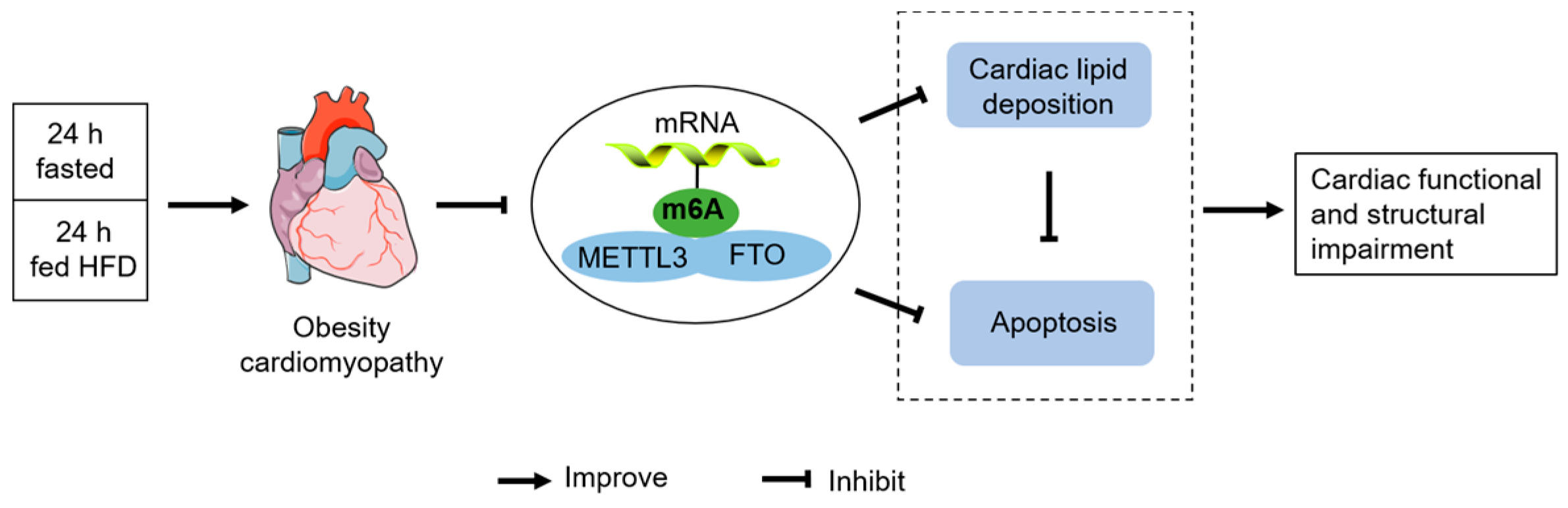
Intermittent Fasting Improves High-Fat Diet-Induced Obesity Cardiomyopathy via Alleviating Lipid Deposition and Apoptosis and Decreasing m6A Methylation in the Heart
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal and Diets
2.2. Echocardiography
2.3. Biochemical Parameters
2.4. m6A RNA Methylation Quantification
2.5. Histology and Oil Red O Staining
2.6. Transmission Electron Microscopy (TEM)
2.7. TUNEL Staining
2.8. Quantitative Real-Time PCR (RT-PCR)
2.9. Western Blot
2.10. Statistical Analysis
3. Results
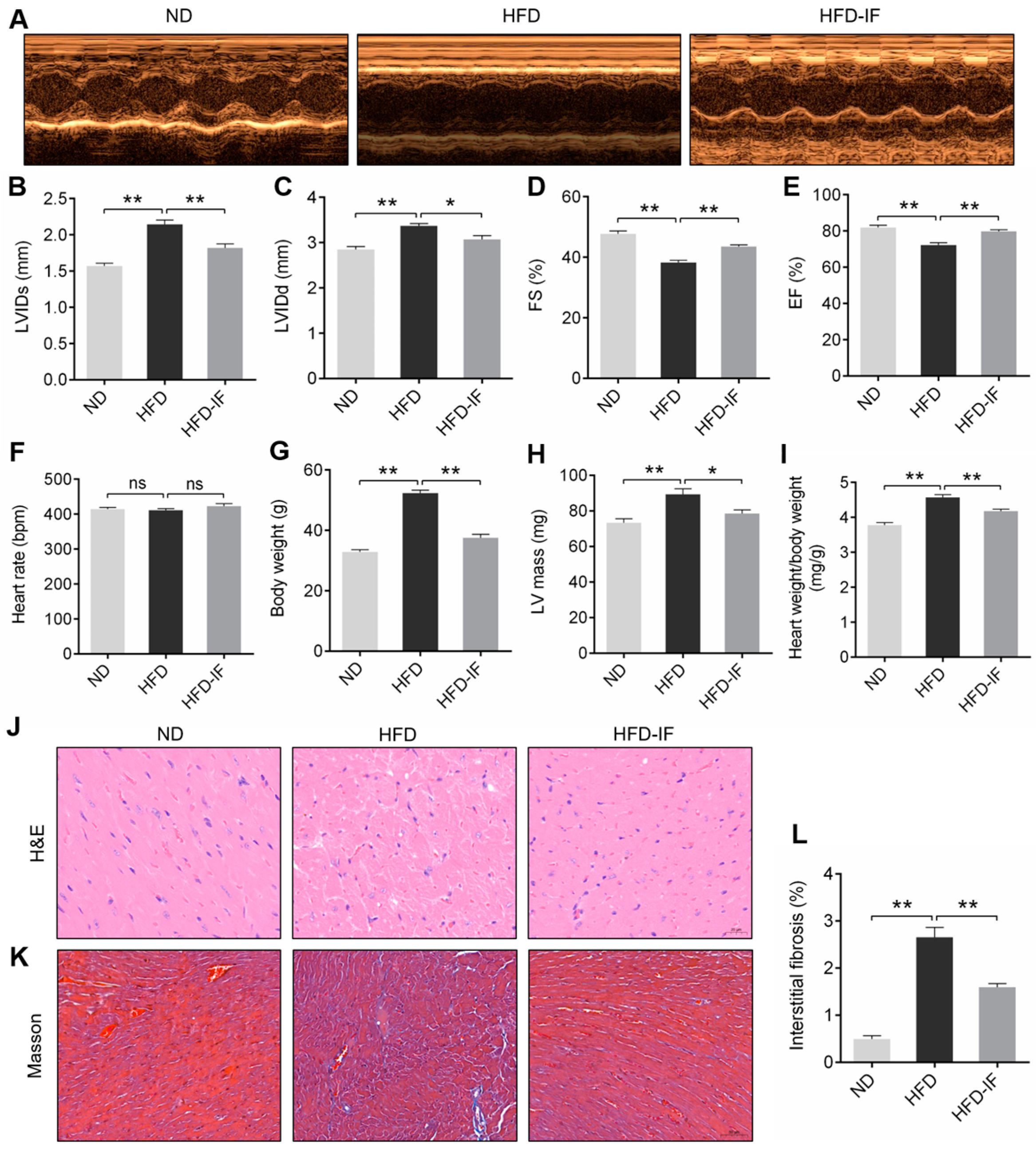
3.1. IF Improves HFD-Induced Mice Obesity Cardiomyopathy
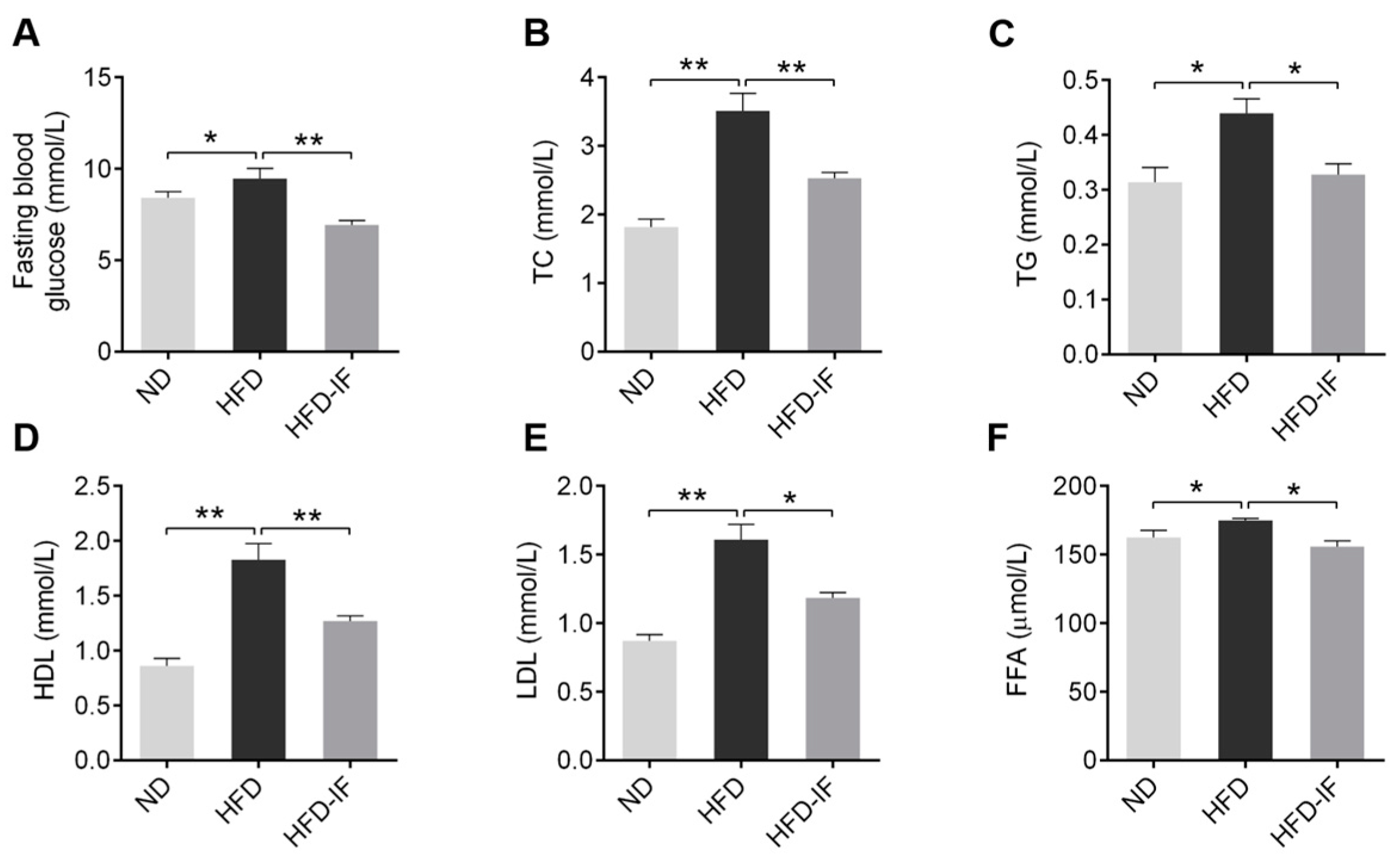
3.2. IF Ameliorates HFD-Induced Serum Lipid Metabolic Disorder
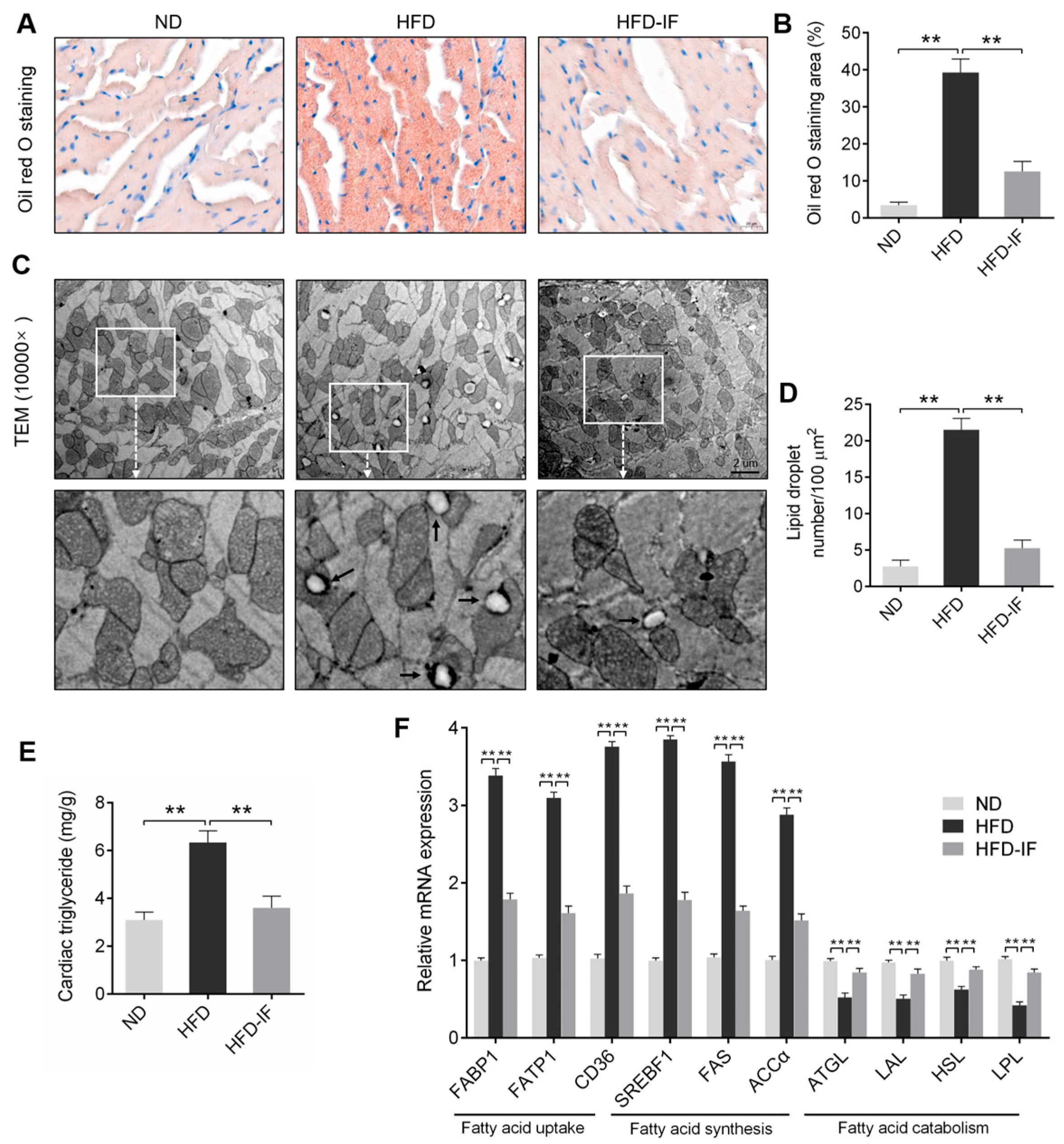
3.3. IF Alleviates HFD-Induced Cardiac Lipid Deposition
3.4. IF Inhibites HFD-Induced Cardiac Apoptosis
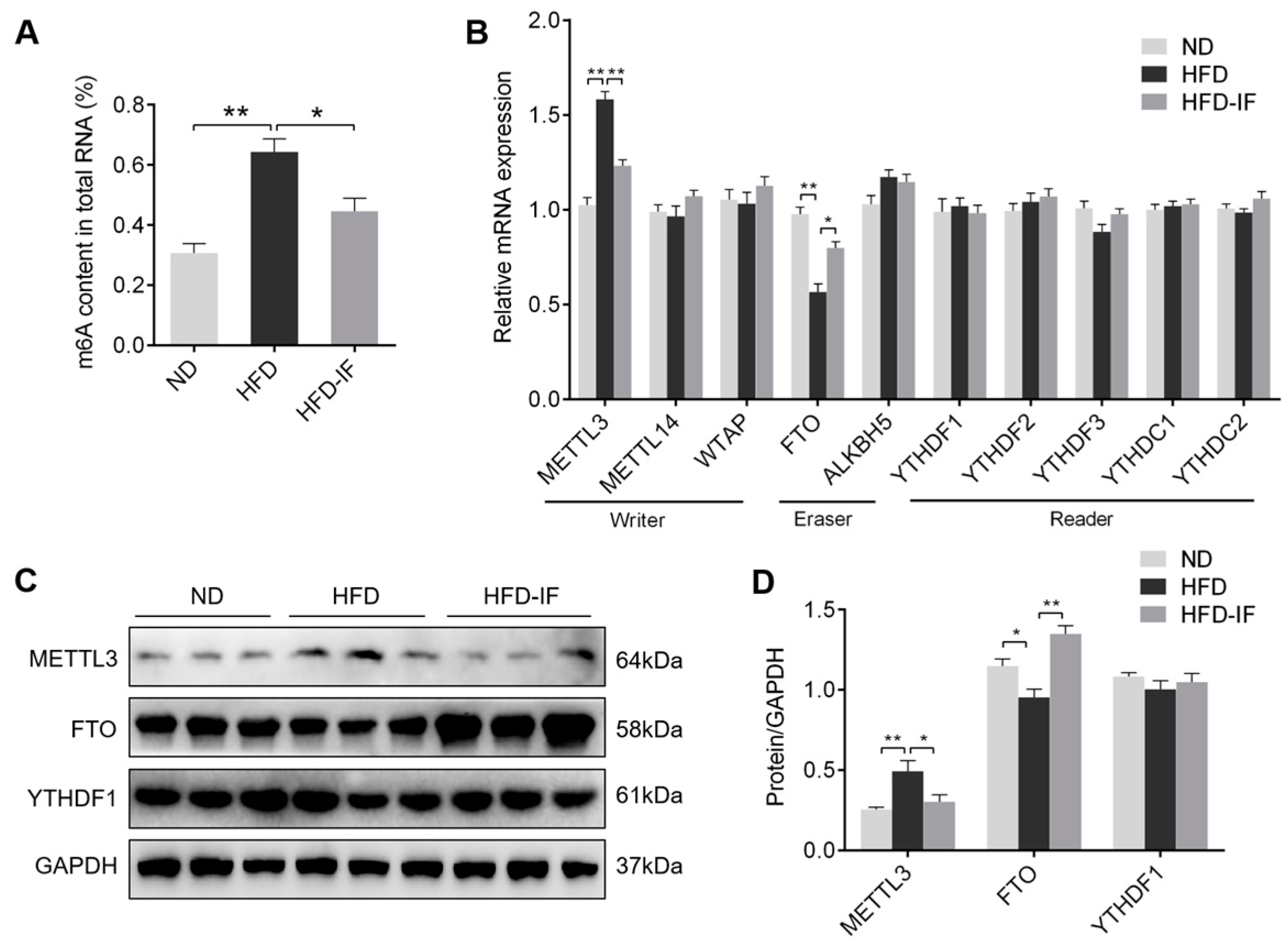
3.5. IF Decreases HFD-Induced Cardiac m6A Methylation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ren, J.; Wu, N.N.; Wang, S.; Sowers, J.R.; Zhang, Y. Obesity cardiomyopathy: Evidence, mechanisms, and therapeutic implications. Physiol. Rev. 2021, 101, 1745–1807. [Google Scholar] [CrossRef]
- Wong, C.; Marwick, T.H. Obesity cardiomyopathy: Pathogenesis and pathophysiology. Nat. Clin. Pract. Cardiovasc. Med. 2007, 4, 436–443. [Google Scholar] [CrossRef]
- Gutierrez-Cuevas, J.; Sandoval-Rodriguez, A.; Meza-Rios, A.; Monroy-Ramirez, H.C.; Galicia-Moreno, M.; Garcia-Banuelos, J.; Santos, A.; Armendariz-Borunda, J. Molecular mechanisms of obesity-linked cardiac dysfunction: An up-date on current knowledge. Cells 2021, 10, 629. [Google Scholar] [CrossRef]
