Digital Biomarkers for Personalized Nutrition: Predicting Meal Moments and Interstitial Glucose with Non-Invasive, Wearable Technologies
Abstract
:1. Introduction
2. Materials and Methods
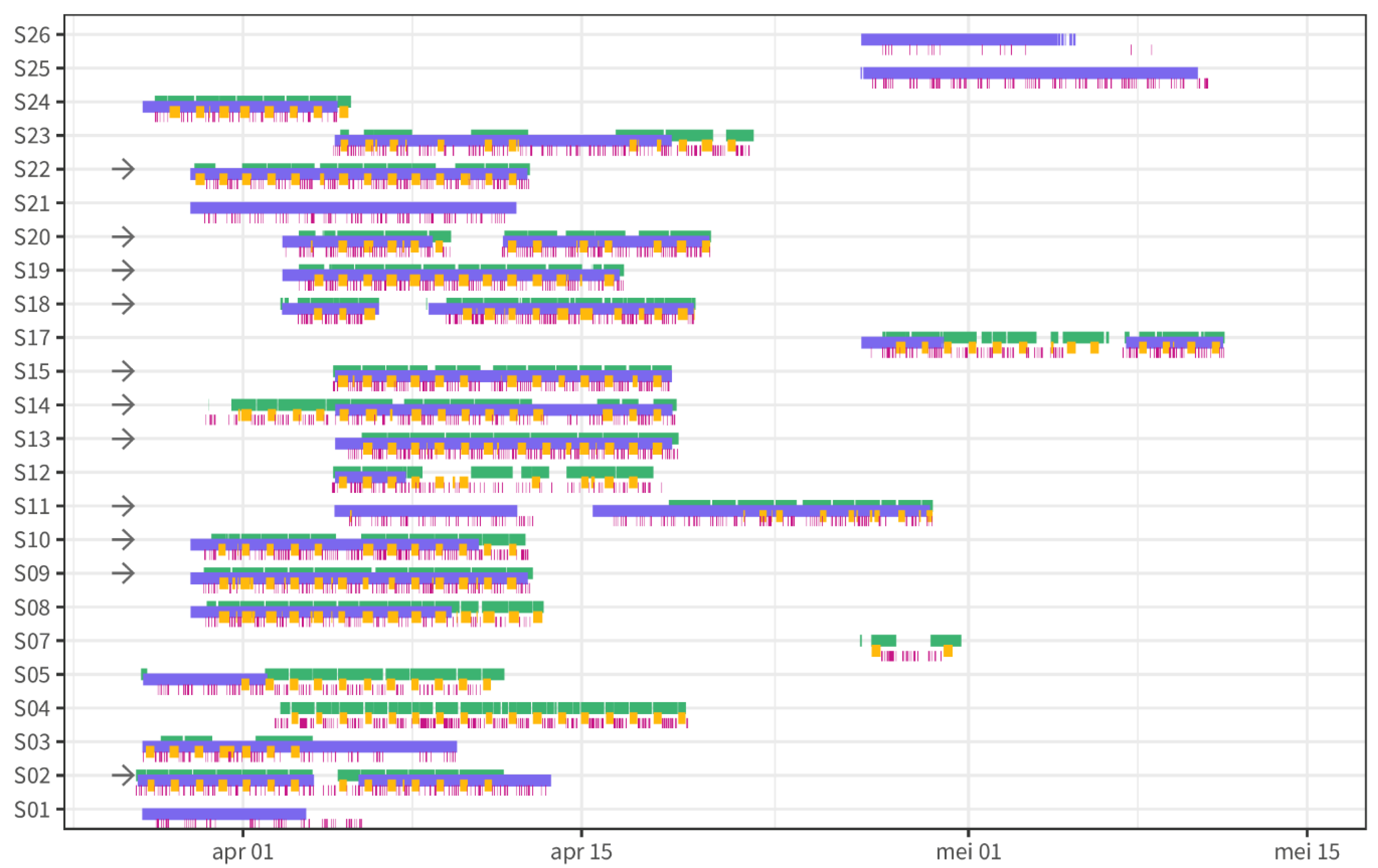
2.1. Study Design and Data Collection
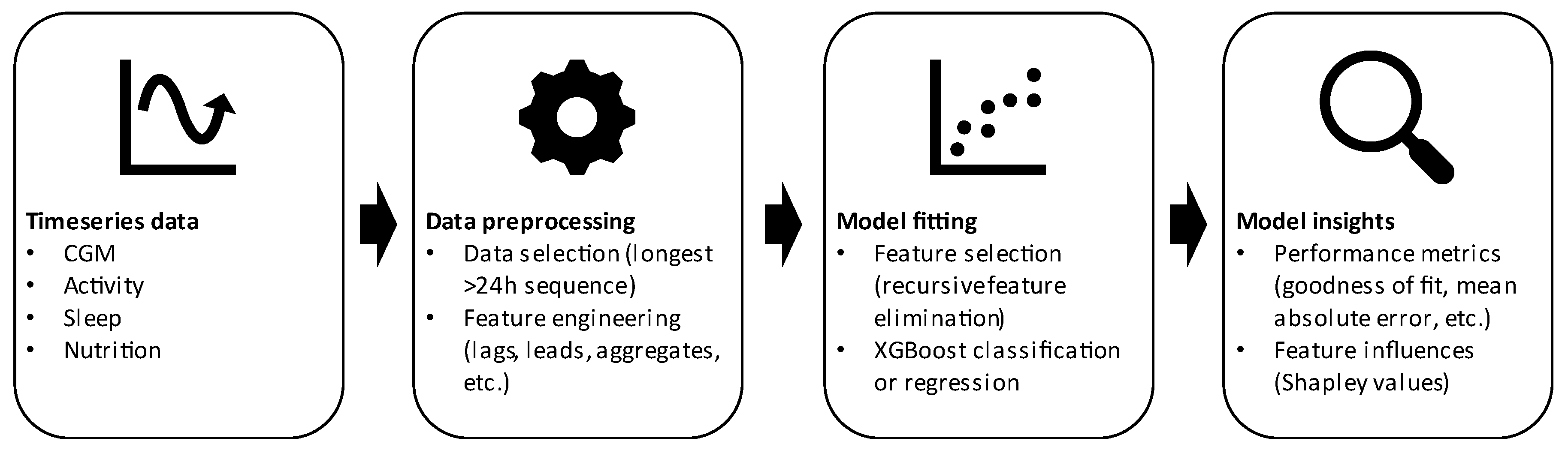
2.2. Data Preprocessing and Feature Engineering
2.3. XGboost for Predicting Eating Moments and Glucose
3. Results
3.1. Baseline Characteristics and Dataset Characteristics
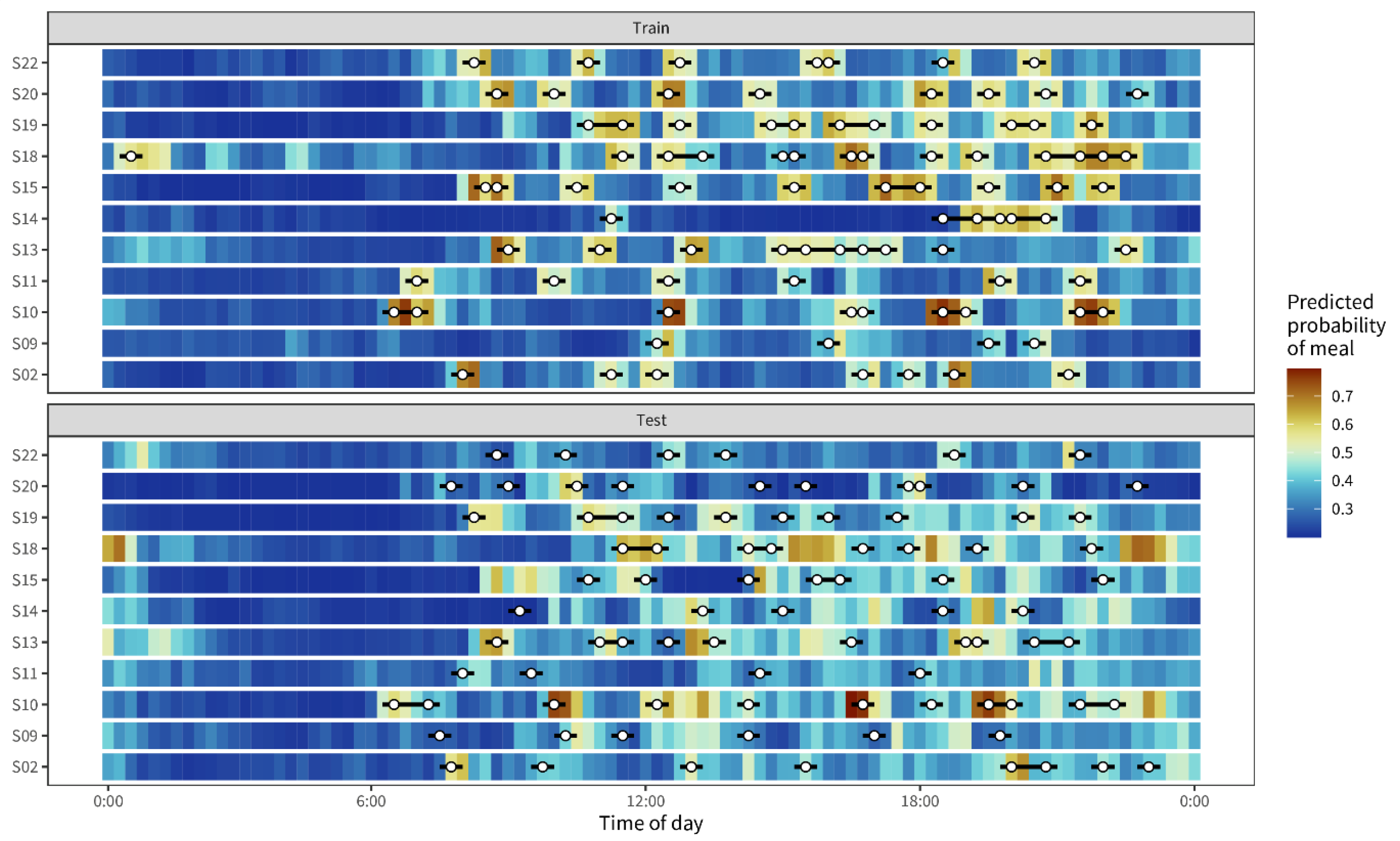
3.2. Detecting Eating Moments Based on Interstitial Glucose Levels
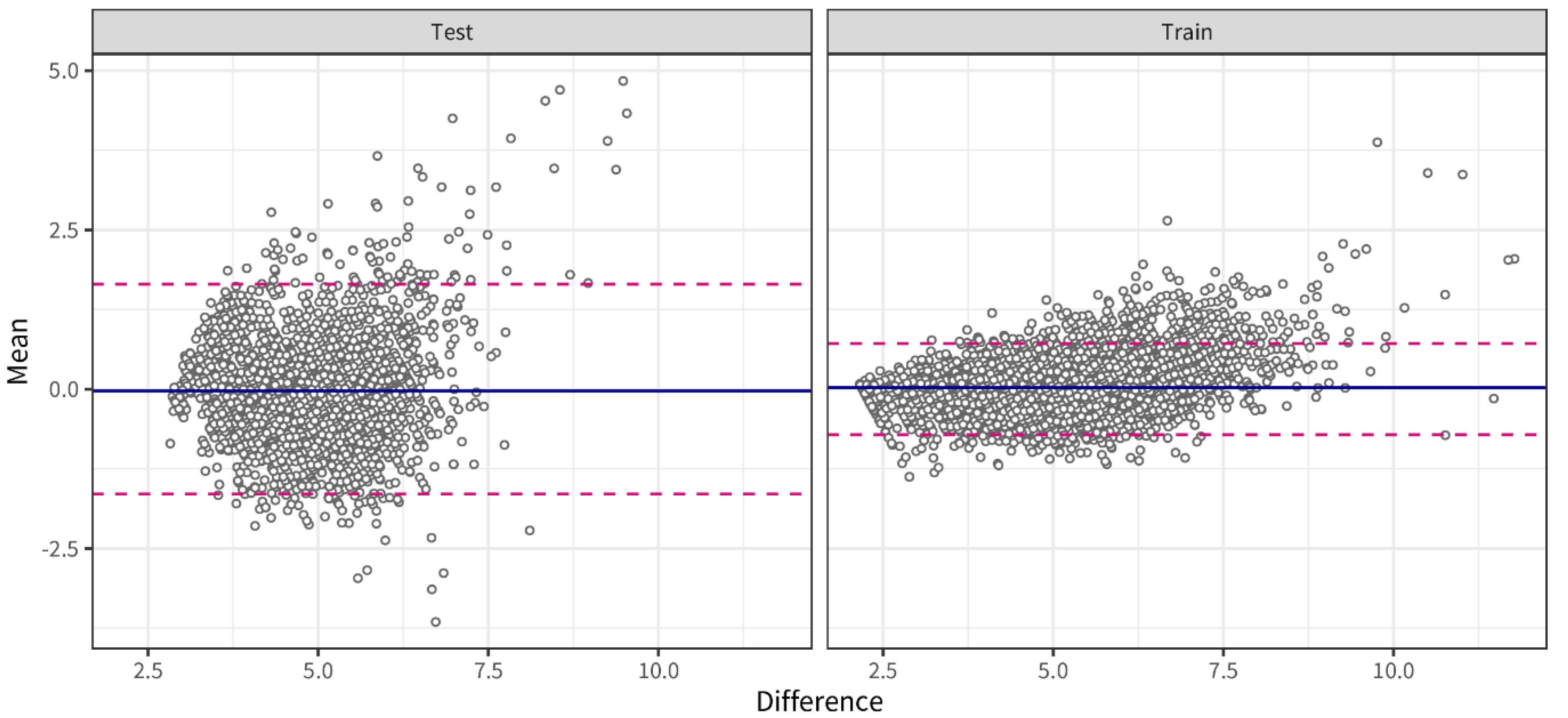
3.3. Predicting Lifestyle Behavior Effects Based on Interstitial Glucose Levels
4. Discussion
4.1. Meal Detection
4.2. Predicting Glucose
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vos, T.; Lim, S.S.; Abbafati, C.; Abbas, K.M.; Abbasi, M.; Abbasifard, M.; Abbasi-Kangevari, M.; Abbastabar, H.; Abd-Allah, F.; Abdelalim, A.; et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef]
- Khan, M.A.B.; Hashim, M.J.; King, J.K.; Govender, R.D.; Mustafa, H.; Al Kaabi, J. Epidemiology of Type 2 Diabetes—Global Burden of Disease and Forecasted Trends. J. Epidemiol. Glob. Health 2020, 10, 107–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Ommen, B.; Wopereis, S.; Van Empelen, P.; Van Keulen, H.M.; Otten, W.; Kasteleyn, M.; Molema, J.J.; De Hoogh, I.M.; Chavannes, N.H.; Numans, M.E.; et al. From diabetes care to diabetes cure-the integration of systems biology, ehealth, and behavioral change. Front. Endocrinol. 2018, 8, 381. [Google Scholar] [CrossRef] [PubMed]
