Mulberrofuran G, a Mulberry Component, Prevents SARS-CoV-2 Infection by Blocking the Interaction between SARS-CoV-2 Spike Protein S1 Receptor-Binding Domain and Human Angiotensin-Converting Enzyme 2 Receptor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. SARS-CoV-2 Spike/ACE2 Inhibitor Screening Assay
2.3. Kinetic Analysis of the Binding between MG and Spike Protein/ACE2 Receptor Based on Biolayer Interferometry
2.4. In Silico Docking Simulation and Pharmacophore Analysis
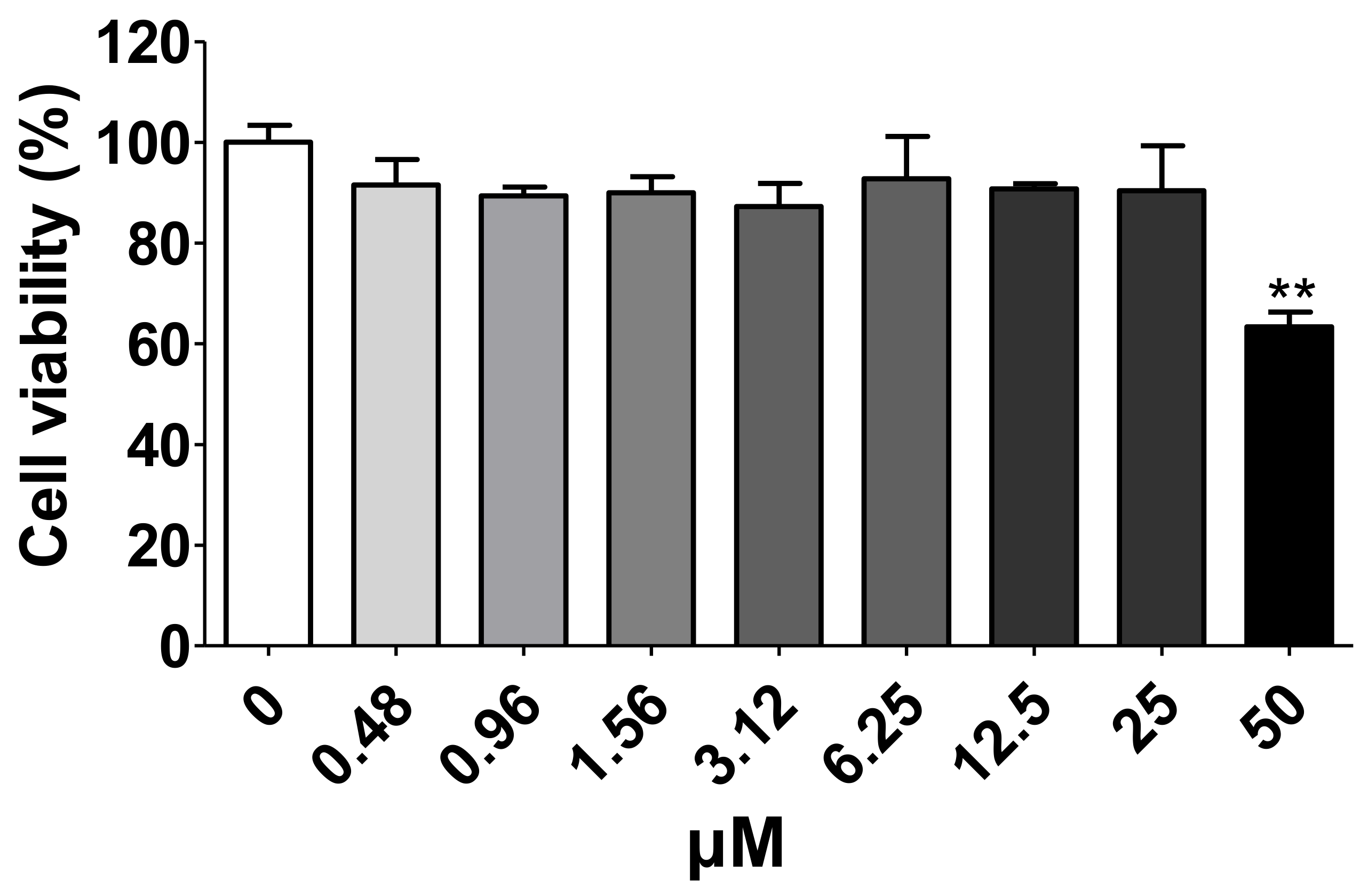
2.5. Cell Viability Assay
2.6. SARS-CoV-2 Lentiviral Pseudovirus Infection Assay
2.7. Hoechst Staining and Immunofluorescence Assay
2.8. Statistical Analysis
3. Results
3.1. The Blockade of Spike S1 RBD:ACE2 Receptor Interaction by MG
3.2. Kinetic Analysis of MG:spike S1 RBD/ACE2 Receptor Interaction
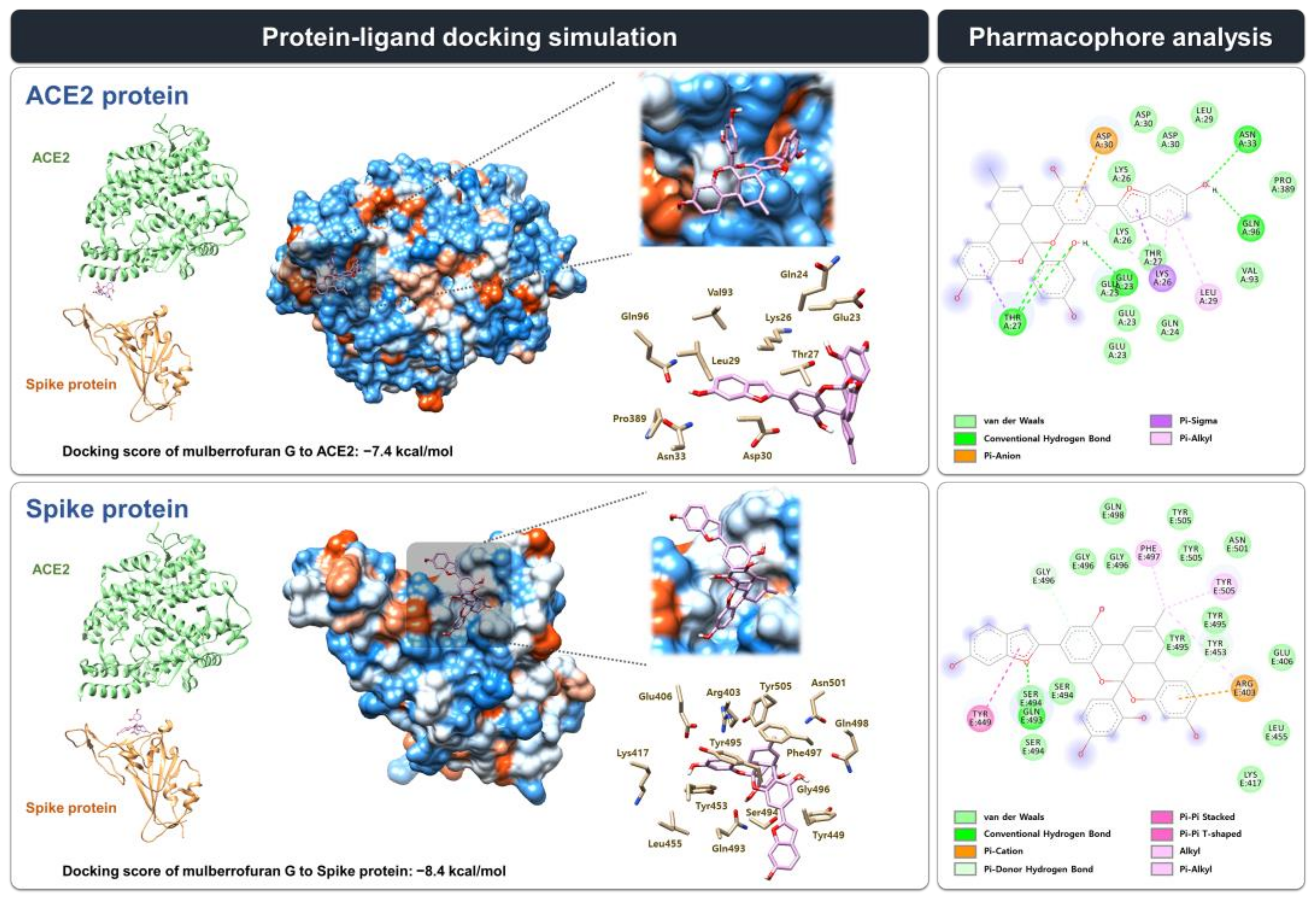
3.3. Molecular Docking Simulation and Pharmacophore Analysis of MG with Spike Protein and ACE2 Receptor
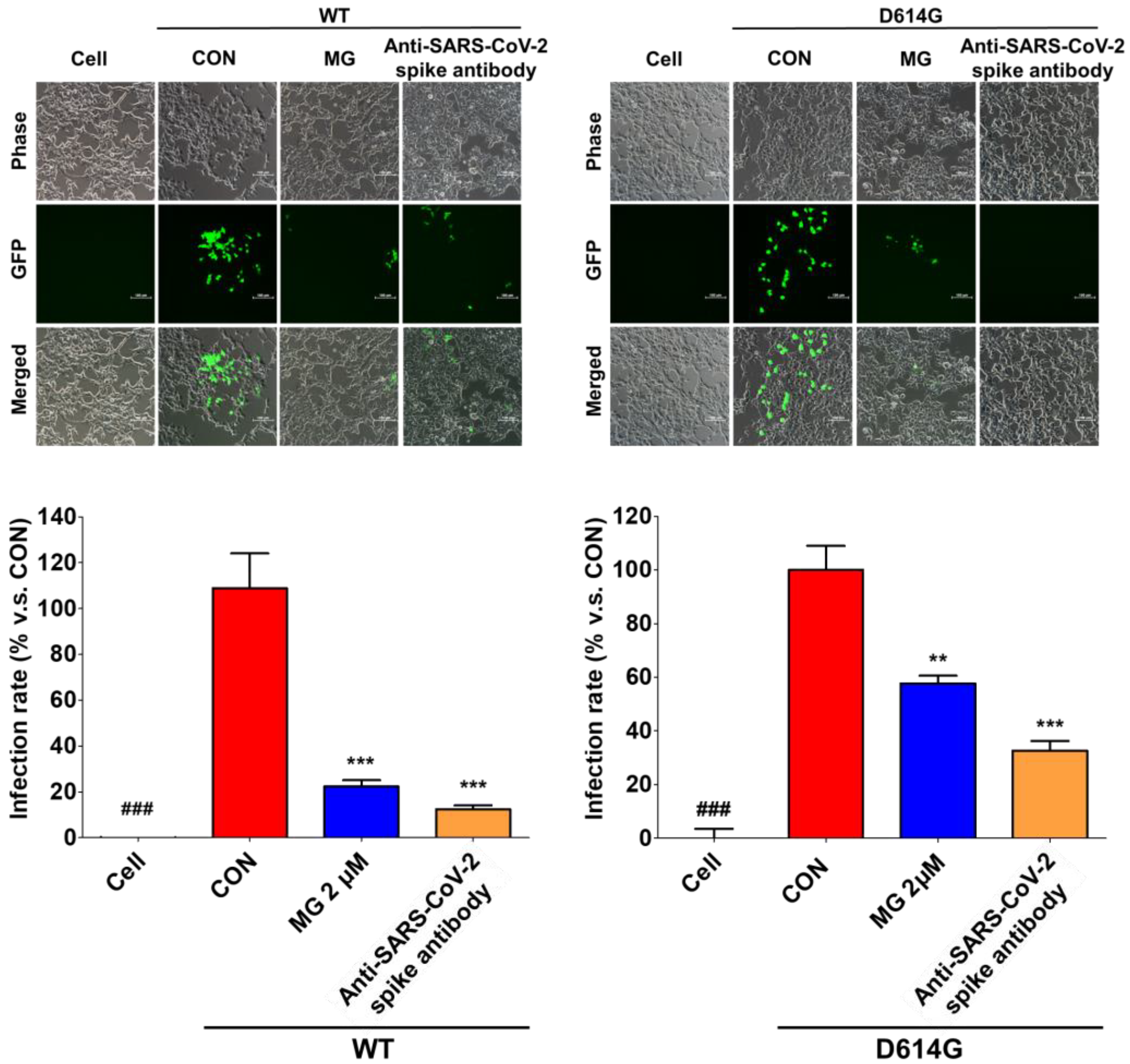
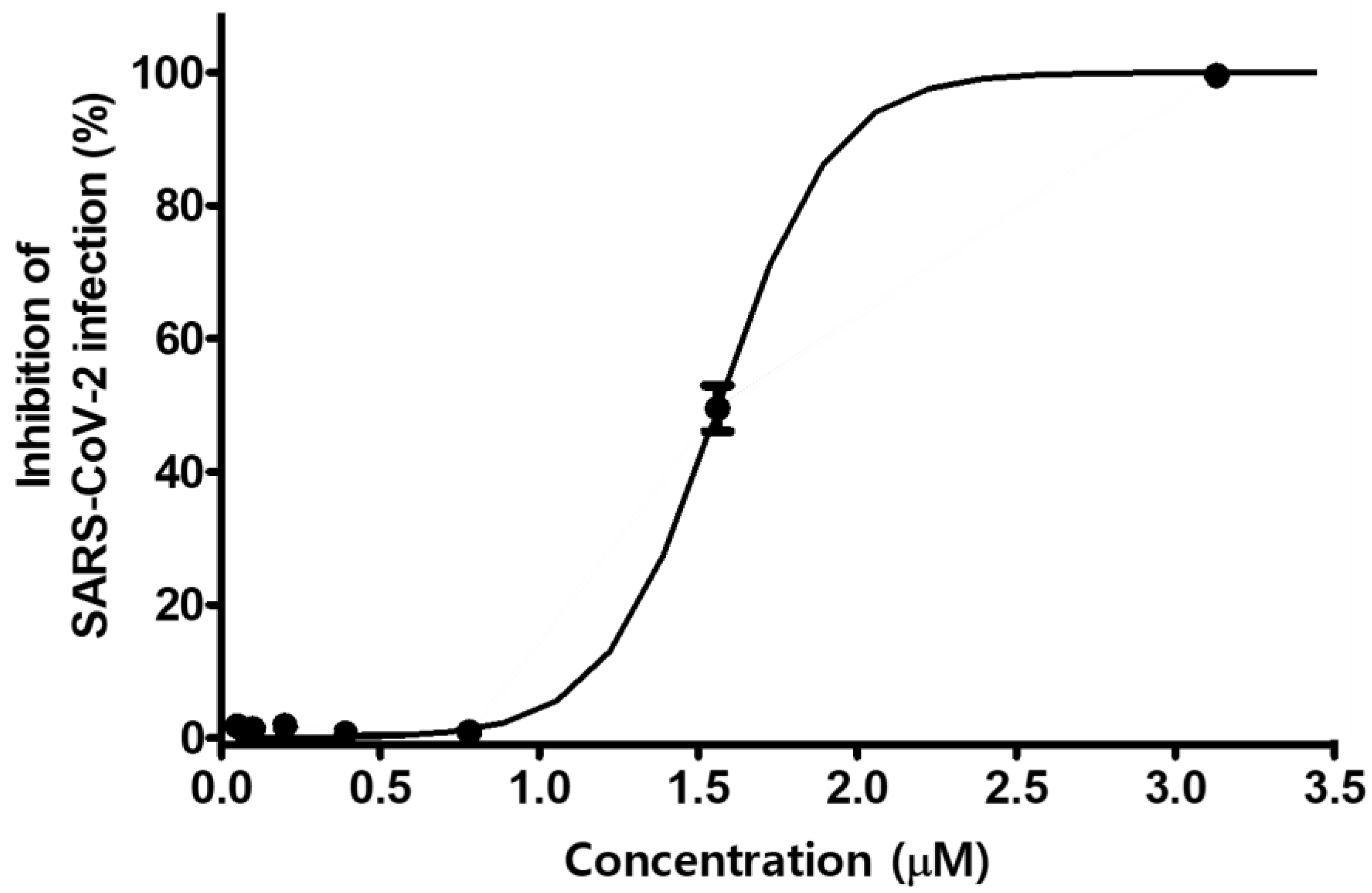
3.4. MG Inhibits SARS-CoV-2 Lentiviral Pseudovirus Infection in HEK293T Cells Overexpressing ACE2/TMPRSS2
3.5. MG Inhibits the Infection of a Clinical Isolate SARS-CoV-2 in Vero Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cucinotta, D.; Vanelli, M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020, 91, 157–160. [Google