- Shao, D.; Kolwicz, S.C., Jr.; Wang, P.; Roe, N.D.; Villet, O.; Nishi, K.; Hsu, Y.A.; Flint, G.V.; Caudal, A.; Wang, W.; et al. Increasing fatty acid oxidation prevents high-fat diet-induced cardiomyopathy through regulating Parkin-mediated mitophagy. Circulation 2020, 142, 983–997. [Google Scholar] [CrossRef]
- Alpert, M.A.; Omran, J.; Bostick, B.P. Effects of obesity on cardiovascular hemodynamics, cardiac morphology, and ventricular function. Curr. Obes. Rep. 2016, 5, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Sletten, A.C.; Peterson, L.R.; Schaffer, J.E. Manifestations and mechanisms of myocardial lipotoxicity in obesity. J. Intern. Med. 2018, 284, 478–491. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, I.J.; Trent, C.M.; Schulze, P.C. Lipid metabolism and toxicity in the heart. Cell Metab. 2012, 15, 805–812. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Ren, J. Role of cardiac steatosis and lipotoxicity in obesity cardiomyopathy. Hypertension 2011, 57, 148–150. [Google Scholar] [CrossRef]
- Dong, T.A.; Sandesara, P.B.; Dhindsa, D.S.; Mehta, A.; Arneson, L.C.; Dollar, A.L.; Taub, P.R.; Sperling, L.S. Intermittent fasting: A heart healthy dietary pattern? Am. J. Med. 2020, 133, 901–907. [Google Scholar] [CrossRef]
- Tinsley, G.M.; Horne, B.D. Intermittent fasting and cardiovascular disease: Current evidence and unresolved questions. Future Cardiol. 2018, 14, 47–54. [Google Scholar] [CrossRef]
- Malinowski, B.; Zalewska, K.; Wesierska, A.; Sokolowska, M.M.; Socha, M.; Liczner, G.; Pawlak-Osinska, K.; Wicinski, M. Intermittent fasting in cardiovascular disorders-an overview. Nutrients 2019, 11, 673. [Google Scholar] [CrossRef] [Green Version]
- Godar, R.J.; Ma, X.; Liu, H.; Murphy, J.T.; Weinheimer, C.J.; Kovacs, A.; Crosby, S.D.; Saftig, P.; Diwan, A. Repetitive stimulation of autophagy-lysosome machinery by intermittent fasting preconditions the myocardium to ischemia-reperfusion injury. Autophagy 2015, 11, 1537–1560. [Google Scholar] [CrossRef] [Green Version]
