The Oral Microbiome Impacts the Link between Sugar Consumption and Caries: A Preliminary Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Clinical Examination and Sampling
2.3. Questionnaire Survey and Free-Sugar Intake Assessment
2.4. Metagenomic Analysis
2.5. Data Visual Exhibition, and Statistical and Bioinformatic Analysis
3. Results
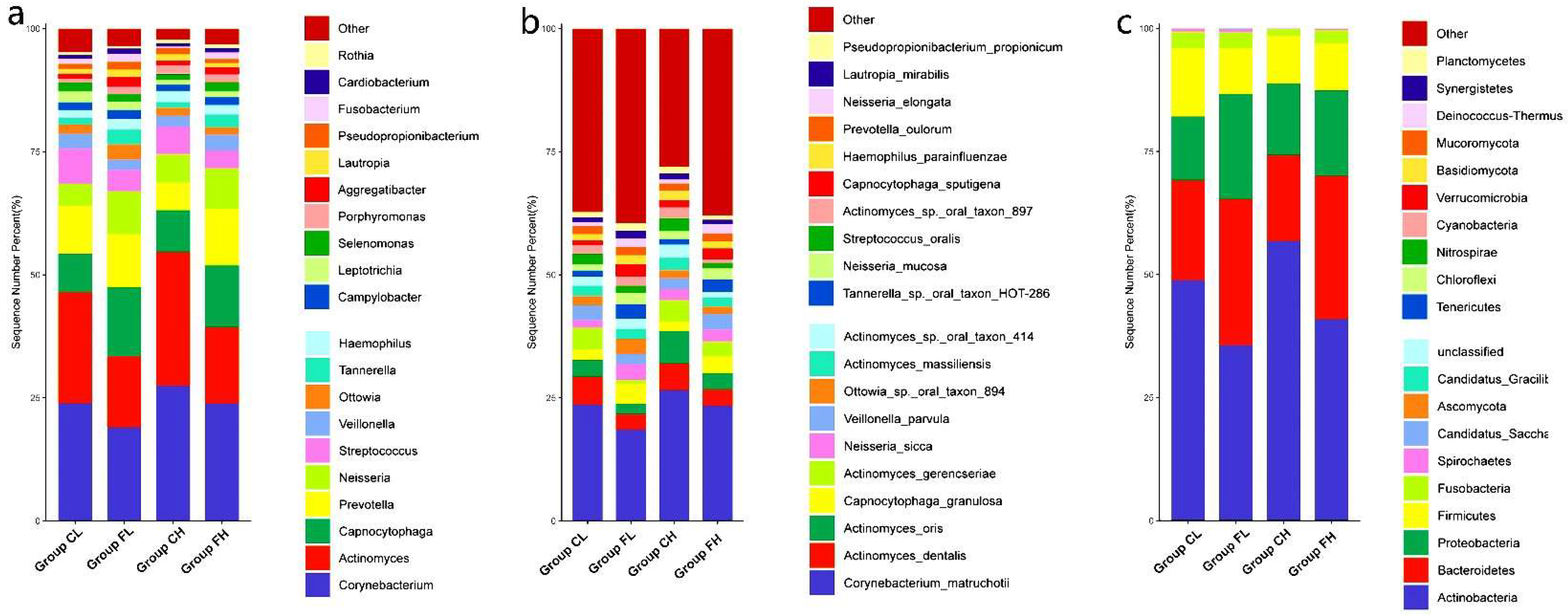
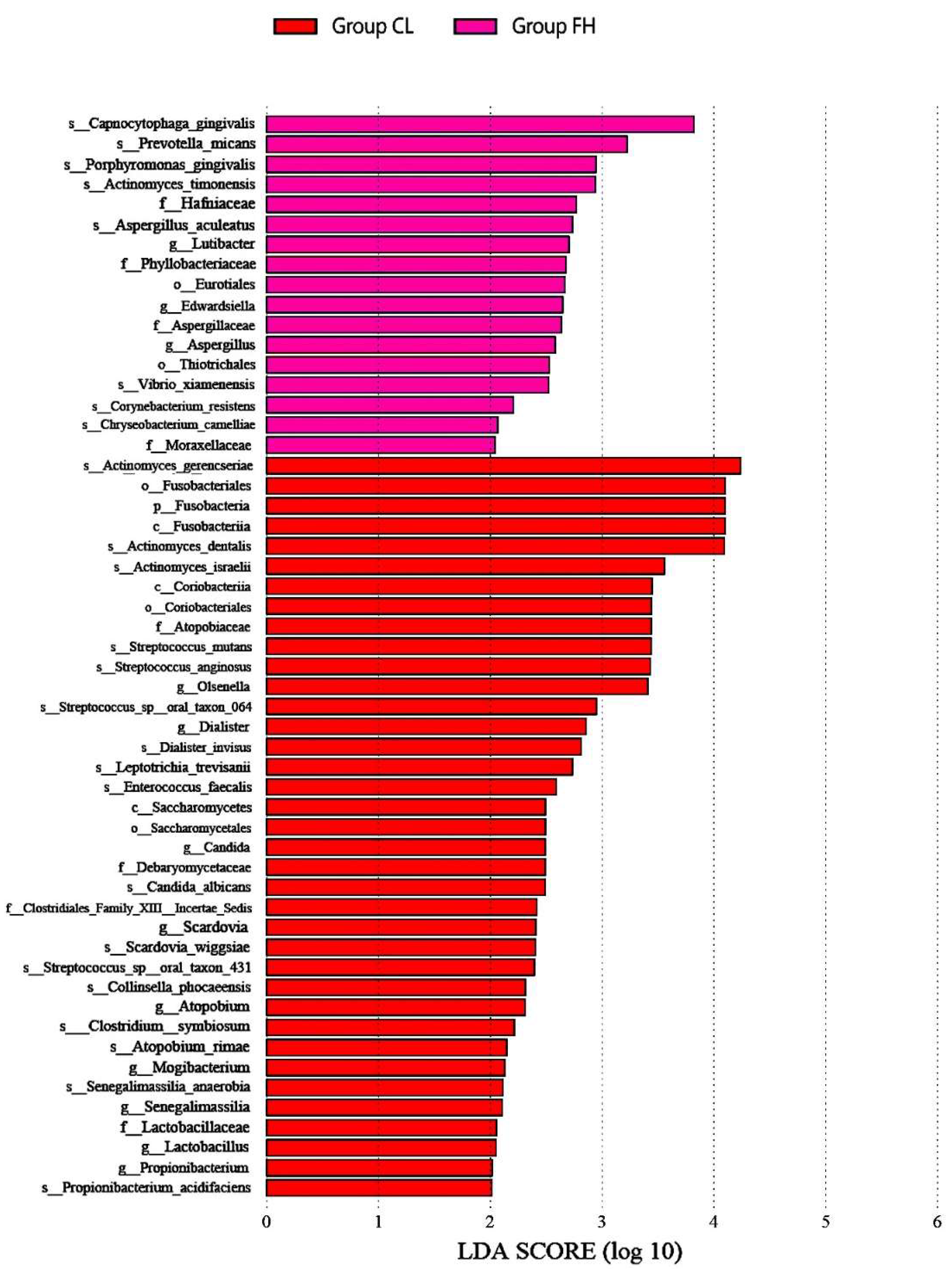
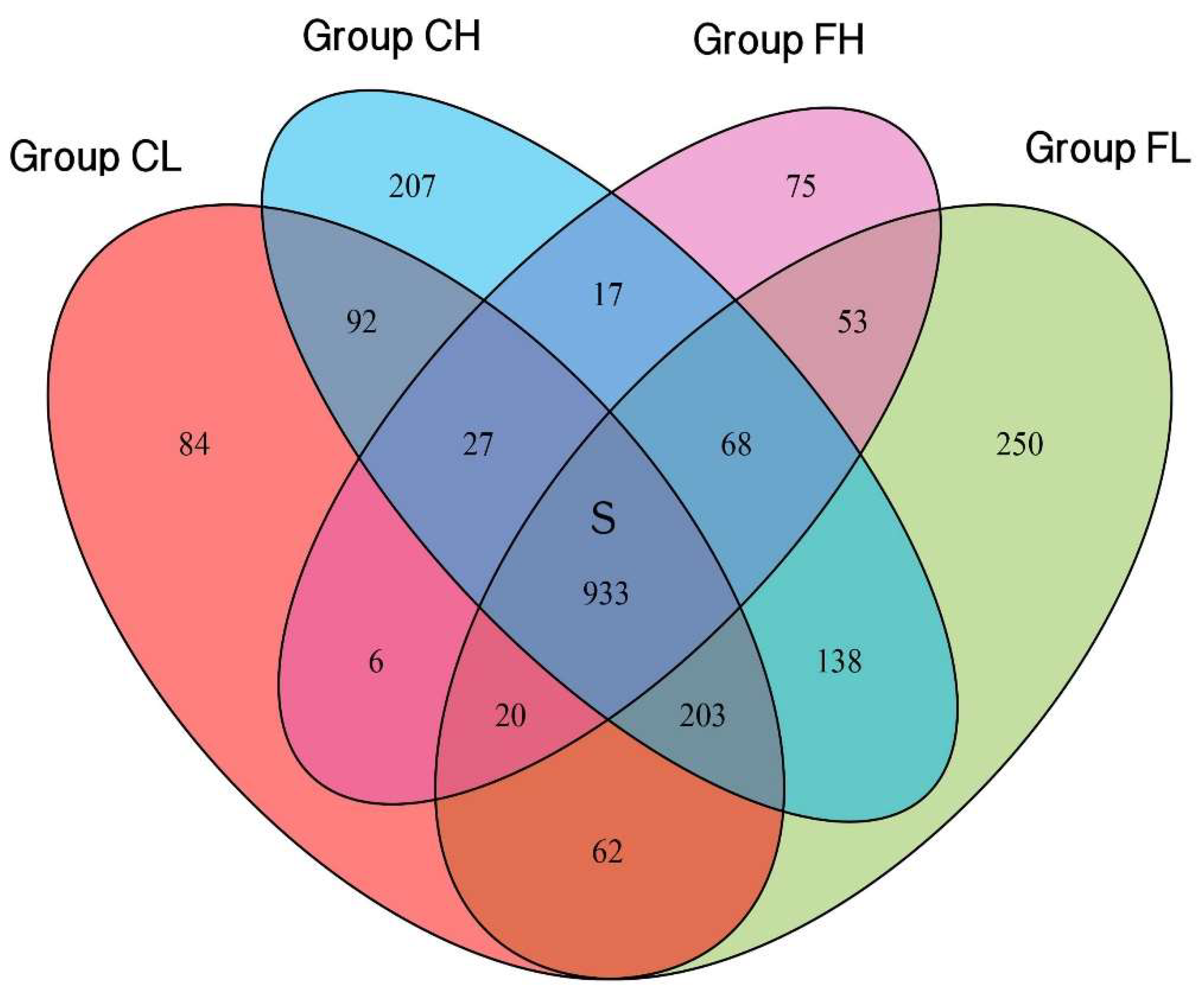
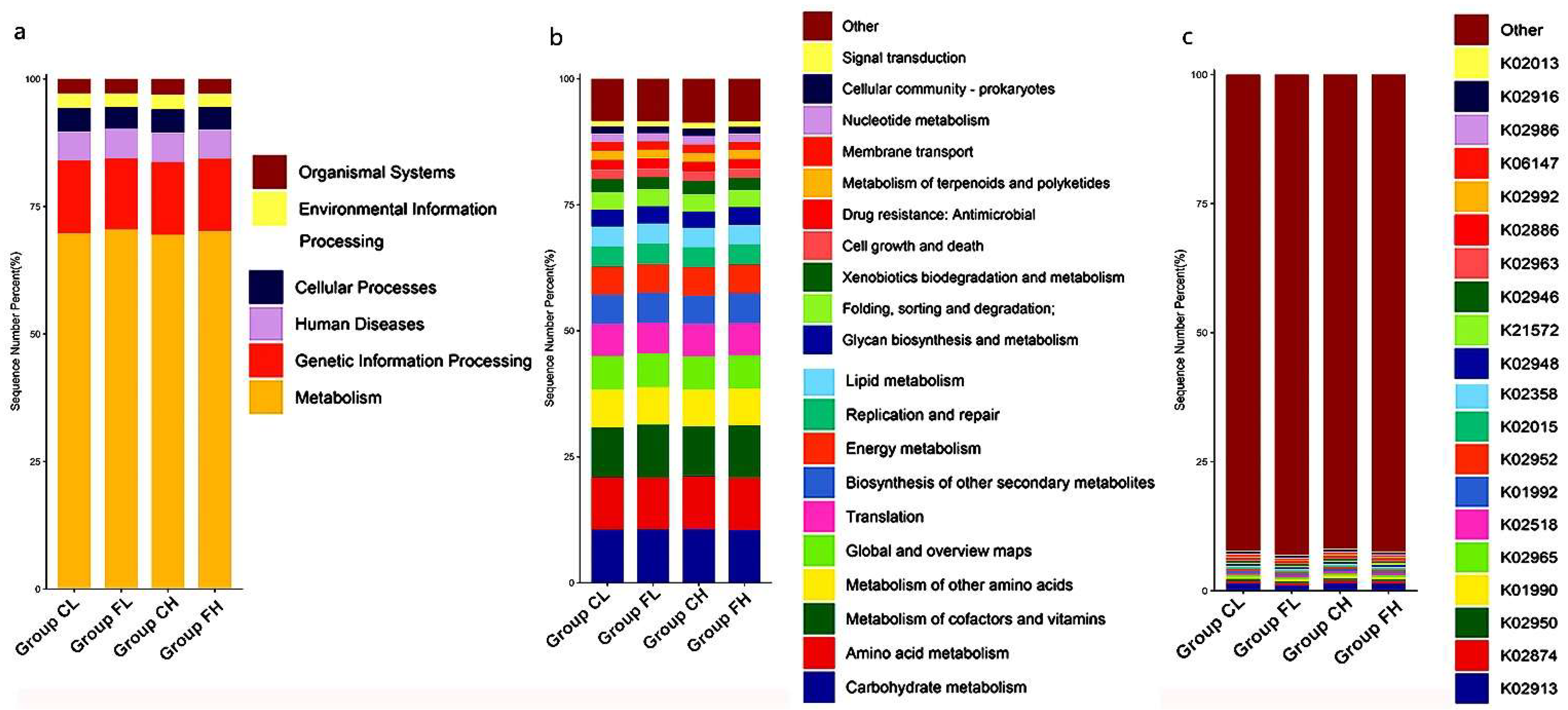
3.1. Microbiota Composition Based on Sugar Consumption and Caries Status
3.2. Microbiota Composition Based on Sugar Consumption and Caries Status
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Selwitz, R.H.; Ismail, A.I.; Pitts, N.B. Dental caries. Lancet 2007, 369, 51–59. [Google Scholar] [CrossRef]
- Armfield, J.M.; Spencer, A.J.; Roberts-Thomson, K.F.; Plastow, K. Water fluoridation and the association of sugar-sweetened beverage consumption and dental caries in Australian children. Am. J. Public Health 2013, 103, 494–500. [Google Scholar] [CrossRef]
- Moynihan, P.J.; Kelly, S.A. Effect on caries of restricting sugars intake: Systematic review to inform WHO guidelines. J. Dent. Res. 2014, 93, 8–18. [Google Scholar] [CrossRef]
- Ha, D.H.; Spencer, A.J.; Moynihan, P.; Thomson, W.M.; Do, L.G. Excess Risk of Dental Caries from Higher Free Sugars Intake Combined with Low Exposure to Water Fluoridation. J. Dent. Res. 2021, 100, 1243–1250. [Google Scholar] [CrossRef]
- Lagerweij, M.; van Loveren, C. Chapter 7: Sugar and Dental Caries. Monogr. Oral Sci. 2020, 28, 68–76. [Google Scholar]