Scholar] [CrossRef]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, S.; Klein, J.; Robertson, A.J.; Pena-Hernandez, M.A.; Lin, M.J.; Roychoudhury, P.; Lu, P.; Fournier, J.; Ferguson, D.; Mohamed Bakhash, S.A.K.; et al. De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: A case report. Nat. Commun. 2022, 13, 1547. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.I.; Kim, J.; Kim, J.Y.; Kwon, O. Acute intake of mulberry leaf aqueous extract affects postprandial glucose response after maltose loading: Randomized double-blind placebo-controlled pilot study. J. Funct. Foods 2013, 5, 1502–1506. [Google Scholar] [CrossRef]
- Enkhmaa, B.; Shiwaku, K.; Katsube, T.; Kitajima, K.; Anuurad, E.; Yamasaki, M.; Yamane, Y. Mulberry (Morus alba L.) leaves and their major flavonol quercetin 3-(6-malonylglucoside) attenuate atherosclerotic lesion development in LDL receptor-deficient mice. J. Nutr. 2005, 135, 729–734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harauma, A.; Murayama, T.; Ikeyama, K.; Sano, H.; Arai, H.; Takano, R.; Kita, T.; Hara, S.; Kamei, K.; Yokode, M. Mulberry leaf powder prevents atherosclerosis in apolipoprotein E-deficient mice. Biochem. Biophys. Res. Commun. 2007, 358, 751–756. [Google Scholar] [CrossRef]