- Ben-Yacov, O.; Godneva, A.; Rein, M.; Shilo, S.; Kolobkov, D.; Koren, N.; Dolev, N.C.; Shmul, T.T.; Wolf, B.C.; Kosower, N.; et al. Personalized Postprandial Glucose Response–Targeting Diet Versus Mediterranean Diet for Glycemic Control in Prediabetes. Diabetes Care 2021, 44, 1980–1991. [Google Scholar] [CrossRef] [PubMed]
- Lean, M.E.J.; Leslie, W.S.; Barnes, A.C.; Brosnahan, N.; Thom, G.; McCombie, L.; Peters, C.; Zhyzhneuskaya, S.; Al-Mrabeh, A.; Hollingsworth, K.G.; et al. Durability of a primary care-led weight-management intervention for remission of type 2 diabetes: 2-year results of the DiRECT open-label, cluster-randomised trial. Lancet Diabetes Endocrinol. 2019, 7, 344–355. [Google Scholar] [CrossRef] [Green Version]
- Steven, S.; Hollingsworth, K.G.; Al-Mrabeh, A.; Avery, L.; Aribisala, B.; Caslake, M.; Taylor, R. Very Low-Calorie Diet and 6 Months of Weight Stability in Type 2 Diabetes: Pathophysiological Changes in Responders and Nonresponders. Diabetes Care 2016, 39, 808–815. [Google Scholar] [CrossRef] [Green Version]