- Loesche, W.J.; Eklund, S.; Earnest, R.; Burt, B. Longitudinal investigation of bacteriology of human fissure decay: Epidemiological studies in molars shortly after eruption. Infect. Immun. 1984, 46, 765–772. [Google Scholar] [CrossRef] [PubMed]
- Touger-Decker, R.; van Loveren, C. Sugars and dental caries. Am. J. Clin. Nutr. 2003, 78, 881S–892S. [Google Scholar] [CrossRef]
- Keller, M.K.; Kressirer, C.A.; Belstrom, D.; Twetman, S.; Tanner, A. Oral microbial profiles of individuals with different levels of sugar intake. J. Oral Microbiol. 2017, 9, 1355207. [Google Scholar] [CrossRef]
- Chen, X.; Hu, X.; Fang, J.; Sun, X.; Zhu, F.; Sun, Y.; Wang, Y. Association of oral microbiota profile with sugar-sweetened beverages consumption in school-aged children. Int. J. Food Sci. Nutr. 2022, 73, 82–92. [Google Scholar] [CrossRef]
- Qudeimat, M.A.; Alyahya, A.; Karched, M.; Behbehani, J.; Salako, N.O. Dental plaque microbiota profiles of children with caries-free and caries-active dentition. J. Dent. 2021, 104, 103539. [Google Scholar] [CrossRef]
- Yang, Q.; Xi, Y.; Liu, H.; Luo, J.; Ouyang, Y.; Sun, M.; Yong, C.; Xiang, C.; Lin, Q. Free Sugars Intake among Chinese Adolescents and Its Association with Dental Caries: A Cross-Sectional Study. Nutrients 2021, 13, 765. [Google Scholar] [CrossRef]
- Takahashi, N.; Nyvad, B. The role of bacteria in the caries process: Ecological perspectives. J. Dent. Res. 2011, 90, 294–303. [Google Scholar] [CrossRef]
- Huson, D.H.; Mitra, S.; Ruscheweyh, H.J.; Weber, N.; Schuster, S.C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011, 21, 1552–1560. [Google Scholar] [CrossRef]