- Hunyadi, A.; Liktor-Busa, E.; Marki, A.; Martins, A.; Jedlinszki, N.; Hsieh, T.J.; Bathori, M.; Hohmann, J.; Zupko, I. Metabolic effects of mulberry leaves: Exploring potential benefits in type 2 diabetes and hyperuricemia. Evid.-Based Complement. Alternat. Med. 2013, 2013, 948627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeszka-Skowron, M.; Flaczyk, E.; Jeszka, J.; Krejpcio, Z.; Król, E.; Buchowski, M.S. Mulberry leaf extract intake reduces hyperglycaemia in streptozotocin (STZ)-induced diabetic rats fed high-fat diet. J. Funct. Foods 2014, 8, 9–17. [Google Scholar] [CrossRef]
- Zhishen, J.; Mengcheng, T.; Jianming, W. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999, 64, 555–559. [Google Scholar] [CrossRef]
- Katsube, T.; Tsurunaga, Y.; Sugiyama, M.; Furuno, T.; Yamasaki, Y. Effect of air-drying temperature on antioxidant capacity and stability of polyphenolic compounds in mulberry (Morus alba L.) leaves. Food Chem. 2009, 113, 964–969. [Google Scholar] [CrossRef]
- Du, J.; He, Z.D.; Jiang, R.W.; Ye, W.C.; Xu, H.X.; But, P.P. Antiviral flavonoids from the root bark of Morus alba L. Phytochemistry 2003, 62, 1235–1238. [Google Scholar] [CrossRef]
- Lee, J.H.; Bae, S.Y.; Oh, M.; Kim, K.H.; Chung, M.S. Antiviral effects of mulberry (Morus alba) juice and its fractions on foodborne viral surrogates. Foodborne Pathog. Dis. 2014, 11, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Thabti, I.; Albert, Q.; Philippot, S.; Dupire, F.; Westerhuis, B.; Fontanay, S.; Risler, A.; Kassab, T.; Elfalleh, W.; Aferchichi, A.; et al. Advances on Antiviral Activity of Morus spp. Plant Extracts: Human Coronavirus and Virus-Related Respiratory Tract Infections in the Spotlight. Molecules 2020, 25, 1876. [Google Scholar] [CrossRef] [PubMed]
