrossRef] [PubMed]
- Lee-Hilz, Y.Y.; Boerboom, A.-M.J.; Westphal, A.H.; van Berkel, W.J.; Aarts, J.M.; Rietjens, I.M. Pro-oxidant activity of flavonoids induces EpRE-mediated gene expression. Chem. Res. Toxicol. 2006, 19, 1499–1505. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Yamada, H. Microbial production of pyrogallol through decarboxylation of gallic acid. Agric. Biol. Chem. 1985, 49, 659–663. [Google Scholar] [CrossRef]
- van der Hooft, J.J.; de Vos, R.C.; Mihaleva, V.; Bino, R.J.; Ridder, L.; de Roo, N.; Jacobs, D.M.; van Duynhoven, J.P.; Vervoort, J. Structural elucidation and quantification of phenolic conjugates present in human urine after tea intake. Anal. Chem. 2012, 84, 7263–7271. [Google Scholar] [CrossRef] [PubMed]
- van der Linden, S.C.; von Bergh, A.R.; van Vught-Lussenburg, B.M.; Jonker, L.R.; Teunis, M.; Krul, C.A.; van der Burg, B. Development of a panel of high-throughput reporter-gene assays to detect genotoxicity and oxidative stress. Mutat. Res./Genet. Toxicol. Environ. Mutagenes. 2014, 760, 23–32. [Google Scholar] [CrossRef]
- Beekmann, K.; Rubió, L.; de Haan, L.H.; Actis-Goretta, L.; van der Burg, B.; van Bladeren, P.J.; Rietjens, I.M. The effect of quercetin and kaempferol aglycones and glucuronides on peroxisome proliferator-activated receptor-gamma (PPAR-γ). Food Funct. 2015, 6, 1098–1107. [Google Scholar] [CrossRef]
- Chen, L.; Wang, W.; Zhang, J.; Cui, H.; Ni, D.; Jiang, H. Dual effects of ascorbic acid on the stability of EGCG by the oxidation product dehydroascorbic acid promoting the oxidation and inhibiting the hydrolysis pathway. Food Chem. 2021, 337, 127639. [Google Scholar] [CrossRef]
- Grzesik, M.; Bartosz, G.; Stefaniuk, I.; Pichla, M.; Namieśnik, J.; Sadowska-Bartosz, I. Dietary antioxidants as a source of hydrogen peroxide. Food Chem. 2019, 278, 692–699. [Google Scholar] [CrossRef]
- Wiechelman, K.J.; Braun, R.D.; Fitzpatrick, J.D. Investigation of the bicinchoninic acid protein assay: Identification of the groups responsible for color formation. Anal. Biochem. 1988, 175, 231–237. [Google Scholar] [CrossRef]
- Batth, T.S.; Tollenaere, M.X.; Rüther, P.; Gonzalez-Franquesa, A.; Prabhakar, B.S.; Bekker-Jensen, S.; Deshmukh, A.S.; Olsen, J.V. Protein aggregation capture on microparticles enables multipurpose proteomics sample preparation. Mol. Cell. Proteom. 2019, 18, 1027–1035. [Google Scholar] [CrossRef]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional regulation by Nrf2. Antioxid. Redox Signal. 2018, 29, 1727–1745. [Google Scholar] [CrossRef] [PubMed]
- Chun, K.-S.; Raut, P.K.; Kim, D.-H.; Surh, Y.-J. Role of chemopreventive phytochemicals in NRF2-mediated redox homeostasis in humans. Free. Radic. Biol. Med. 2021, 172, 699–715. [Google Scholar] [CrossRef]
- Zhan, X.; Li, J.; Zhou, T. Targeting Nrf2-mediated oxidative stress response signaling pathways as new therapeutic strategy for pituitary adenomas. Front. Pharmacol. 2021, 12, 565748. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Dinkova-Kostova, A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014, 39, 199–218. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.N.; Shankar, S.; Srivastava, R.K. Green tea catechin, epigallocatechin-3-gallate (EGCG): Mechanisms, perspectives and clinical applications. Biochem. Pharmacol. 2011, 82, 1807–1821. [Google Scholar] [CrossRef]
- Cai, Y.-Z.; Sun, M.; Xing, J.; Luo, Q.; Corke, H. Structure–radical scavenging activity relationships of phenolic compounds from traditional Chinese medicinal plants. Life Sci. 2006, 78, 2872–2888. [Google Scholar] [CrossRef]
- Dey, P.; Olmstead, B.D.; Sasaki, G.Y.; Vodovotz, Y.; Yu, Z.; Bruno, R.S. Epigallocatechin gallate but not catechin prevents nonalcoholic steatohepatitis in mice similar to green tea extract while differentially affecting the gut microbiota. J. Nutr. Biochem. 2020, 84, 108455. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zheng, Y.; Chow, M.S.; Zuo, Z. Investigation of intestinal absorption and disposition of green tea catechins by Caco-2 monolayer model. Int. J. Pharm. 2004, 287, 1–12. [Google Scholar] [CrossRef]
- Liu, Z.; Bruins, M.E.; Ni, L.; Vincken, J.-P. Green and black tea phenolics: Bioavailability, transformation by colonic microbiota, and modulation of colonic microbiota. J. Agric. Food Chem. 2018, 66, 8469–8477. [Google Scholar] [CrossRef]
- Na, H.-K.; Surh, Y.-J. Modulation of Nrf2-mediated antioxidant and detoxifying enzyme induction by the green tea polyphenol EGCG. Food Chem. Toxicol. 2008, 46, 1271–1278. [Google Scholar] [CrossRef]