- Ried-Larsen, M.; Johansen, M.Y.; Macdonald, C.S.; Hansen, K.B.; Christensen, R.; Wedell-Neergaard, A.; Pilmark, N.S.; Langberg, H.; Vaag, A.A.; Pedersen, B.K.; et al. Type 2 diabetes remission 1 year after an intensive lifestyle intervention: A secondary analysis of a randomized clinical trial. Diabetes Obes. Metab. 2019, 21, 2257–2266. [Google Scholar]
- Knowler, W.C.; Barrett-Connor, E.; Fowler, S.E.; Hamman, R.F.; Lachin, J.M.; Walker, E.A.; Nathan, D.M.; Diabetes Prevention Program Research Group. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N. Engl. J. Med. 2002, 346, 393–403. [Google Scholar]
- Ahlqvist, E.; Storm, P.; Käräjämäki, A.; Martinell, M.; Dorkhan, M.; Carlsson, A.; Vikman, P.; Prasad, R.B.; Aly, D.M.; Almgren, P.; et al. Novel subgroups of adult-onset diabetes and their association with outcomes: A data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 2018, 6, 361–369. [Google Scholar] [CrossRef] [Green Version]
- Trouwborst, I.; Bowser, S.M.; Goossens, G.; Blaak, E.E. Ectopic Fat Accumulation in Distinct Insulin Resistant Phenotypes; Targets for Personalized Nutritional Interventions. Front. Nutr. 2018, 5, 77. [Google Scholar]
- Blaak, E.E. Current metabolic perspective on malnutrition in obesity: Towards more subgroup-based nutritional approaches? Proc. Nutr. Soc. 2020, 32, 331–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanco-Rojo, R.; Alcala-Diaz, J.F.; Wopereis, S.; Perez-Martinez, P.; Quintana-Navarro, G.M.; Marin, C.; Ordovas, J.M.; Van Ommen, B.; Perez-Jimenez, F.; Delgado-Lista, J.; et