- Shakya, A.; Chikhale, R.V.; Bhat, H.R.; Alasmary, F.A.; Almutairi, T.M.; Ghosh, S.K.; Alhajri, H.M.; Alissa, S.A.; Nagar, S.; Islam, M.A. Pharmacoinformatics-based identification of transmembrane protease serine-2 inhibitors from Morus Alba as SARS-CoV-2 cell entry inhibitors. Mol. Divers. 2022, 26, 265–278. [Google Scholar] [CrossRef]
- Geng, C.A.; Ma, Y.B.; Zhang, X.M.; Yao, S.Y.; Xue, D.Q.; Zhang, R.P.; Chen, J.J. Mulberrofuran G and isomulberrofuran G from Morus alba L.: Anti-hepatitis B virus activity and mass spectrometric fragmentation. J. Agric. Food Chem. 2012, 60, 8197–8202. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Kwon, J.; Kim, D.W.; Lee, H.J.; Lee, D.; Mar, W. Mulberrofuran G Protects Ischemic Injury-induced Cell Death via Inhibition of NOX4-mediated ROS Generation and ER Stress. Phytother. Res. 2017, 31, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.W.; Ko, W.M.; Park, J.H.; Seo, K.H.; Oh, E.J.; Lee, D.Y.; Lee, D.S.; Kim, Y.C.; Lim, D.W.; Han, D.; et al. Isoprenylated flavonoids from the root bark of Morus alba and their hepatoprotective and neuroprotective activities. Arch. Pharm. Res. 2015, 38, 2066–2075. [Google Scholar] [CrossRef] [PubMed]
