Effect of Curcumin on Attenuation of Liver Cirrhosis via Genes/Proteins and Pathways: A System Pharmacology Study
Abstract
:1. Introduction
2. Methods
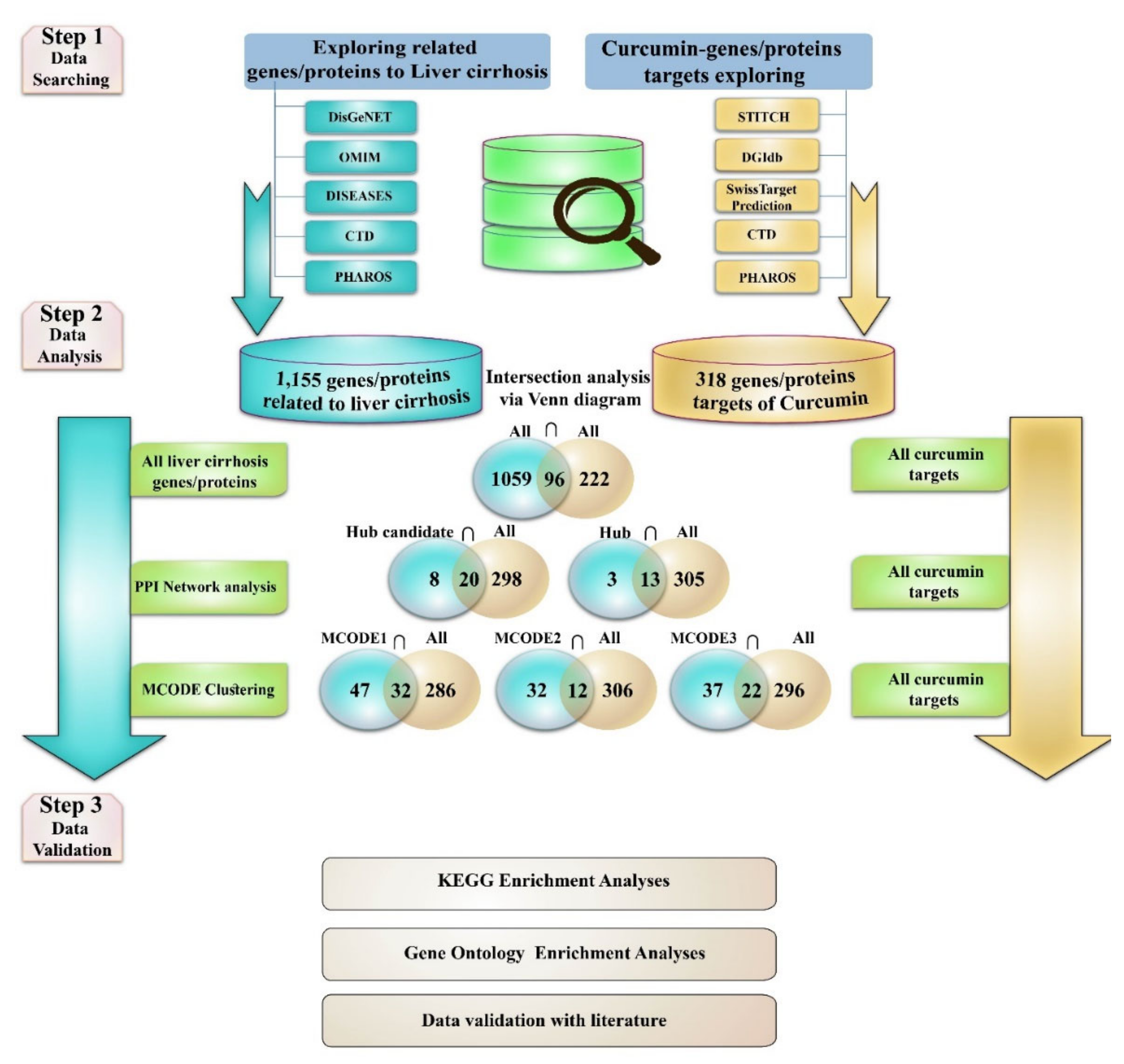
2.1. Exploring Liver Cirrhosis-Protein/Gene Associations
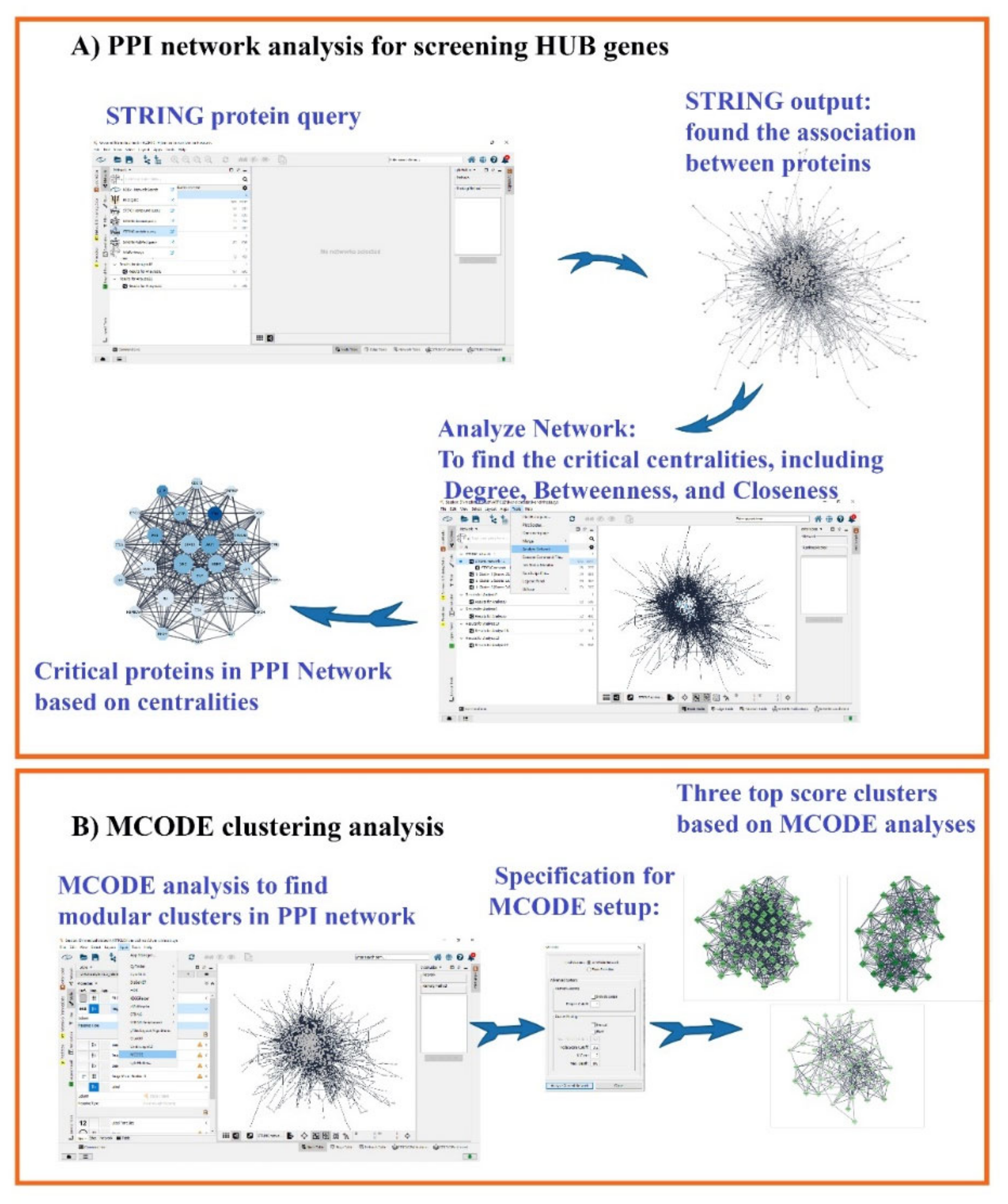
2.2. Protein–Protein Interaction (PPI) Network and MCODE Analysis of Proteins/Genes Related to Liver Cirrhosis
2.3. Curcumin and Target Search
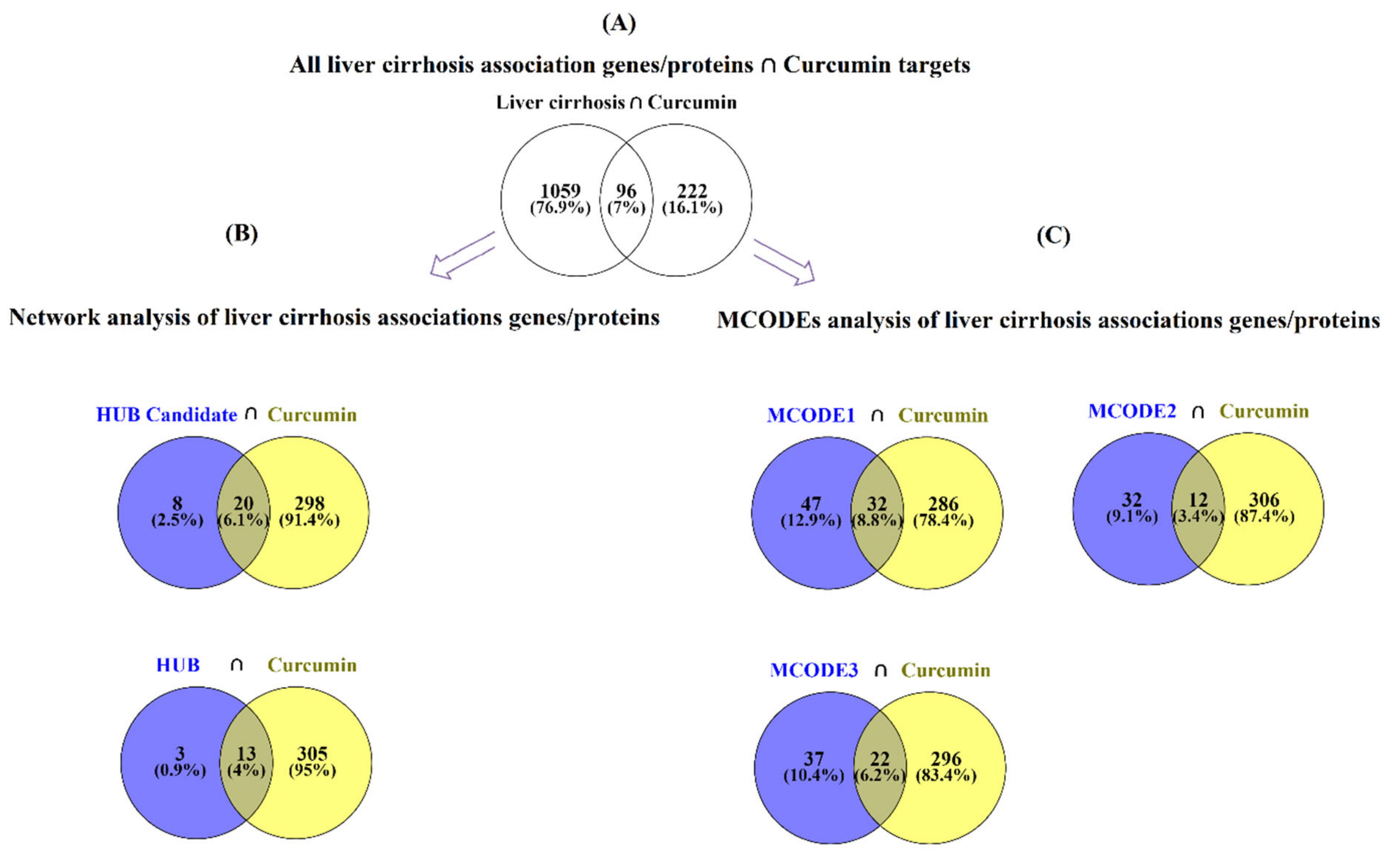
2.4. Estimation of Curcumin Targets with the Genes/Proteins Connected with LIVER Cirrhosis
- 1
- Evaluation of the curcumin targets with all achieved proteins/genes associated with liver cirrhosis (All liver cirrhosis connection genes ∩ curcumin targets).