al. The insulin resistance phenotype (muscle or liver) interacts with the type of diet to determine changes in disposition index after 2 years of intervention: The CORDIOPREV-DIAB randomised clinical trial. Diabetologia 2015, 59, 67–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasman, W.J.; Memelink, R.G.; Bosch, J.D.V.-V.D.; Begieneman, M.P.V.; Brink, W.J.V.D.; Weijs, P.J.M.; Wopereis, S. Obese Older Type 2 Diabetes Mellitus Patients with Muscle Insulin Resistance Benefit from an Enriched Protein Drink during Combined Lifestyle Intervention: The PROBE Study. Nutrients 2020, 12, 2979. [Google Scholar] [CrossRef] [PubMed]
- Celis-Morales, C.A.; Livingstone, K.M.; Marsaux, C.F.M.; Macready, A.L.; Fallaize, R.; O’Donovan, C.B.; Woolhead, C.; Forster, H.; Walsh, M.C.; Navas-Carretero, S.; et al. Effect of personalized nutrition on health-related behaviour change: Evidence from the Food4me European randomized controlled trial. Int. J. Epidemiol. 2017, 46, 578–588. [Google Scholar] [CrossRef] [Green Version]
- Doets, E.L.; de Hoogh, I.M.; Holthuysen, N.; Wopereis, S.; Verain, M.C.; Puttelaar, J.V.D.; Hogenelst, K.; Boorsma, A.; Bouwman, E.P.; Timmer, M.; et al. Beneficial effect of personalized lifestyle advice compared to generic advice on wellbeing among Dutch seniors—An explorative study. Physiol. Behav. 2019, 210, 112642. [Google Scholar] [CrossRef]