- Pang, L.; Wang, Y.; Ye, Y.; Zhou, Y.; Zhi, Q.; Lin, H. Metagenomic Analysis of Dental Plaque on Pit and Fissure Sites with and Without Caries Among Adolescents. Front. Cell. Infect. Microbiol. 2021, 11, 740981. [Google Scholar] [CrossRef]
- Valm, A.M. The Structure of Dental Plaque Microbial Communities in the Transition from Health to Dental Caries and Periodontal Disease. J Mol Biol. 2019, 431, 2957–2969. [Google Scholar] [CrossRef] [PubMed]
- Gross, E.L.; Leys, E.J.; Gasparovich, S.R.; Firestone, N.D.; Schwartzbaum, J.A.; Janies, D.A.; Asnani, K.; Griffen, A.L. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J. Clin. Microbiol. 2010, 48, 4121–4128. [Google Scholar] [CrossRef] [PubMed]
- Caufield, P.W.; Schon, C.N.; Saraithong, P.; Li, Y.; Argimon, S. Oral Lactobacilli and Dental Caries: A Model for Niche Adaptation in Humans. J. Dent. Res. 2015, 94 (Suppl. 9), 110S–118S. [Google Scholar] [CrossRef] [PubMed]
- Falsetta, M.L.; Klein, M.I.; Colonne, P.M.; Scott-Anne, K.; Gregoire, S.; Pai, C.H.; Gonzalez-Begne, M.; Watson, G.; Krysan, D.J.; Bowen, W.H.; et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect. Immun. 2014, 82, 1968–1981. [Google Scholar] [CrossRef]
- Garcia, B.A.; Acosta, N.C.; Tomar, S.L.; Roesch, L.; Lemos, J.A.; Mugayar, L.; Abranches, J. Association of Candida albicans and Cbp(+) Streptococcus mutans with early childhood caries recurrence. Sci. Rep. 2021, 11, 10802. [Google Scholar] [CrossRef]