- 2
- Evaluation of the curcumin targets with important proteins/genes obtained based on PPI network analysis (hub/candidate hub genes connection liver cirrhosis ∩ curcumin targets).
- 3
- Evaluation of the curcumin targets with important proteins/genes obtained based on MCODE analysis (MCODEs connection with liver cirrhosis ∩ curcumin targets). The intersection across these lists was conducted with online Venn diagram software (https://bioinfogp.cnb.csic.es/tools/venny; accessed on 24 April 2022).
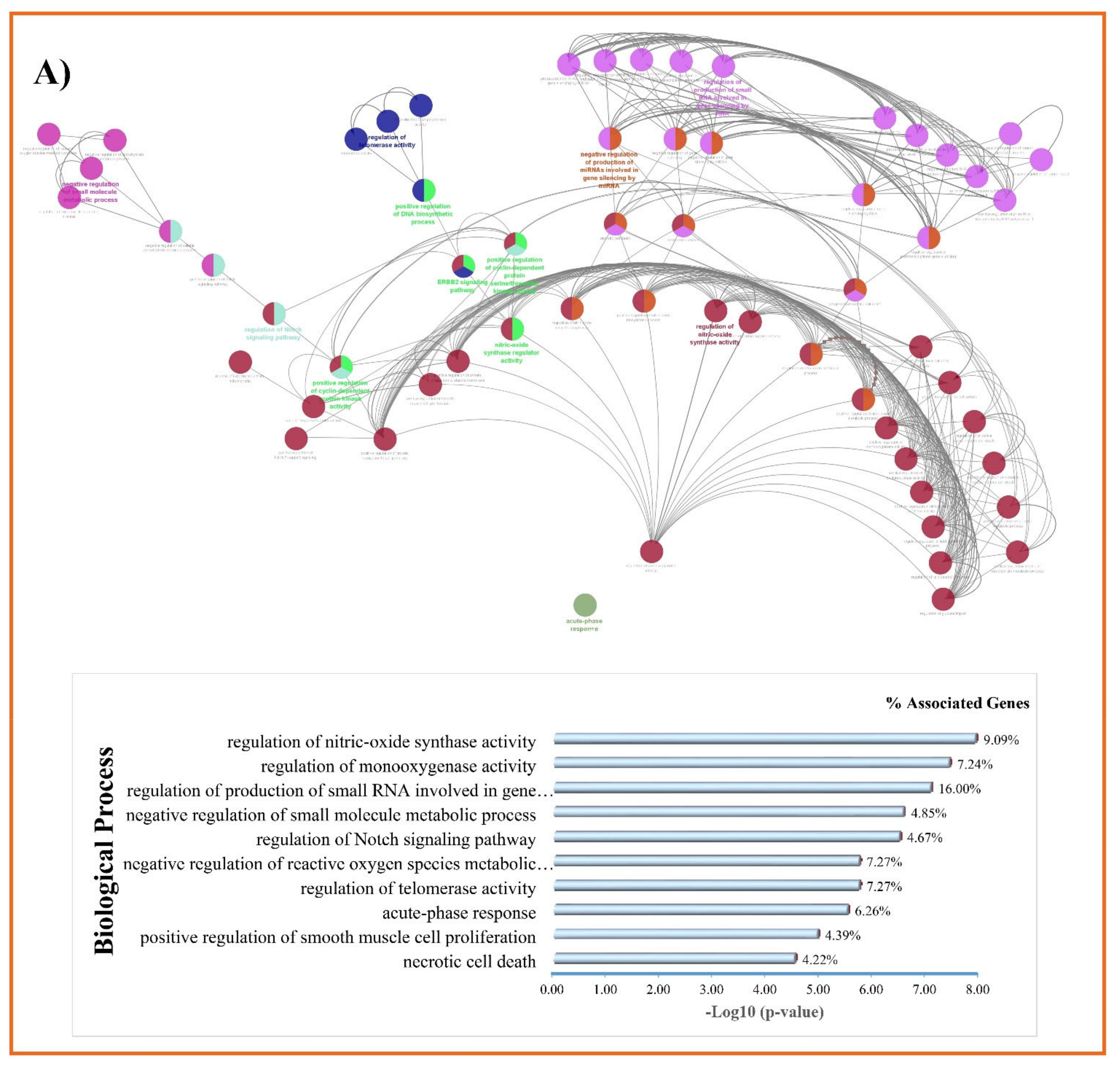
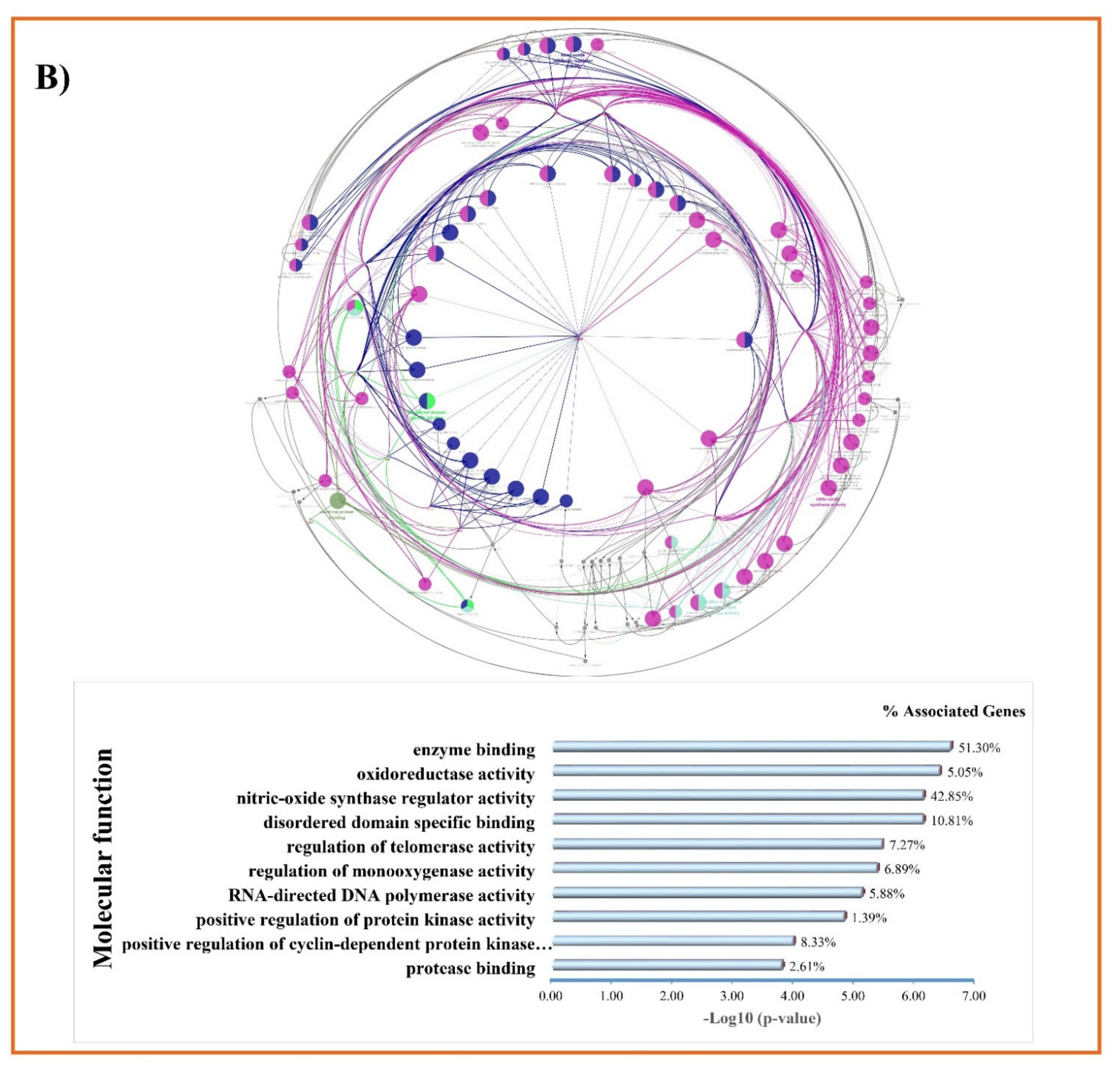
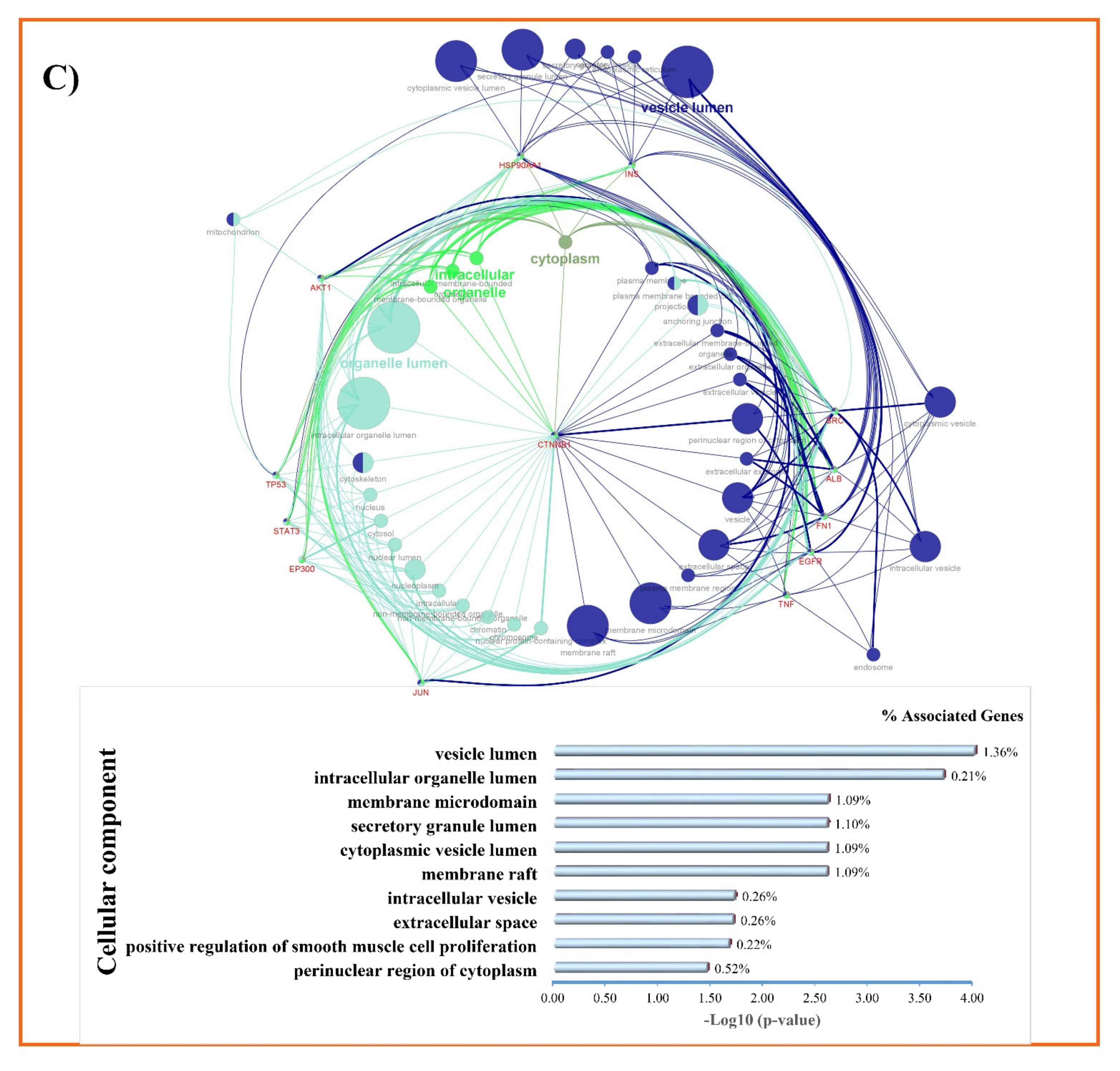
2.5. Gene Ontology and KEGG Pathways Enrichment Analysis
3. Results
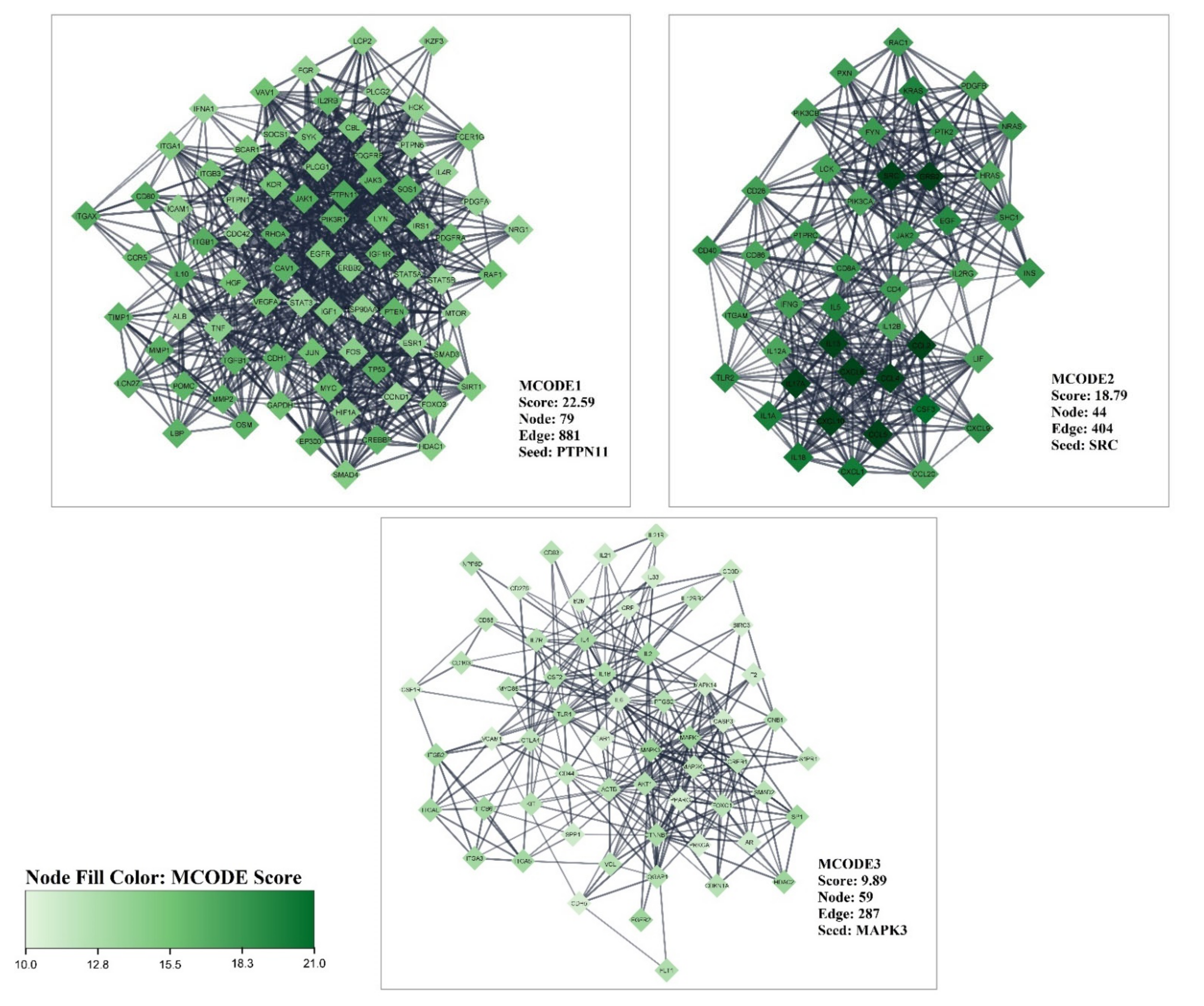
3.1. Gene Related to Liver Cirrhosis and PPI Network Analysis
3.2. Obtained Curcumin Targets and Exploring the Impact of Curcumin on Proteins/Genes Related to Liver Cirrhosis
3.3. GO and KEGG Enrichment Analyses of Shared Proteins
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Friedman, S.L. Mechanisms of hepatic fibrogenesis. Gastroenterology 2008, 134, 1655–1669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nouri-Vaskeh, M.; Malek Mahdavi, A. Effect of curcumin supplementation on disease severity in patients with liver cirrhosis: A randomized controlled trial. Phytother. Res. 2020, 34, 1446–1454. [Google Scholar] [CrossRef] [PubMed]
- Moon, A.M.; Singal, A.G.; Tapper, E.B. Contemporary Epidemiology of Chronic Liver Disease and Cirrhosis. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2020, 18, 2650–2666. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.R.; Machado, M.V. New Insights About Albumin and Liver Disease. Ann. Hepatol. 2018, 17, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Tahashi, Y.; Matsuzaki, K.; Date, M.; Yoshida, K.; Furukawa, F.; Sugano, Y.; Matsushita, M.; Himeno, Y.; Inagaki, Y.; Inoue, K. Differential regulation of TGF-beta signal in hepatic stellate cells between acute and chronic rat liver injury. Hepatology 2002, 35, 49–61. [Google Scholar] [CrossRef]
- Iredale, J.P. Hepatic stellate cell behavior during resolution of liver injury. Semin. Liver Dis. 2001, 21, 427–436. [Google Scholar] [CrossRef]
- Ueki, T.; Okamoto, E.; Takeuchi, M.; Fujimoto, J. Persectives on postgenome medicine: Gene therapy for liver cirrhosis. Nippon Rinsho 2001, 59, 152–156. [Google Scholar]
- Bataller, R.; North, K.E.; Brenner, D.A. Genetic polymorphisms and the progression of liver fibrosis: A critical appraisal. Hepatology 2003, 37, 493–503. [Google Scholar] [CrossRef]
- Carulli, L. Telomere shortening as genetic risk factor of liver cirrhosis. World J. Gastroenterol. 2015, 21, 379–383. [Google Scholar] [CrossRef] [Green Version]
- Scorza, M.; Elce, A.; Zarrilli, F.; Liguori, R.; Amato, F.; Castaldo, G. Genetic Diseases That Predispose to Early Liver Cirrhosis. Int. J. Hepatol. 2014, 2014, 713754. [Google Scholar] [CrossRef] [Green Version]
- Tessari, P. Protein metabolism in liver cirrhosis: From albumin to muscle myofibrils. Curr. Opin. Clin. Nutr. Metab. Care 2003, 6, 79–85. [Google Scholar] [CrossRef]
- Ginès, P.; Krag, A.; Abraldes, J.G.; Solà, E.; Fabrellas, N.; Kamath, P.S. Liver cirrhosis. Lancet (Lond. Engl.) 2021, 398, 1359–1376. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, Z.; Li, B.; Zhang, B.; Du, Y.; Liu, Y.; He, Y.; Chen, X. Curcumin attenuates renal interstitial fibrosis of obstructive nephropathy by suppressing epithelial-mesenchymal transition through inhibition of the TLR4/NF-kB and PI3K/AKT signalling pathways. Pharm. Biol. 2020, 58, 828–837. [Google Scholar] [CrossRef]
- Kong, D.; Zhang, Z.; Chen, L.; Huang, W.; Zhang, F.; Wang, L.; Wang, Y.; Cao, P.; Zheng, S. Curcumin blunts epithelial-mesenchymal transition of hepatocytes to alleviate hepatic fibrosis through regulating oxidative stress and autophagy. Redox Biol. 2020, 36, 101600. [Google Scholar] [CrossRef]
- Biswas, S.; Chen, S.; Liang, G.; Feng, B.; Cai, L.; Khan, Z.A.; Chakrabarti, S. Curcumin Analogs Reduce Stress and Inflammation Indices in Experimental Models of Diabetes. Front. Endocrinol. 2019, 10, 887. [Google Scholar] [CrossRef] [Green Version]
- Mahmoudi, A.; Atkin, S.L.; Nikiforov, N.G.; Sahebkar, A. Therapeutic Role of Curcumin in Diabetes: An Analysis Based on Bioinformatic Findings. Nutrients 2022, 14, 3244. [Google Scholar] [CrossRef]
- Mahmoudi, A.; Kesharwani, P.; Majeed, M.; Teng, Y.; Sahebkar, A. Recent advances in nanogold as a promising nanocarrier for curcumin delivery. Colloids Surf. B Biointerfaces 2022, 215, 112481. [Google Scholar] [CrossRef]
- Radbakhsh, S.; Momtazi-Borojeni, A.A.; Mahmoudi, A.; Sarborji, M.R.; Hatamipour, M.; Moallem, S.A.; Atkin, S.L.; Sahebkar, A. Investigation of the Effects of Difluorinated Curcumin on Glycemic Indices in Streptozotocin-Induced Diabetic Rats. In Natural Products and Human Diseases: Pharmacology, Molecular Targets, and Therapeutic Benefits; Sahebkar, A., Sathyapalan, T., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 131–141. [Google Scholar] [CrossRef]
- Rodrigues, F.C.; Anil Kumar, N.V.; Thakur, G. Developments in the anticancer activity of structurally modified curcumin: An up-to-date review. Eur. J. Med. Chem. 2019, 177, 76–104. [Google Scholar] [CrossRef]
- Shakeri, A.; Cicero, A.F.G.; Panahi, Y.; Mohajeri, M.; Sahebkar, A. Curcumin: A naturally occurring autophagy modulator. J. Cell. Physiol. 2019, 234, 5643–5654. [Google Scholar] [CrossRef]