- Koirala, P.; Seong, S.H.; Zhou, Y.; Shrestha, S.; Jung, H.A.; Choi, J.S. Structure(-)Activity Relationship of the Tyrosinase Inhibitors Kuwanon G, Mulberrofuran G, and Albanol B from Morus Species: A Kinetics and Molecular Docking Study. Molecules 2018, 23, 1413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosseini, F.S.; Motamedi, M.R. Mulberrofuran G, a potent inhibitor of spike protein of SARS corona virus 2. J. Pharm. Care 2021, 9, 74–81. [Google Scholar] [CrossRef]
- Alamri, M.A.; Altharawi, A.; Alabbas, A.B.; Alossaimi, M.A.; Alqahtani, S.M. Structure-based virtual screening and molecular dynamics of phytochemicals derived from Saudi medicinal plants to identify potential COVID-19 therapeutics. Arab. J. Chem. 2020, 13, 7224–7234. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, S.; Corbett, Y.; Parapini, S.; Perego, F.; Cavicchini, L.; Signorini, L.; Delbue, S.; Perego, C.; Ferrante, P.; Taramelli, D.; et al. Malaria pigment accelerates MTT - formazan exocytosis in human endothelial cells. Parasitology 2019, 146, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Isidori, A.M.; Giannetta, E.; Pofi, R.; Venneri, M.A.; Gianfrilli, D.; Campolo, F.; Mastroianni, C.M.; Lenzi, A.; d’Ettorre, G. Targeting the NO-cGMP-PDE5 pathway in COVID-19 infection. The DEDALO project. Andrology 2021, 9, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Joshi, T.; Joshi, T.; Mathpal, S.; Maiti, P.; Nand, M.; Chandra, S.; Tamta, S. In silico screening of natural