- Ahmet, I.; Wan, R.; Mattson, M.P.; Lakatta, E.G.; Talan, M. Cardioprotection by intermittent fasting in rats. Circulation 2005, 112, 3115–3121. [Google Scholar] [CrossRef] [Green Version]
- Antoni, R.; Johnston, K.L.; Collins, A.L.; Robertson, M.D. Effects of intermittent fasting on glucose and lipid metabolism. Proc. Nutr. Soc. 2017, 76, 361–368. [Google Scholar] [CrossRef]
- Wilson, R.A.; Deasy, W.; Stathis, C.G.; Hayes, A.; Cooke, M.B. Intermittent fasting with or without exercise prevents weight gain and improves lipids in diet-induced obese mice. Nutrients 2018, 10, 346. [Google Scholar] [CrossRef] [Green Version]
- Camelo, L.; Marinho, T.S.; Aguila, M.B.; Souza-Mello, V.; Barbosa-da-Silva, S. Intermittent fasting exerts beneficial metabolic effects on blood pressure and cardiac structure by modulating local renin-angiotensin system in the heart of mice fed high-fat or high-fructose diets. Nutr. Res. 2019, 63, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Liu, W.; Wang, J.; Yu, J.; Yang, L.Q. Intermittent fasting improves lipid metabolism through changes in gut microbiota in diet-induced obese mice. Med. Sci. Monit. 2020, 26, e926789. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef]
- Yang, C.; Hu, Y.; Zhou, B.; Bao, Y.; Li, Z.; Gong, C.; Yang, H.; Wang, S.; Xiao, Y. The role of m(6)A modification in physiology and disease. Cell Death Dis. 2020, 11, 960. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, S.; Wu, X.; Zhou, X. m(6)A RNA methylation in cardiovascular diseases. Mol. Ther. 2020, 28, 2111–2119. [Google Scholar] [CrossRef]
- Zhang, B.; Jiang, H.; Dong, Z.; Sun, A.; Ge, J. The critical roles of m6A modification in metabolic abnormality and cardiovascular diseases. Genes Dis. 2021, 8, 746–758. [Google Scholar] [CrossRef]