- Bagherniya, M.; Nobili, V.; Blesso, C.N.; Sahebkar, A. Medicinal plants and bioactive natural compounds in the treatment of non-alcoholic fatty liver disease: A clinical review. Pharmacol. Res. 2018, 130, 213–240. [Google Scholar] [CrossRef]
- Farhood, B.; Mortezaee, K.; Goradel, N.H.; Khanlarkhani, N.; Salehi, E.; Nashtaei, M.S.; Najafi, M.; Sahebkar, A. Curcumin as an anti-inflammatory agent: Implications to radiotherapy and chemotherapy. J. Cell. Physiol. 2019, 234, 5728–5740. [Google Scholar] [CrossRef]
- Panahi, Y.; Khalili, N.; Sahebi, E.; Namazi, S.; Reiner, Ž.; Majeed, M.; Sahbekar, A. Curcuminoids modify lipid profile in type 2 diabetes mellitus: A randomized controlled trial. Complement. Ther. Med. 2017, 33, 1–5. [Google Scholar] [CrossRef]
- Parsamanesh, N.; Moossavi, M.; Bahrami, A.; Butler, A.E.; Sahebkar, A. Therapeutic potential of curcumin in diabetic complications. Pharmacol. Res. 2018, 136, 181–193. [Google Scholar] [CrossRef]
- Sahebkar, A. Molecular mechanisms for curcumin benefits against ischemic injury. Fertil. Steril. 2010, 94, e75–e76. [Google Scholar] [CrossRef]
- Afshari, A.R.; Jalili-Nik, M.; Abbasinezhad-Moud, F.; Javid, H.; Karimi, M.; Mollazadeh, H.; Jamialahmadi, T.; Sathyapalan, T.; Sahebkar, A. Anti-tumor effects of curcuminoids in glioblastoma multiforme: An updated literature review. Curr. Med. Chem. 2021, 28, 8116–8138. [Google Scholar] [CrossRef]
- Gorabi, A.M.; Kiaie, N.; Hajighasemi, S.; Jamialahmadi, T.; Majeed, M.; Sahebkar, A. The effect of curcumin on the differentiation of mesenchymal stem cells into mesodermal lineage. Molecules 2019, 24, 4029. [Google Scholar] [CrossRef] [Green Version]
- Heidari, Z.; Daei, M.; Boozari, M.; Jamialahmadi, T.; Sahebkar, A. Curcumin supplementation in pediatric patients: A systematic review of current clinical evidence. Phytother. Res. 2022, 36, 1442–1458. [Google Scholar] [CrossRef]
- Vahedian-Azimi, A.; Abbasifard, M.; Rahimi-Bashar, F.; Guest, P.C.; Majeed, M.; Mohammadi, A.; Banach, M.; Jamialahmadi, T.; Sahebkar, A. Effectiveness of Curcumin on Outcomes of Hospitalized COVID-19 Patients: A Systematic Review of Clinical Trials. Nutrients 2022, 14, 256. [Google Scholar] [CrossRef]
- Alidadi, M.; Jamialahmadi, T.; Cicero, A.F.G.; Bianconi, V.; Pirro, M.; Banach, M.; Sahebkar, A. The potential role of lant-derived natural products in improving arterial stiffness: A review of dietary intervention. Trends Food Sci. Technol. 2020, 99, 426–440. [Google Scholar] [CrossRef]
- Iranshahi, M.; Sahebkar, A.; Hosseini, S.T.; Takasaki, M.; Konoshima, T.; Tokuda, H. Cancer chemopreventive activity of diversin from Ferula diversivittata in vitro and in vivo. Phytomedicine 2010, 17, 269–273. [Google Scholar] [CrossRef]
- Iranshahi, M.; Sahebkar, A.; Takasaki, M.; Konoshima, T.; Tokuda, H. Cancer chemopreventive activity of the prenylated coumarin, umbelliprenin, in vivo. Eur. J. Cancer Prev. 2009, 18, 412–415. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, E.S.; El-Beih, N.M.; El-Hussieny, E.A.; El-Ahwany, E.; Hassan, M.; Zoheiry, M. Effects of free and nanoparticulate curcumin on chemically induced liver carcinoma in an animal model. Arch. Med. Sci. 2021, 17, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Fatima Zaidi, S.N.; Mahboob, T. Hepatoprotective role of curcumin in rat liver cirrhosis. Pak. J. Pharma. Sci. 2020, 33, 1519–1525. [Google Scholar] [CrossRef]
- Abo-Zaid, M.A.; Shaheen, E.S.; Ismail, A.H. Immunomodulatory effect of curcumin on hepatic cirrhosis in experimental rats. J. Food Biochem. 2020, 44, e13219. [Google Scholar] [CrossRef]
- Kyung, E.J.; Kim, H.B.; Hwang, E.S.; Lee, S.; Choi, B.K.; Kim, J.W.; Kim, H.J.; Lim, S.M.; Kwon, O.I.; Woo, E.J. Evaluation of hepatoprotective effect of curcumin on liver cirrhosis using a combination of biochemical analysis and magnetic resonance-based electrical conductivity imaging. Mediat. Inflamm. 2018, 2018, 5491797. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Lu, D.; Zou, Y.; Zhou, C.; Liu, H.; Tu, C.; Li, F.; Liu, L.; Zhang, S. Curcumin Protects Against Intestinal Origin Endotoxemia in Rat Liver Cirrhosis by Targeting PCSK9. J. Food. Sci. 2017, 82, 772–780. [Google Scholar] [CrossRef]
- Abd-Allah, G.A.; El-Bakry, K.A.; Bahnasawy, M.H.; El-Khodary, E.S.R. Protective effects of curcumin and ginger on liver cirrhosis induced by carbon tetrachloride in rats. Int. J. Pharmacol. 2016, 12, 361–369. [Google Scholar] [CrossRef] [Green Version]
- Nouri-Vaskeh, M.; Afshan, H.; Malek Mahdavi, A.; Alizadeh, L.; Fan, X.; Zarei, M. Curcumin ameliorates health-related quality of life in patients with liver cirrhosis: A randomized, double-blind placebo-controlled trial. Complement. Ther. Med. 2020, 49, 102351. [Google Scholar] [CrossRef]
- Shishodia, S. Molecular mechanisms of curcumin action: Gene expression. BioFactors 2013, 39, 37–55. [Google Scholar] [CrossRef]
- Mahmoudi, A.; Butler, A.E.; Majeed, M.; Banach, M.; Sahebkar, A. Investigation of the Effect of Curcumin on Protein Targets in NAFLD Using Bioinformatic Analysis. Nutrients 2022, 14, 1331. [Google Scholar] [CrossRef]
- Mahmoudi, A.; Butler, A.E.; Jamialahmadi, T.; Sahebkar, A. Target Deconvolution of Fenofibrate in Nonalcoholic Fatty Liver Disease Using Bioinformatics Analysis. Biomed. Res. Int. 2021, 2021, 3654660. [Google Scholar] [CrossRef]
- Likić, V.A.; McConville, M.J.; Lithgow, T.; Bacic, A. Systems biology: The next frontier for bioinformatics. Adv. Bioinform. 2010, 2010, 268925. [Google Scholar] [CrossRef] [Green Version]