- Xiao, J.; Moon, Y.; Li, L.; Rustchenko, E.; Wakabayashi, H.; Zhao, X.; Feng, C.; Gill, S.R.; McLaren, S.; Malmstrom, H.; et al. Candida albicans Carriage in Children with Severe Early Childhood Caries (S-ECC) and Maternal Relatedness. PLoS ONE 2016, 11, e164242. [Google Scholar] [CrossRef]
- Fragkou, S.; Balasouli, C.; Tsuzukibashi, O.; Argyropoulou, A.; Menexes, G.; Kotsanos, N.; Kalfas, S. Streptococcus mutans, Streptococcus sobrinus and Candida albicans in oral samples from caries-free and caries-active children. Eur. Arch. Paediatr. Dent. 2016, 17, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Tanner, A.; Kressirer, C.A.; Rothmiller, S.; Johansson, I.; Chalmers, N.I. The Caries Microbiome: Implications for Reversing Dysbiosis. Adv. Dent. Res. 2018, 29, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, L.; Lif, H.P.; Esberg, A.; Johansson, I. Microbial Complexes and Caries in 17-Year-Olds with and without Streptococcus mutans. J. Dent. Res. 2018, 97, 275–282. [Google Scholar] [CrossRef]
- Kameda, M.; Abiko, Y.; Washio, J.; Tanner, A.; Kressirer, C.A.; Mizoguchi, I.; Takahashi, N. Sugar Metabolism of Scardovia wiggsiae, a Novel Caries-Associated Bacterium. Front. Microbiol. 2020, 11, 479. [Google Scholar] [CrossRef] [PubMed]
- Kressirer, C.A.; Smith, D.J.; King, W.F.; Dobeck, J.M.; Starr, J.R.; Tanner, A. Scardovia wiggsiae and its potential role as a caries pathogen. J. Oral Biosci. 2017, 59, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Wolff, D.; Frese, C.; Maier-Kraus, T.; Krueger, T.; Wolff, B. Bacterial biofilm composition in caries and caries-free subjects. Caries Res. 2013, 47, 69–77. [Google Scholar] [CrossRef]