- Kumari, R.; Ranjan, P.; Suleiman, Z.G.; Goswami, S.K.; Li, J.; Prasad, R.; Verma, S.K. mRNA modifications in cardiovascular biology and disease: With a focus on m6A modification. Cardiovasc. Res. 2021; in press. [Google Scholar] [CrossRef]
- Li, G.; Xie, C.; Lu, S.; Nichols, R.G.; Tian, Y.; Li, L.; Patel, D.; Ma, Y.; Brocker, C.N.; Yan, T.; et al. Intermittent fasting promotes white adipose browning and decreases obesity by shaping the gut microbiota. Cell Metab. 2017, 26, 672–685. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Li, Z.; Cai, M.; Xi, Y.; Xu, Z.; Zhang, Z.; Li, H.; Zhu, W.; Tian, Z. Aerobic exercise alleviates oxidative stress-induced apoptosis in kidneys of myocardial infarction mice by inhibiting ALCAT1 and activating FNDC5/Irisin signaling pathway. Free Radic. Biol. Med. 2020, 158, 171–180. [Google Scholar] [CrossRef]
- Battiprolu, P.K.; Hojayev, B.; Jiang, N.; Wang, Z.V.; Luo, X.; Iglewski, M.; Shelton, J.M.; Gerard, R.D.; Rothermel, B.A.; Gillette, T.G.; et al. Metabolic stress-induced activation of FoxO1 triggers diabetic cardiomyopathy in mice. J. Clin. Investig. 2012, 122, 1109–1118. [Google Scholar] [CrossRef]
- Hsu, H.C.; Chen, C.Y.; Lee, B.C.; Chen, M.F. High-fat diet induces cardiomyocyte apoptosis via the inhibition of autophagy. Eur. J. Nutr. 2016, 55, 2245–2254. [Google Scholar] [CrossRef]
- Zuo, A.; Zhao, X.; Li, T.; Li, J.; Lei, S.; Chen, J.; Xu, D.; Song, C.; Liu, T.; Li, C.; et al. CTRP9 knockout exaggerates lipotoxicity in cardiac myocytes and high-fat diet-induced cardiac hypertrophy through inhibiting the LKB1/AMPK pathway. J. Cell Mol. Med. 2020, 24, 2635–2647. [Google Scholar] [CrossRef]