- Mahmoudi, A.; Heydari, S.; Markina, Y.V.; Barreto, G.E.; Sahebkar, A. Role of statins in regulating molecular pathways following traumatic brain injury: A system pharmacology study. Biomed. Pharmacother. 2022, 153, 113304. [Google Scholar] [CrossRef]
- Amberger, J.S.; Bocchini, C.A.; Schiettecatte, F.; Scott, A.F.; Hamosh, A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015, 43, D789–D798. [Google Scholar] [CrossRef] [Green Version]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; King, B.L.; McMorran, R.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. The Comparative Toxicogenomics Database: Update 2017. Nucleic Acids Res. 2017, 45, D972–D978. [Google Scholar] [CrossRef] [Green Version]
- Pletscher-Frankild, S.; Pallejà, A.; Tsafou, K.; Binder, J.X.; Jensen, L.J. DISEASES: Text mining and data integration of disease–gene associations. Methods 2015, 74, 83–89. [Google Scholar] [CrossRef]
- Nguyen, D.T.; Mathias, S.; Bologa, C.; Brunak, S.; Fernandez, N.; Gaulton, A.; Hersey, A.; Holmes, J.; Jensen, L.J.; Karlsson, A.; et al. Pharos: Collating protein information to shed light on the druggable genome. Nucleic Acids Res. 2017, 45, D995–D1002. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Santos, A.; von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef]
- Freshour, S.L.; Kiwala, S.; Cotto, K.C.; Coffman, A.C.; McMichael, J.F.; Song, J.J.; Griffith, M.; Griffith, O.L.; Wagner, A.H. Integration of the Drug-Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021, 49, D1144–D1151. [Google Scholar] [CrossRef]
- Gfeller, D.; Grosdidier, A.; Wirth, M.; Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014, 42, W32–W38. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive. Methods Mol. Biol. 2017, 1607, 627–641. [Google Scholar] [CrossRef] [Green Version]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Yin, B.; Bi, Y.M.; Fan, G.J. Molecular Mechanism of the Effect of Huanglian Jiedu Decoction on Type 2 Diabetes Mellitus Based on Network Pharmacology and Molecular Docking. J. Diabetes Res. 2020, 2020, 5273914. [Google Scholar] [CrossRef]
- Cheng, Y.; Liu, P.; Hou, T.L.; Maimaitisidike, M.; Ababaikeli, R.; Abudureyimu, A. Mechanisms of Huangqi Decoction Granules (黄芪汤颗粒剂) on Hepatitis B Cirrhosis Patients Based on RNA-Sequencing. Chin. J. Integr. Med. 2019, 25, 507–514. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [Green Version]
- Palasca, O.; Santos, A.; Stolte, C.; Gorodkin, J.; Jensen, L.J. TISSUES 2.0: An integrative web resource on mammalian tissue expression. Database 2018, 2018, bay003. [Google Scholar] [CrossRef]
- Baghy, K.; Iozzo, R.V.; Kovalszky, I. Decorin-TGFβ axis in hepatic fibrosis and cirrhosis. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2012, 60, 262–268. [Google Scholar] [CrossRef]
- Kershenobich Stalnikowitz, D.; Weissbrod, A.B. Liver fibrosis and inflammation. A review. Ann. Hepatol. 2003, 2, 159–163. [Google Scholar] [CrossRef]
- Kovalszky, I.; Dudás, J.; Gallai, M.; Hollósi, P.; Tátrai, P.; Tátrai, E.; Schaff, Z. Proteoglycans in the liver. Magy. Onkol. 2004, 48, 207–213. [Google Scholar] [PubMed]
- Tai, C.J.; Choong, C.Y.; Lin, Y.C.; Shi, Y.C.; Tai, C.J. The anti-hepatic fibrosis activity of ergosterol depended on upregulation of PPARgamma in HSC-T6 cells. Food Funct. 2016, 7, 1915–1923. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.-K.; Yu, L.; Cheng, M.-L.; Che, P.; Lu, Y.-Y.; Zhang, Q.; Mu, M.; Li, H.; Zhu, L.-L.; Zhu, J.-J.; et al. Focal Adhesion Kinase Regulates Hepatic Stellate Cell Activation and Liver Fibrosis. Sci. Rep. 2017, 7, 4032. [Google Scholar] [CrossRef] [PubMed]
- Qiao, M.; Yang, J.; Zhu, Y.; Zhao, Y.; Hu, J. Transcriptomics and proteomics analysis of system-level mechanisms in the liver of apigenin-treated fibrotic rats. Life Sci. 2020, 248, 117475. [Google Scholar] [CrossRef]
- Gressner, A.M.; Krull, N.; Bachem, M.G. Regulation of proteoglycan expression in fibrotic liver and cultured fat-storing cells. Pathol. Res. Pract. 1994, 190, 864–882. [Google Scholar] [CrossRef]
- Biernacka, A.; Dobaczewski, M.; Frangogiannis, N.G. TGF-β signaling in fibrosis. Growth Factors 2011, 29, 196–202. [Google Scholar] [CrossRef] [Green Version]
- Nault, J.C.; Guyot, E.; Laguillier, C.; Chevret, S.; Ganne-Carrie, N.; N’Kontchou, G.; Beaugrand, M.; Seror, O.; Trinchet, J.C.; Coelho, J.; et al. Serum proteoglycans as prognostic biomarkers of hepatocellular carcinoma in patients with alcoholic cirrhosis. Cancer Epidemiol. Biomark. Prev. 2013, 22, 1343–1352. [Google Scholar] [CrossRef] [Green Version]
- Kovalszky, I.; Nagy, P.; Szende, B.; Lapis, K.; Szalay, F.; Jeney, A.; Schaff, Z. Experimental and human liver fibrogenesis. Scand. J. Gastroenterol. Suppl. 1998, 33, 51–55. [Google Scholar] [CrossRef]
- Chin, K.Y. The spice for joint inflammation: Anti-inflammatory role of curcumin in treating osteoarthritis. Drug Des. Dev. Ther. 2016, 10, 3029–3042. [Google Scholar] [CrossRef]
- Buhrmann, C.; Mobasheri, A.; Matis, U.; Shakibaei, M. Curcumin mediated suppression of nuclear factor-κB promotes chondrogenic differentiation of mesenchymal stem cells in a high-density co-culture microenvironment. Arthritis Res. Ther. 2010, 12, R127. [Google Scholar] [CrossRef] [Green Version]