compounds to inhibit interaction of human ACE2 receptor and spike protein of SARS-CoV-2 for the prevention of COVID-19. J. Biomol. Struct. Dyn. 2021, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lapaillerie, D.; Charlier, C.; Guyonnet-Duperat, V.; Murigneux, E.; Fernandes, H.S.; Martins, F.G.; Magalhaes, R.P.; Vieira, T.F.; Richetta, C.; Subra, F.; et al. Selection of Bis-Indolyl Pyridines and Triphenylamines as New Inhibitors of SARS-CoV-2 Cellular Entry by Modulating the Spike Protein/ACE2 Interfaces. Antimicrob. Agents Chemother. 2022, 66, e0008322. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Zhou, H.; Guo, L.; Xie, D.; He, H.; Zhang, H.; Liu, Y.; Peng, L.; Zheng, L.; Lu, W.; et al. Potential inhibitors for blocking the interaction of the coronavirus SARS-CoV-2 spike protein and its host cell receptor ACE2. J. Transl. Med. 2022, 20, 314. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, Y.; Yao, S.; Ge, H.; Zhu, Y.; Chen, K.; Chen, W.Z.; Zhang, Y.; Zhu, W.; Wang, H.Y.; et al. Discovery of potential small molecular SARS-CoV-2 entry blockers targeting the spike protein. Acta Pharmacol. Sin. 2022, 43, 788–796. [Google Scholar] [CrossRef] [PubMed]
- Puhl, A.C.; Mottin, M.; Sacramento, C.Q.; Tavella, T.A.; Dias, G.G.; Fintelman-Rodrigues, N.; Temerozo, J.R.; Dias, S.S.G.; Ramos, P.; Merten, E.M.; et al. Computational and Experimental Approaches Identify Beta-Blockers as Potential SARS-CoV-2 Spike Inhibitors. ACS Omega 2022, 7, 27950–27958. [Google Scholar] [CrossRef] [PubMed]