- Lin, Y.Y.; Hsieh, P.S.; Cheng, Y.J.; Cheng, S.M.; Chen, C.J.; Huang, C.Y.; Kuo, C.H.; Kao, C.L.; Shyu, W.C.; Lee, S.D. Anti-apoptotic and pro-survival effects of food restriction on high-fat diet-induced obese hearts. Cardiovasc. Toxicol. 2017, 17, 163–174. [Google Scholar] [CrossRef]
- Klempel, M.C.; Kroeger, C.M.; Varady, K.A. Alternate day fasting (ADF) with a high-fat diet produces similar weight loss and cardio-protection as ADF with a low-fat diet. Metabolism 2013, 62, 137–143. [Google Scholar] [CrossRef]
- Varady, K.A.; Hudak, C.S.; Hellerstein, M.K. Modified alternate-day fasting and cardioprotection: Relation to adipose tissue dynamics and dietary fat intake. Metabolism 2009, 58, 803–811. [Google Scholar] [CrossRef]
- Ge, C.X.; Xu, M.X.; Qin, Y.T.; Gu, T.T.; Lou, D.S.; Li, Q.; Hu, L.F.; Wang, B.C.; Tan, J. Endoplasmic reticulum stress-induced iRhom2 up-regulation promotes macrophage-regulated cardiac inflammation and lipid deposition in high fat diet (HFD)-challenged mice: Intervention of fisetin and metformin. Free Radic. Biol. Med. 2019, 141, 67–83. [Google Scholar] [CrossRef]
- Wu, J.; Li, Y.; Yu, J.; Gan, Z.; Wei, W.; Wang, C.; Zhang, L.; Wang, T.; Zhong, X. Resveratrol attenuates high-fat diet induced hepatic lipid homeostasis disorder and decreases m(6)A RNA methylation. Front. Pharmacol. 2020, 11, 568006. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, D.; Tang, L.; Li, H.; Liu, Z.; Gao, J.; Edin, M.L.; Zhang, H.; Zhang, K.; Chen, J.; et al. Soluble epoxide hydrolase deficiency attenuates lipotoxic cardiomyopathy via upregulation of AMPK-mTORC mediated autophagy. J. Mol. Cell Cardiol. 2021, 154, 80–91. [Google Scholar] [CrossRef]
- Costantino, S.; Akhmedov, A.; Melina, G.; Mohammed, S.A.; Othman, A.; Ambrosini, S.; Wijnen, W.J.; Sada, L.; Ciavarella, G.M.; Liberale, L.; et al. Obesity-induced activation of JunD promotes myocardial lipid accumulation and metabolic cardiomyopathy. Eur Heart J. 2019, 40, 997–1008. [Google Scholar] [CrossRef] [Green Version]