- Hsu, W.H.; Lee, B.H.; Hsu, Y.W.; Pan, T.M. Peroxisome proliferator-activated receptor-γ activators monascin and rosiglitazone attenuate carboxymethyllysine-induced fibrosis in hepatic stellate cells through regulating the oxidative stress pathway but independent of the receptor for advanced glycation end products signaling. J. Agric. Food Chem. 2013, 61, 6873–6879. [Google Scholar] [CrossRef]
- Smedsrød, B.; Melkko, J.; Araki, N.; Sano, H.; Horiuchi, S. Advanced glycation end products are eliminated by scavenger-receptor-mediated endocytosis in hepatic sinusoidal Kupffer and endothelial cells. Biochem. J. 1997, 322 Pt 2, 567–573. [Google Scholar] [CrossRef] [Green Version]
- He, Y.L.; Zhu, J.Q.; Huang, Y.Q.; Gao, H.; Zhao, Y.R. Advanced glycation end product (AGE)-induced hepatic stellate cell activation via autophagy contributes to hepatitis C-related fibrosis. Acta Diabetol. 2015, 52, 959–969. [Google Scholar] [CrossRef]
- Lin, J.; Tang, Y.; Kang, Q.; Feng, Y.; Chen, A. Curcumin inhibits gene expression of receptor for advanced glycation end-products (RAGE) in hepatic stellate cells in vitro by elevating PPARγ activity and attenuating oxidative stress. Br. J. Pharmacol. 2012, 166, 2212–2227. [Google Scholar] [CrossRef] [Green Version]
- Hazra, S.; Xiong, S.; Wang, J.; Rippe, R.A.; Krishna, V.; Chatterjee, K.; Tsukamoto, H. Peroxisome proliferator-activated receptor gamma induces a phenotypic switch from activated to quiescent hepatic stellate cells. J. Biol. Chem. 2004, 279, 11392–11401. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Chen, A. Curcumin eliminates the effect of advanced glycation end-products (AGEs) on the divergent regulation of gene expression of receptors of AGEs by interrupting leptin signaling. Lab. Investig. J. Tech. Methods Pathol. 2014, 94, 503–516. [Google Scholar] [CrossRef] [Green Version]
- Xie, T.; Chen, X.; Chen, W.; Huang, S.; Peng, X.; Tian, L.; Wu, X.; Huang, Y. Curcumin is a Potential Adjuvant to Alleviates Diabetic Retinal Injury via Reducing Oxidative Stress and Maintaining Nrf2 Pathway Homeostasis. Front. Pharmacol. 2021, 12, 796565. [Google Scholar] [CrossRef]
- Leube, R.E.; Moch, M.; Windoffer, R. Intermediate filaments and the regulation of focal adhesion. Curr. Opin. Cell Biol. 2015, 32, 13–20. [Google Scholar] [CrossRef]
- Mishra, Y.G.; Manavathi, B. Focal adhesion dynamics in cellular function and disease. Cell. Signal. 2021, 85, 110046. [Google Scholar] [CrossRef]
- Paluch, E.K.; Aspalter, I.M.; Sixt, M. Focal Adhesion-Independent Cell Migration. Annu. Rev. Cell Dev. Biol. 2016, 32, 469–490. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.; Lieberthal, T.J.; Zhou, V.X.; Lopez-Ichikawa, M.; Armas-Phan, M.; Bond, T.K.; Yoshida, M.C.; Choi, W.T.; Chang, T.T. Liver epithelial focal adhesion kinase modulates fibrogenesis and hedgehog signaling. JCI Insight 2020, 5, e141217. [Google Scholar] [CrossRef] [PubMed]
- Sathe, G.; Pinto, S.M.; Syed, N.; Nanjappa, V.; Solanki, H.S.; Renuse, S.; Chavan, S.; Khan, A.A.; Patil, A.H.; Nirujogi, R.S.; et al. Phosphotyrosine profiling of curcumin-induced signaling. Clin. Proteom. 2016, 13, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ray, S.; Chattopadhyay, N.; Mitra, A.; Siddiqi, M.; Chatterjee, A. Curcumin exhibits antimetastatic properties by modulating integrin receptors, collagenase activity, and expression of Nm23 and E-cadherin. J. Environ. Pathol. Toxicol. Oncol. Off. Organ Int. Soc. Environ. Toxicol. Cancer 2003, 22, 49–58. [Google Scholar]
- Mitra, A.; Chakrabarti, J.; Banerji, A.; Chatterjee, A.; Das, B.R. Curcumin, a potential inhibitor of MMP-2 in human laryngeal squamous carcinoma cells HEp2. J. Environ. Pathol. Toxicol. Oncol. 2006, 25, 679–689. [Google Scholar] [CrossRef]
- Gressner, O.A.; Weiskirchen, R.; Gressner, A.M. Evolving concepts of liver fibrogenesis provide new diagnostic and therapeutic options. Comp. Hepatol. 2007, 6, 7. [Google Scholar] [CrossRef] [Green Version]
- Pugh, C.W.; Ratcliffe, P.J. Regulation of angiogenesis by hypoxia: Role of the HIF system. Nat. Med. 2003, 9, 677–684. [Google Scholar] [CrossRef]
- Rankin, E.B.; Giaccia, A.J. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008, 15, 678–685. [Google Scholar] [CrossRef] [Green Version]
- Copple, B.L.; Bai, S.; Burgoon, L.D.; Moon, J.O. Hypoxia-inducible factor-1α regulates the expression of genes in hypoxic hepatic stellate cells important for collagen deposition and angiogenesis. Liver Int. Off. J. Int. Assoc. Study Liver 2011, 31, 230–244. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Huang, Y.; Guan, F.; Xiao, Y.; Deng, J.; Chen, H.; Chen, X.; Li, J.; Huang, H.; Shi, C. Hypoxia-inducible factor-1alpha and MAPK co-regulate activation of hepatic stellate cells upon hypoxia stimulation. PLoS ONE 2013, 8, e74051. [Google Scholar] [CrossRef] [Green Version]
- Moczydlowska, J.; Miltyk, W.; Hermanowicz, A.; Lebensztejn, D.M.; Palka, J.A.; Debek, W. HIF-1 α as a Key Factor in Bile Duct Ligation-Induced Liver Fibrosis in Rats. J. Investig. Surg. 2017, 30, 41–46. [Google Scholar] [CrossRef]
- Nath, B.; Szabo, G. Hypoxia and hypoxia inducible factors: Diverse roles in liver diseases. Hepatology 2012, 55, 622–633. [Google Scholar] [CrossRef] [Green Version]
- Tu, W.; Ye, J.; Wang, Z.J. Embryonic liver fordin is involved in glucose glycolysis of hepatic stellate cell by regulating PI3K/Akt signaling. World J. Gastroenterol. 2016, 22, 8519–8527. [Google Scholar] [CrossRef]