- Tang, G.; Yip, H.K.; Samaranayake, L.P.; Luo, G.; Lo, E.C.; Teo, C.S. Actinomyces spp. in supragingival plaque of ethnic Chinese preschool children with and without active dental caries. Caries Res. 2003, 37, 381–390. [Google Scholar] [CrossRef]
- Shi, C.; Cai, L.; Xun, Z.; Zheng, S.; Shao, F.; Wang, B.; Zhu, R.; He, Y. Metagenomic analysis of the salivary microbiota in patients with caries, periodontitis and comorbid diseases. J. Dent. Sci. 2021, 16, 1264–1273. [Google Scholar] [CrossRef]
- Iwano, Y.; Sugano, N.; Matsumoto, K.; Nishihara, R.; Iizuka, T.; Yoshinuma, N.; Ito, K. Salivary microbial levels in relation to periodontal status and caries development. J. Periodontal Res. 2010, 45, 165–169. [Google Scholar] [CrossRef]
- Tu, Y.; Ling, X.; Chen, Y.; Wang, Y.; Zhou, N.; Chen, H. Effect of S. Mutans and S. Sanguinis on Growth and Adhesion of P. Gingivalis and Their Ability to Adhere to Different Dental Materials. Med. Sci. Monit. 2017, 23, 4539–5445. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.Y.; Alvarez, P.; Hong, J.; Kuramitsu, H.K. Periodontal pathogens interfere with quorum-sensing-dependent virulence properties in Streptococcus mutans. J. Periodontal Res. 2011, 46, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Muras, A.; Mallo, N.; Otero-Casal, P.; Pose-Rodriguez, J.M.; Otero, A. Quorum sensing systems as a new target to prevent biofilm-related oral diseases. Oral Dis. 2022, 28, 307–313. [Google Scholar] [CrossRef]
- Relman, D.A. The human microbiome: Ecosystem resilience and health. Nutr. Rev. 2012, 70, S2–S9. [Google Scholar] [CrossRef] [PubMed]