- Feng, W.; Lei, T.; Wang, Y.; Feng, R.; Yuan, J.; Shen, X.; Wu, Y.; Gao, J.; Ding, W.; Lu, Z. GCN2 deficiency ameliorates cardiac dysfunction in diabetic mice by reducing lipotoxicity and oxidative stress. Free Radic. Biol. Med. 2019, 130, 128–139. [Google Scholar] [CrossRef]
- Lee, S.D.; Shyu, W.C.; Cheng, I.S.; Kuo, C.H.; Chan, Y.S.; Lin, Y.M.; Tasi, C.Y.; Tsai, C.H.; Ho, T.J.; Huang, C.Y. Effects of exercise training on cardiac apoptosis in obese rats. Nutr. Metab. Cardiovasc. Dis. 2013, 23, 566–573. [Google Scholar] [CrossRef]
- Wu, J.; Frazier, K.; Zhang, J.; Gan, Z.; Wang, T.; Zhong, X. Emerging role of m(6) A RNA methylation in nutritional physiology and metabolism. Obes. Rev. 2020, 21, e12942. [Google Scholar] [CrossRef]
- Sun, M.; Zhang, X. Epigenetic regulation of N6-methyladenosine modifications in obesity. J. Diabetes Investig. 2021, 12, 1306–1315. [Google Scholar] [CrossRef]
- Guo, J.; Ren, W.; Li, A.; Ding, Y.; Guo, W.; Su, D.; Hu, C.; Xu, K.; Chen, H.; Xu, X.; et al. Fat mass and obesity-associated gene enhances oxidative stress and lipogenesis in nonalcoholic fatty liver disease. Dig. Dis. Sci. 2013, 58, 1004–1009. [Google Scholar] [CrossRef]
- Xie, W.; Ma, L.L.; Xu, Y.Q.; Wang, B.H.; Li, S.M. METTL3 inhibits hepatic insulin sensitivity via N6-methyladenosine modification of Fasn mRNA and promoting fatty acid metabolism. Biochem. Biophys. Res. Commun. 2019, 518, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Liu, C.; Xu, L.; Yuan, Y.; Zhao, J.; Zhao, W.; Chen, Y.; Qiu, J.; Meng, M.; Zheng, Y.; et al. N(6)-methyladenosine reader protein YT521-B homology domain-containing 2 suppresses liver steatosis by regulation of mRNA stability of lipogenic genes. Hepatology 2021, 73, 91–103. [Google Scholar] [CrossRef]