- Deng, J.; Huang, Q.; Wang, Y.; Shen, P.; Guan, F.; Li, J.; Huang, H.; Shi, C. Hypoxia-inducible factor-1alpha regulates autophagy to activate hepatic stellate cells. Biochem. Biophys. Res. Commun. 2014, 454, 328–334. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, Y.J.; Song, C.H.; Ahn, Y.K.; Han, H.J. Role of FAK phosphorylation in hypoxia-induced hMSCS migration: Involvement of VEGF as well as MAPKS and eNOS pathways. Am. J. Physiology. Cell Physiol. 2010, 298, C847–C856. [Google Scholar] [CrossRef]
- Alisi, A.; Arciello, M.; Petrini, S.; Conti, B.; Missale, G.; Balsano, C. Focal adhesion kinase (FAK) mediates the induction of pro-oncogenic and fibrogenic phenotypes in hepatitis C virus (HCV)-infected cells. PLoS ONE 2012, 7, e44147. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, B.; Shi, Q.; Wang, X.; Wang, D.; Zhu, L. Brusatol inhibits HIF-1 signaling pathway and suppresses glucose uptake under hypoxic conditions in HCT116 cells. Sci. Rep. 2016, 6, 39123. [Google Scholar] [CrossRef] [Green Version]
- Rahban, M.; Habibi-Rezaei, M.; Mazaheri, M.; Saso, L.; Moosavi-Movahedi, A.A. Anti-Viral Potential and Modulation of Nrf2 by Curcumin: Pharmacological Implications. Antioxidants 2020, 9, 1228. [Google Scholar] [CrossRef]
- Hasima, N.; Aggarwal, B.B. Targeting proteasomal pathways by dietary curcumin for cancer prevention and treatment. Curr. Med. Chem. 2014, 21, 1583–1594. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.; Yan, Y.; Jia, P.; Yang, K.; Guo, C.; Chen, H.; Qi, J.; Qian, N.; Xu, X.; Wang, F.; et al. Desferrioxamine reduces ultrahigh-molecular-weight polyethylene-induced osteolysis by restraining inflammatory osteoclastogenesis via heme oxygenase-1. Cell Death Dis. 2016, 7, e2435. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, Y.; Zou, J.; Yan, L.; Du, W.; Zhang, Y.; Sun, H.; Lu, P.; Geng, S.; Gu, R.; et al. Tetrahydrocurcumin induces mesenchymal-epithelial transition and suppresses angiogenesis by targeting HIF-1α and autophagy in human osteosarcoma. Oncotarget 2017, 8, 91134–91149. [Google Scholar] [CrossRef]
- Seo, H.Y.; Lee, S.H.; Lee, J.H.; Kang, Y.N.; Hwang, J.S.; Park, K.G.; Kim, M.K.; Jang, B.K. Src Inhibition Attenuates Liver Fibrosis by Preventing Hepatic Stellate Cell Activation and Decreasing Connetive Tissue Growth Factor. Cells 2020, 9, 558. [Google Scholar] [CrossRef] [Green Version]
- Shao, W.H.; Cohen, P.L. The role of tyrosine kinases in systemic lupus erythematosus and their potential as therapeutic targets. Expert Rev. Clin. Immunol. 2014, 10, 573–582. [Google Scholar] [CrossRef]
- Zhao, P.-W.; Zhang, J.-W.; Liu, Y.; Liu, Y.; Liu, J.-W.; Huang, J.-Z. SRC-1 and Twist1 are prognostic indicators of liver cancer and are associated with cell viability, invasion, migration and epithelial-mesenchymal transformation of hepatocellular carcinoma cells. Transl Cancer Res 2020, 9, 603–612. [Google Scholar] [CrossRef]
- Masaki, T.; Tokuda, M.; Ohnishi, M.; Itano, T.; Matsui, H.; Watanabe, S.; Arima, K.; Kohno, K.; Maeba, T.; Tai, Y.; et al. Increases in src-related protein tyrosine kinases in human liver cirrhosis and hepatocellular carcinoma. Int. Hepatol. Commun. 1995, 4, 54–60. [Google Scholar] [CrossRef]
- Ding, Q.; Tian, X.G.; Li, Y.; Wang, Q.Z.; Zhang, C.Q. Carvedilol may attenuate liver cirrhosis by inhibiting angiogenesis through the VEGF-Src-ERK signaling pathway. World J. Gastroenterol. 2015, 21, 9566–9576. [Google Scholar] [CrossRef] [PubMed]
- Leu, T.H.; Su, S.L.; Chuang, Y.C.; Maa, M.C. Direct inhibitory effect of curcumin on Src and focal adhesion kinase activity. Biochem. Pharmacol. 2003, 66, 2323–2331. [Google Scholar] [CrossRef]
- Yang, W.H.; Kuo, M.Y.; Liu, C.M.; Deng, Y.T.; Chang, H.H.; Chang, J.Z. Curcumin inhibits TGFβ1-induced CCN2 via Src, JNK, and Smad3 in gingiva. J. Dent. Res. 2013, 92, 629–634. [Google Scholar] [CrossRef]
- Saini, S.; Arora, S.; Majid, S.; Shahryari, V.; Chen, Y.; Deng, G.; Yamamura, S.; Ueno, K.; Dahiya, R. Curcumin modulates microRNA-203-mediated regulation of the Src-Akt axis in bladder cancer. Cancer Prev. Res. (Phila) 2011, 4, 1698–1709. [Google Scholar] [CrossRef] [Green Version]
- Dillon, R.L.; White, D.E.; Muller, W.J. The phosphatidyl inositol 3-kinase signaling network: Implications for human breast cancer. Oncogene 2007, 26, 1338–1345. [Google Scholar] [CrossRef]
- Reyes-Gordillo, K.; Shah, R.; Arellanes-Robledo, J.; Cheng, Y.; Ibrahim, J.; Tuma, P.L. Akt1 and Akt2 Isoforms Play Distinct Roles in Regulating the Development of Inflammation and Fibrosis Associated with Alcoholic Liver Disease. Cells 2019, 8, 1337. [Google Scholar] [CrossRef] [PubMed]
- Fortier, A.M.; Cadrin, M. Simple epithelial keratins K8 and K18: From structural to regulatory protein. In Keratin: Structure, Properties and Applications; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2012; pp. 1–36. [Google Scholar]
- Coant, N.; Simon-Rudler, M.; Gustot, T.; Fasseu, M.; Gandoura, S.; Ragot, K.; Abdel-Razek, W.; Thabut, D.; Lettéron, P.; Ogier-Denis, E.; et al. Glycogen synthase kinase 3 involvement in the excessive proinflammatory response to LPS in patients with decompensated cirrhosis. J. Hepatol. 2011, 55, 784–793. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.Z.; Deng, G.; Liu, X.Y.; Luo, C.Q. Akt gene therapy for cirrhotic rats with portal hypertension. J. Cent. South Univ. Med. Sci. 2008, 33, 31–37. [Google Scholar]