| Characteristics | CL (n = 9) | FL (n = 14) | CH (n = 11) | FH (n = 6) | p |
|---|---|---|---|---|---|
| Frequency of tooth brushing | 0.75 | ||||
| <2 time a day | 4 | 9 | 7 | 3 | |
| ≥2 time a day | 5 | 5 | 4 | 3 | |
| Toothpaste containing fluoride or not | 0.12 | ||||
| Yes | 8 | 10 | 11 | 6 | |
| No | 1 | 4 | 0 | 0 | |
| Floss or not | 0.32 | ||||
| Yes | 0 | 3 | 2 | 0 | |
| No | 9 | 11 | 9 | 6 | |
| Plaque index | 0.91 | ||||
| 1–2 | 5 | 9 | 7 | 3 | |
| 0–1 | 4 | 5 | 4 | 3 | |
| BMI status (kg/m2) | 16.96 ± 1.29 | 18.86 ± 2.4 | 19.69 ± 5.9 | 17.44 ± 1.26 | |
| Free sugar consumption (g/day) | 27.28 ± 20.67 | 33.85 ± 23.33 | 221.77 ± 113.87 | 246.74 ± 261.83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pang, L.; Zhi, Q.; Jian, W.; Liu, Z.; Lin, H. The Oral Microbiome Impacts the Link between Sugar Consumption and Caries: A Preliminary Study. Nutrients 2022, 14, 3693. https://doi.org/10.3390/nu14183693
Pang L, Zhi Q, Jian W, Liu Z, Lin H. The Oral Microbiome Impacts the Link between Sugar Consumption and Caries: A Preliminary Study. Nutrients. 2022; 14(18):3693. https://doi.org/10.3390/nu14183693
Chicago/Turabian StylePang, Liangyue, Qinghui Zhi, Wenting Jian, Zhuoying Liu, and Huancai Lin. 2022. "The Oral Microbiome Impacts the Link between Sugar Consumption and Caries: A Preliminary Study" Nutrients 14, no. 18: 3693. https://doi.org/10.3390/nu14183693
APA StylePang, L., Zhi, Q., Jian, W., Liu, Z., & Lin, H. (2022). The Oral Microbiome Impacts the Link between Sugar Consumption and Caries: A Preliminary Study. Nutrients, 14(18), 3693. https://doi.org/10.3390/nu14183693