- Mathiyalagan, P.; Adamiak, M.; Mayourian, J.; Sassi, Y.; Liang, Y.; Agarwal, N.; Jha, D.; Zhang, S.; Kohlbrenner, E.; Chepurko, E.; et al. FTO-dependent N(6)-methyladenosine regulates cardiac function during remodeling and repair. Circulation 2019, 139, 518–532. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Feng, X.; Zhang, H.; Luo, Y.; Huang, J.; Lin, M.; Jin, J.; Ding, X.; Wu, S.; Huang, H.; et al. METTL3 and ALKBH5 oppositely regulate m(6)A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy 2019, 15, 1419–1437. [Google Scholar] [CrossRef] [Green Version]
- Asif, S.; Morrow, N.M.; Mulvihill, E.E.; Kim, K.H. Understanding dietary intervention-mediated epigenetic modifications in metabolic diseases. Front. Genet. 2020, 11, 590369. [Google Scholar] [CrossRef] [PubMed]
- Gensous, N.; Franceschi, C.; Santoro, A.; Milazzo, M.; Garagnani, P.; Bacalini, M.G. The impact of caloric restriction on the epigenetic signatures of aging. Int. J. Mol. Sci. 2019, 20, 2022. [Google Scholar] [CrossRef] [Green Version]


| Genes | Primer Sequences | |
|---|---|---|
| Forward (5′–3′) | Reverse (5′–3′) | |
| FABP1 | CCATGACTGGGGAAAAAGTC | GCCTTTGAAAGTTGTCACCAT |
| FATP1 | TGCACAGCAGGTACTACCGCAT | TGCGCAGTACCACCGTCAAC |
| CD36 | ATTGGTCAAGCCAGCT | TGTAGGCTCATCCACTAC |
| SREBP1c | AATCAGGACCATGCCG | CTCAACCTATGAAAATAAAGTTTGC |
| FAS | GCGGGTTCGTGAAACTGATAA | CAGGTTGGCATGGTTGACAG |
| ACCα | GCCTCCGTCAGCTCAGATAC | ATGTGAAAGGCCAAACCATC |
| ATGL | TGTTTCAGACGGAGAGAACG | GGAGGGGTGGAGGAATGAGG |
| LAL | TGGAGGGACAAACCACTGA | AAGGGAATCGGACCACTTG |
| HSL | CTTCTCCCTCTCGTCTGCTG | AATGGTCCTCTGCCTCTGTC |
| LPL | GATCCGAGTGAAAGCCGGAG | TTGTTTGTCCAGTGTCAGCCA |
| METTL3 | CTGGGCACTTGGATTTAAGGAA | GTATCCCATCCAGTTGGTTTC |