- Hanikoglu, A.; Kucuksayan, E.; Hanikoglu, F.; Ozben, T.; Menounou, G.; Sansone, A.; Chatgilialoglu, C.; Di Bella, G.; Ferreri, C. Effects of somatostatin, curcumin, and quercetin on the fatty acid profile of breast cancer cell membranes. Can. J. Physiol. Pharmacol. 2020, 98, 131–138. [Google Scholar] [CrossRef]
- Doukas, S.G.; Doukas, P.G.; Sasaki, C.T.; Vageli, D. The in vivo preventive and therapeutic properties of curcumin in bile reflux-related oncogenesis of the hypopharynx. J. Cell. Mol. Med. 2020, 24, 10311–10321. [Google Scholar] [CrossRef]
- Tian, B.; Zhao, Y.; Liang, T.; Ye, X.; Li, Z.; Yan, D.; Fu, Q.; Li, Y. Curcumin inhibits urothelial tumor development by suppressing IGF2 and IGF2-mediated PI3K/AKT/mTOR signaling pathway. J. Drug Target. 2017, 25, 626–636. [Google Scholar] [CrossRef]
- Han, X.; Yang, C.; Guo, C.; Xu, Y.; Liu, X.; Xie, R.; Meng, X.; Cheng, Z.; Fu, X. Bioinformatics Analysis to Screen Key Targets of Curcumin against Colorectal Cancer and the Correlation with Tumor-Infiltrating Immune Cells. Evid.-Based Complement. Altern. Med. 2021, 2021, 9132608. [Google Scholar] [CrossRef]
- Komposch, K.; Sibilia, M. EGFR signaling in liver diseases. Int. J. Mol. Sci. 2016, 17, 30. [Google Scholar] [CrossRef]
- Fuchs, B.C.; Hoshida, Y.; Fujii, T.; Wei, L.; Yamada, S.; Lauwers, G.Y.; McGinn, C.M.; DePeralta, D.K.; Chen, X.; Kuroda, T.; et al. Epidermal growth factor receptor inhibition attenuates liver fibrosis and development of hepatocellular carcinoma. Hepatology 2014, 59, 1577–1590. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Kim, J.Y.; Park, S.Y.; Jeong, W.T.; Kim, J.M.; Bae, S.H.; Kim, G.J. Activation of the EGFR-PI3K-CaM pathway by PRL-1-overexpressing placenta-derived mesenchymal stem cells ameliorates liver cirrhosis via ER stress-dependent calcium. Stem Cell Res. Ther. 2021, 12, 551. [Google Scholar] [CrossRef]
- Liang, D.; Chen, H.; Zhao, L.; Zhang, W.; Hu, J.; Liu, Z.; Zhong, P.; Wang, W.; Wang, J.; Liang, G. Inhibition of EGFR attenuates fibrosis and stellate cell activation in diet-induced model of nonalcoholic fatty liver disease. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 133–142. [Google Scholar] [CrossRef]
- Jiang, Y.X.; Yu, W.J.; Lin, D.L.; Li, H.; Li, Y.J. Significance of expression of EGFR, Connexin43 and E-cadherin in primary hepatocellular carcinoma. World Chin. J. Dig. 2013, 21, 2185–2191. [Google Scholar] [CrossRef]
- Liang, Y.; Zhao, J.; Zou, H.; Zhang, J.; Zhang, T. In vitro and in silico evaluation of EGFR targeting activities of curcumin and its derivatives. Food. Funct. 2021, 12, 10667–10675. [Google Scholar] [CrossRef]
- Xu, Y.Y.; Cao, Y.; Ma, H.; Li, H.Q.; Ao, G.Z. Design, synthesis and molecular docking of α,β-unsaturated cyclohexanone analogous of curcumin as potent EGFR inhibitors with antiproliferative activity. Bioorg. Med. Chem. 2013, 21, 388–394. [Google Scholar] [CrossRef]
- Yadav, I.S.; Nandekar, P.P.; Shrivastava, S.; Sangamwar, A.; Chaudhury, A.; Agarwal, S.M. Ensemble docking and molecular dynamics identify knoevenagel curcumin derivatives with potent anti-EGFR activity. Gene 2014, 539, 82–90. [Google Scholar] [CrossRef]
- Shaik, N.A.; Al-Kreathy, H.M.; Ajabnoor, G.M.; Verma, P.K.; Banaganapalli, B. Molecular designing, virtual screening and docking study of novel curcumin analogue as mutation (S769L and K846R) selective inhibitor for EGFR. Saudi J. Biol. Sci. 2019, 26, 439–448. [Google Scholar] [CrossRef]
- Lee, J.Y.; Lee, Y.M.; Chang, G.C.; Yu, S.L.; Hsieh, W.Y.; Chen, J.J.; Chen, H.W.; Yang, P.C. Curcumin induces EGFR degradation in lung adenocarcinoma and modulates p38 activation in intestine: The versatile adjuvant for gefitinib therapy. PLoS ONE 2011, 6, e23756. [Google Scholar] [CrossRef] [Green Version]
- Chiu, Y.-J.; Yang, J.-S.; Tsai, F.-J.; Chiu, H.-Y.; Juan, Y.-N.; Lo, Y.-H.; Chiang, J.-H. Curcumin suppresses cell proliferation and triggers apoptosis in vemurafenib-resistant melanoma cells by downregulating the EGFR signaling pathway. Environ. Toxicol. 2022, 37, 868–879. [Google Scholar] [CrossRef]
- Zhen, L.; Fan, D.; Yi, X.; Cao, X.; Chen, D.; Wang, L. Curcumin inhibits oral squamous cell carcinoma proliferation and invasion via EGFR signaling pathways. Int. J. Clin. Exp. Pathol. 2014, 7, 6438–6446. [Google Scholar]
- Wada, K.; Lee, J.-Y.; Hung, H.-Y.; Shi, Q.; Lin, L.; Zhao, Y.; Goto, M.; Yang, P.-C.; Kuo, S.-C.; Chen, H.-W.; et al. Novel curcumin analogs to overcome EGFR-TKI lung adenocarcinoma drug resistance and reduce EGFR-TKI-induced GI adverse effects. Bioorg. Med. Chem. 2015, 23, 1507–1514. [Google Scholar] [CrossRef] [Green Version]
- Qiu, P.; Xu, L.; Gao, L.; Zhang, M.; Wang, S.; Tong, S.; Sun, Y.; Zhang, L.; Jiang, T. Exploring pyrimidine-substituted curcumin analogues: Design, synthesis and effects on EGFR signaling. Bioorg. Med. Chem. 2013, 21, 5012–5020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Qi, Y.-F.; Yu, Y.-R. STAT3: A key regulator in liver fibrosis. Ann. Hepatol. 2021, 21, 100224. [Google Scholar] [CrossRef] [PubMed]
- Horiguchi, N.; Ishac, E.J.N.; Gao, B. Liver regeneration is suppressed in alcoholic cirrhosis: Correlation with decreased STAT3 activation. Alcohol 2007, 41, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Stärkel, P.; De Saeger, C.; Leclercq, I.; Strain, A.; Horsmans, Y. Deficient Stat3 DNA-binding is associated with high Pias3 expression and a positive anti-apoptotic balance in human end-stage alcoholic and hepatitis C cirrhosis. J. Hepatol. 2005, 43, 687–695. [Google Scholar] [CrossRef]