- Chow, H.S.; Cai, Y.; Alberts, D.S.; Hakim, I.; Dorr, R.; Shahi, F.; Crowell, J.A.; Yang, C.S.; Hara, Y. Phase I pharmacokinetic study of tea polyphenols following single-dose administration of epigallocatechin gallate and polyphenon E. Cancer Epidemiol. Prev. Biomark. 2001, 10, 53–58. [Google Scholar]
- Naumovski, N.; Blades, B.L.; Roach, P.D. Food inhibits the oral bioavailability of the major green tea antioxidant epigallocatechin gallate in humans. Antioxidants 2015, 4, 373–393. [Google Scholar] [CrossRef]
- Chow, H.S.; Cai, Y.; Hakim, I.A.; Crowell, J.A.; Shahi, F.; Brooks, C.A.; Dorr, R.T.; Hara, Y.; Alberts, D.S. Pharmacokinetics and safety of green tea polyphenols after multiple-dose administration of epigallocatechin gallate and polyphenon E in healthy individuals. Clin. Cancer Res. 2003, 9, 3312–3319. [Google Scholar] [PubMed]
- Chow, H.S.; Hakim, I.A.; Vining, D.R.; Crowell, J.A.; Ranger-Moore, J.; Chew, W.M.; Celaya, C.A.; Rodney, S.R.; Hara, Y.; Alberts, D.S. Effects of dosing condition on the oral bioavailability of green tea catechins after single-dose administration of Polyphenon E in healthy individuals. Clin. Cancer Res. 2005, 11, 4627–4633. [Google Scholar] [CrossRef]
- Li, J.; Sapper, T.N.; Mah, E.; Rudraiah, S.; Schill, K.E.; Chitchumroonchokchai, C.; Moller, M.V.; McDonald, J.D.; Rohrer, P.R.; Manautou, J.E. Green tea extract provides extensive Nrf2-independent protection against lipid accumulation and NFκB pro-inflammatory responses during nonalcoholic steatohepatitis in mice fed a high-fat diet. Mol. Nutr. Food Res. 2016, 60, 858–870. [Google Scholar] [CrossRef] [PubMed]
- Schwedhelm, E.; Maas, R.; Troost, R.; Böger, R.H. Clinical pharmacokinetics of antioxidants and their impact on systemic oxidative stress. Clin. Pharmacokinet. 2003, 42, 437–459. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.; Siegel, D. The diverse functionality of NQO1 and its roles in redox control. Redox Biol. 2021, 41, 101950. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.; Siegel, D. Functions of NQO1 in cellular protection and CoQ10 metabolism and its potential role as a redox sensitive molecular switch. Front. Physiol. 2017, 8, 595. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Talalay, P. NAD (P) H: Quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch. Biochem. Biophys. 2010, 501, 116–123. [Google Scholar] [CrossRef]
- Wu, K.C.; Cui, J.Y.; Klaassen, C.D. Beneficial role of Nrf2 in regulating NADPH generation and consumption. Toxicol. Sci. 2011, 123, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Ju, H.-Q.; Lin, J.-F.; Tian, T.; Xie, D.; Xu, R.-H. NADPH homeostasis in cancer: Functions, mechanisms and therapeutic implications. Signal. Transduct. Target. Ther. 2020, 5, 231. [Google Scholar] [CrossRef] [PubMed]
- Ge, T.; Yang, J.; Zhou, S.; Wang, Y.; Li, Y.; Tong, X. The role of the pentose phosphate pathway in diabetes and cancer. Front. Endocrinol. 2020, 11, 365. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.S.; Cha, Y.H.; Kim, H.S.; Kim, N.H.; Yook, J.I. The pentose phosphate pathway as a potential target for cancer therapy. Biomol. Ther. 2018, 26, 29–38. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Boeren, S.; Miro Estruch, I.; Rietjens, I.M.C.M. The Gut Microbial Metabolite Pyrogallol Is a More Potent Inducer of Nrf2-Associated Gene Expression Than Its Parent Compound Green Tea (-)-Epigallocatechin Gallate. Nutrients 2022, 14, 3392. https://doi.org/10.3390/nu14163392
Liu C, Boeren S, Miro Estruch I, Rietjens IMCM. The Gut Microbial Metabolite Pyrogallol Is a More Potent Inducer of Nrf2-Associated Gene Expression Than Its Parent Compound Green Tea (-)-Epigallocatechin Gallate. Nutrients. 2022; 14(16):3392. https://doi.org/10.3390/nu14163392
Chicago/Turabian StyleLiu, Chen, Sjef Boeren, Ignacio Miro Estruch, and Ivonne M. C. M. Rietjens. 2022. "The Gut Microbial Metabolite Pyrogallol Is a More Potent Inducer of Nrf2-Associated Gene Expression Than Its Parent Compound Green Tea (-)-Epigallocatechin Gallate" Nutrients 14, no. 16: 3392. https://doi.org/10.3390/nu14163392
APA StyleLiu, C., Boeren, S., Miro Estruch, I., & Rietjens, I. M. C. M. (2022). The Gut Microbial Metabolite Pyrogallol Is a More Potent Inducer of Nrf2-Associated Gene Expression Than Its Parent Compound Green Tea (-)-Epigallocatechin Gallate. Nutrients, 14(16), 3392. https://doi.org/10.3390/nu14163392