| METTL14 | CTGAGAGTGCGGATAGCATTG | GAGCAGATGTATCATAGGAAGCC |
| WTAP | TAGACCCAGCGATCAACTTGT | CCTGTTTGGCTATCAGGCGTA |
| FTO | TTCATGCTGGATGACCTCAATG | GCCAACTGACAGCGTTCTAAG |
| ALKBH5 | GCATACGGCCTCAGGACATTA | TTCCAATCGCGGTGCATCTAA |
| YTHDF1 | ACAGTTACCCCTCGATGAGTG | GGTAGTGAGATACGGGATGGGA |
| YTHDF2 | GAGCAGAGACCAAAAGGTCAAG | CTGTGGGCTCAAGTAAGGTTC |
| YTHDF3 | GATCAGCCTATGCCATATCTGAC | CCCCTGGTTGACTAAAAACACC |
| YTHDC1 | GGAAGCACCCAGTGTATAGGA | GGAAGCACCCAGTGTATAGGA |
| YTHDC2 | GAAGATCGCCGTCAACATCG | GCTCTTTCCGTACTGGTCAAA |
| GAPDH | GCAAGGACACTGAGCAAGA | GGATGGAAATTGTGAGGGAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Qin, Y.; Lv, B.; Tian, Z.; Zhang, B. Intermittent Fasting Improves High-Fat Diet-Induced Obesity Cardiomyopathy via Alleviating Lipid Deposition and Apoptosis and Decreasing m6A Methylation in the Heart. Nutrients 2022, 14, 251. https://doi.org/10.3390/nu14020251
Xu Z, Qin Y, Lv B, Tian Z, Zhang B. Intermittent Fasting Improves High-Fat Diet-Induced Obesity Cardiomyopathy via Alleviating Lipid Deposition and Apoptosis and Decreasing m6A Methylation in the Heart. Nutrients. 2022; 14(2):251. https://doi.org/10.3390/nu14020251
Chicago/Turabian StyleXu, Zujie, Ying Qin, Binbin Lv, Zhenjun Tian, and Bing Zhang. 2022. "Intermittent Fasting Improves High-Fat Diet-Induced Obesity Cardiomyopathy via Alleviating Lipid Deposition and Apoptosis and Decreasing m6A Methylation in the Heart" Nutrients 14, no. 2: 251. https://doi.org/10.3390/nu14020251
APA StyleXu, Z., Qin, Y., Lv, B., Tian, Z., & Zhang, B. (2022). Intermittent Fasting Improves High-Fat Diet-Induced Obesity Cardiomyopathy via Alleviating Lipid Deposition and Apoptosis and Decreasing m6A Methylation in the Heart. Nutrients, 14(2), 251. https://doi.org/10.3390/nu14020251