- Stärkel, P.; Bishop, K.; Horsmans, Y.; Straint, A.J. Expression and DNA-binding activity of signal transducer and activator of transcription 3 in alcoholic cirrhosis compared to normal liver and primary biliary cirrhosis in humans. Am. J. Pathol. 2003, 162, 587–596. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.; Jung, H.J.; Kim, M.W.; Kang, J.H.; Shin, D.; Jang, Y.S.; Yoon, Y.S.; Oh, S.H. A novel STAT3 inhibitor, STX-0119, attenuates liver fibrosis by inactivating hepatic stellate cells in mice. Biochem. Biophys. Res. Commun. 2019, 513, 49–55. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.; Zeng, S.; Zhang, X.; Zhao, J.; Zhang, X.; Chen, X.; Yang, W.; Yang, Y.; Dong, Z.; et al. The natural polyphenol curcumin induces apoptosis by suppressing STAT3 signaling in esophageal squamous cell carcinoma 06 Biological Sciences 0601 Biochemistry and Cell Biology 11 Medical and Health Sciences 1112 Oncology and Carcinogenesis. J. Exp. Clin. Cancer Res. 2018, 37, 303. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Liu, Y.L.; Liu, G.X.; Chen, X.; Yang, K.; Yang, Y.X.; Xie, Q.; Gan, H.K.; Huang, X.L.; Gan, H.T. Curcumin ameliorates dextran sulfate sodium-induced experimental colitis by blocking STAT3 signaling pathway. Int. Immunopharmacol. 2013, 17, 314–320. [Google Scholar] [CrossRef]
- Mahata, S.; Behera, S.K.; Kumar, S.; Sahoo, P.K.; Sarkar, S.; Fazil, M.H.U.T.; Nasare, V.D. In-silico and in-vitro investigation of STAT3-PIM1 heterodimeric complex: Its mechanism and inhibition by curcumin for cancer therapeutics. Int. J. Biol. Macromol. 2022, 208, 356–366. [Google Scholar] [CrossRef]
- Hahn, Y.I.; Kim, S.J.; Choi, B.Y.; Cho, K.C.; Bandu, R.; Kim, K.P.; Kim, D.H.; Kim, W.; Park, J.S.; Han, B.W.; et al. Curcumin interacts directly with the Cysteine 259 residue of STAT3 and induces apoptosis in H-Ras transformed human mammary epithelial cells. Sci. Rep. 2018, 8, 6409. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Li, N.; Grivennikov, S.I.; Karin, M. The unholy trinity: Inflammation, cytokines, and STAT3 shape the cancer microenvironment. Cancer cell 2011, 19, 429–431. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.Q.; Ahmed, E.I.; Elareer, N.; Fathima, H.; Prabhu, K.S.; Siveen, K.S.; Kulinski, M.; Azizi, F.; Dermime, S.; Ahmad, A.; et al. Curcumin-mediated apoptotic cell death in papillary thyroid cancer and cancer stem-like cells through targeting of the JAK/STAT3 signaling pathway. Int. J. Mol. Sci. 2020, 21, 438. [Google Scholar] [CrossRef]
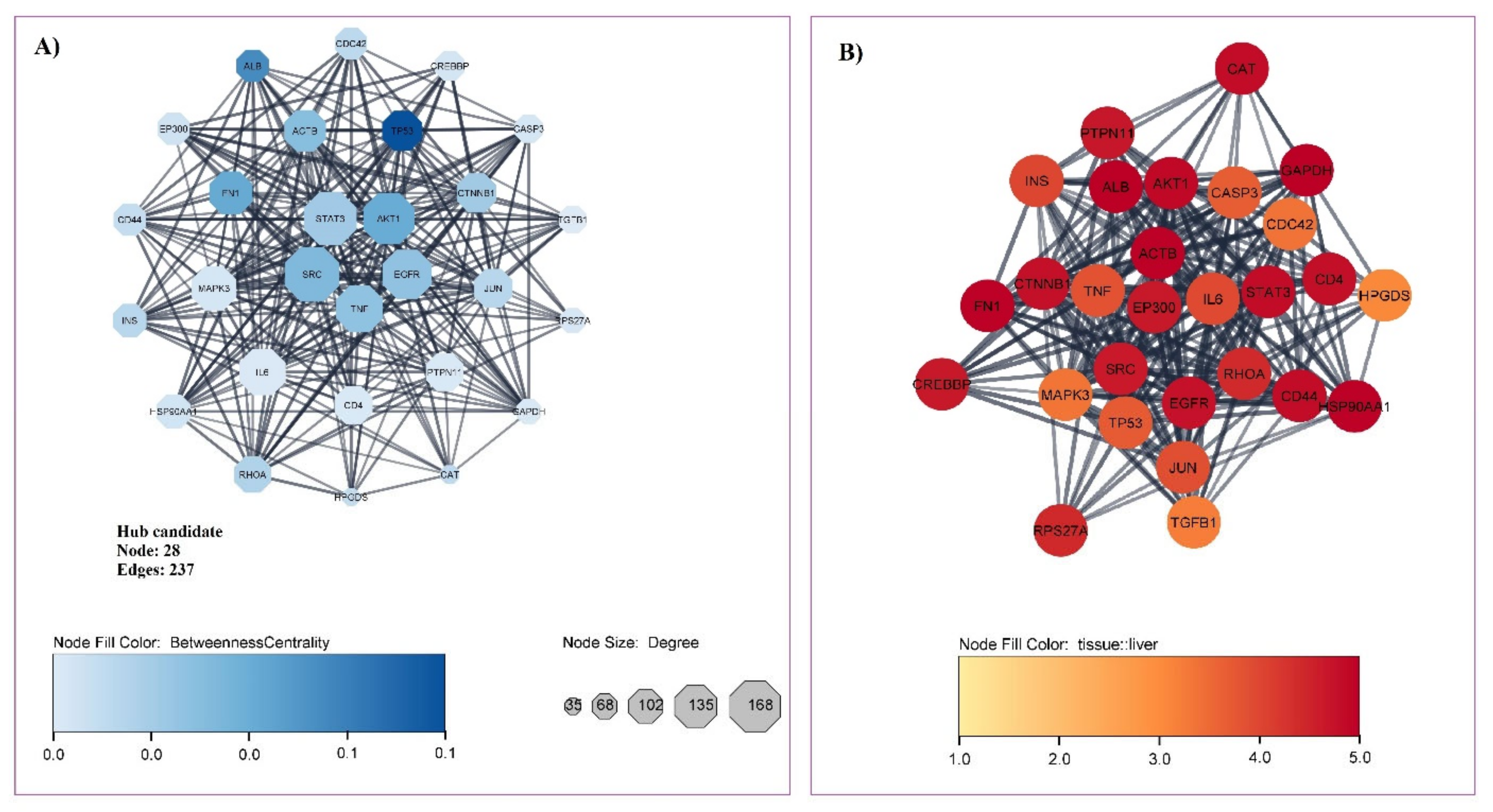



| Ensembl Gene ID | Gene Symbol | Gene Full Name | Network Analyzer | ||||
|---|---|---|---|---|---|---|---|
| Protein Class | Liver Specificity | Degree | Betweenness | Closeness | |||
| ENSG00000075624 | ACTB | Actin beta | Plasma proteins | 4.98 | 121 | 0.0332 | 0.4504 |
| ENSG00000142208 | AKT1 | serine/threonine kinase 1 | Enzymes | 4.88 | 156 | 0.0403 | 0.4619 |
| ENSG00000163631 | ALB | Albumin | Plasma proteins | 5.00 | 88 | 0.0493 | 0.4336 |
| ENSG00000026508 | CD44 | CD44 Molecule | Blood group antigen proteins | 4.83 | 88 | 0.0197 | 0.4219 |
| ENSG00000168036 | CTNNB1 | Catenin beta 1 | Plasma proteins | 4.77 | 113 | 0.0276 | 0.4401 |
| ENSG00000146648 | EGFR | Epidermal growth factor receptor | RAS pathway related proteins | 4.75 | 149 | 0.0311 | 0.4498 |
| ENSG00000100393 | EP300 | E1A binding protein p300 | Metabolic proteins | 4.65 | 88 | 0.0172 | 0.4166 |
| ENSG00000115414 | FN1 | Fibronectin 1 | Plasma proteins | 4.97 | 127 | 0.0408 | 0.4293 |
| ENSG00000080824 | HSP90AA1 | Heat shock protein 90 alpha family class A member 1 | Enzymes | 4.98 | 95 | 0.0166 | 0.4285 |
| ENSG00000254647 | INS | Insulin | RAS pathway related proteins | 4.00 | 92 | 0.0219 | 0.4295 |
| ENSG00000177606 | JUN | Jun proto-oncogene | Transcription factors | 3.88 | 111 | 0.0224 | 0.4395 |
| ENSG00000067560 | RHOA | Ras homolog family member A | Enzymes | 4.40 | 103 | 0.0239 | 0.4256 |
| ENSG00000197122 | SRC | non-receptor tyrosine kinase | Enzymes | 4.64 | 168 | 0.0350 | 0.4612 |
| ENSG00000168610 | STAT3 | Signal transducer and activator of transcription 3 | Transcription factors | 4.84 | 162 | 0.0279 | 0.4491 |
| ENSG00000232810 | TNF | Tumor necrosis factor | Plasma proteins | 3.82 | 140 | 0.0314 | 0.4449 |
| ENSG00000141510 | TP53 | Tumor protein p53 | Transcription factors | 3.72 | 115 | 0.0658 | 0.4412 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahmoudi, A.; Atkin, S.L.; Jamialahmadi, T.; Banach, M.; Sahebkar, A. Effect of Curcumin on Attenuation of Liver Cirrhosis via Genes/Proteins and Pathways: A System Pharmacology Study. Nutrients 2022, 14, 4344. https://doi.org/10.3390/nu14204344
Mahmoudi A, Atkin SL, Jamialahmadi T, Banach M, Sahebkar A. Effect of Curcumin on Attenuation of Liver Cirrhosis via Genes/Proteins and Pathways: A System Pharmacology Study. Nutrients. 2022; 14(20):4344. https://doi.org/10.3390/nu14204344
Chicago/Turabian StyleMahmoudi, Ali, Stephen L. Atkin, Tannaz Jamialahmadi, Maciej Banach, and Amirhossein Sahebkar. 2022. "Effect of Curcumin on Attenuation of Liver Cirrhosis via Genes/Proteins and Pathways: A System Pharmacology Study" Nutrients 14, no. 20: 4344. https://doi.org/10.3390/nu14204344
APA StyleMahmoudi, A., Atkin, S. L., Jamialahmadi, T., Banach, M., & Sahebkar, A. (2022). Effect of Curcumin on Attenuation of Liver Cirrhosis via Genes/Proteins and Pathways: A System Pharmacology Study. Nutrients, 14(20), 4344. https://doi.org/10.3390/nu14204344