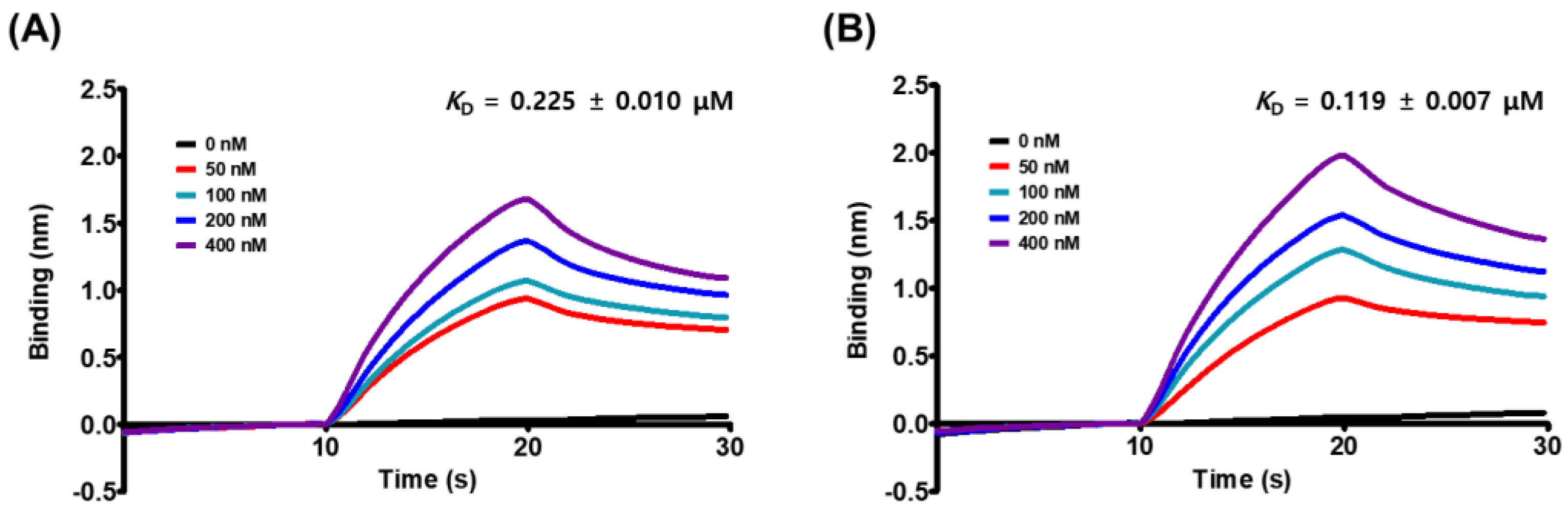

| KD (μM) | ka (μM−1 s−1) | kd (s−1) | R2 | |
|---|---|---|---|---|
| ACE2 receptor | 2.25 × 10−1 | 2.43 × 10−1 | 5.46 × 10−2 | 0.9943 |
| Spike S1 RBD | 1.19 × 10−1 | 3.90 × 10−1 | 4.63 × 10−2 | 0.9958 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.S.; Kim, B.; Kwon, E.-B.; Chung, H.-S.; Choi, J.-G. Mulberrofuran G, a Mulberry Component, Prevents SARS-CoV-2 Infection by Blocking the Interaction between SARS-CoV-2 Spike Protein S1 Receptor-Binding Domain and Human Angiotensin-Converting Enzyme 2 Receptor. Nutrients 2022, 14, 4170. https://doi.org/10.3390/nu14194170
Kim YS, Kim B, Kwon E-B, Chung H-S, Choi J-G. Mulberrofuran G, a Mulberry Component, Prevents SARS-CoV-2 Infection by Blocking the Interaction between SARS-CoV-2 Spike Protein S1 Receptor-Binding Domain and Human Angiotensin-Converting Enzyme 2 Receptor. Nutrients. 2022; 14(19):4170. https://doi.org/10.3390/nu14194170
Chicago/Turabian StyleKim, Young Soo, Buyun Kim, Eun-Bin Kwon, Hwan-Suck Chung, and Jang-Gi Choi. 2022. "Mulberrofuran G, a Mulberry Component, Prevents SARS-CoV-2 Infection by Blocking the Interaction between SARS-CoV-2 Spike Protein S1 Receptor-Binding Domain and Human Angiotensin-Converting Enzyme 2 Receptor" Nutrients 14, no. 19: 4170. https://doi.org/10.3390/nu14194170