- De Hoogh, I.M.; Winters, B.L.; Nieman, K.M.; Bijlsma, S.; Krone, T.; van den Broek, T.J.; Anderson, B.D.; Caspers, M.P.; Anthony, J.C.; Wopereis, S. A novel personalized systems nutrition program improves dietary patterns, lifestyle behaviors and health-related outcomes: Results from the habit study. Nutrients 2021, 13, 1763. [Google Scholar] [CrossRef]
- Price, N.D.; Magis, A.T.; Earls, J.C.; Glusman, G.; Levy, R.; Lausted, C.; McDonald, D.T.; Kusebauch, U.; Moss, C.L.; Zhou, Y.; et al. A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat. Biotechnol. 2017, 35, 747–756. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, D.E.; El-Sohemy, A. Disclosure of Genetic Information and Change in Dietary Intake: A Randomized Controlled Trial. PLoS ONE 2014, 9, e112665. [Google Scholar] [CrossRef]
- Taylor, R.; Al-Mrabeh, A.; Zhyzhneuskaya, S.; Peters, C.; Barnes, A.C.; Aribisala, B.S.; Hollingsworth, K.G.; Mathers, J.C.; Sattar, N.; Lean, M.E. Remission of human type 2 diabetes requires decrease in liver and pancreas fat content but is dependent upon capacity for beta-cell recovery. Cell Metab. 2018, 28, 547–556.e3. [Google Scholar] [CrossRef] [Green Version]
- de Hoogh, I.M.; Oosterman, J.E.; Otten, W.; Krijger, A.-M.; Berbée-Zadelaar, S.; Pasman, W.J.; van Ommen, B.; Pijl, H.; Wopereis, S. The Effect of a Lifestyle Intervention on Type 2 Diabetes Pathophysiology and Remission: The Stevenshof Pilot Study. Nutrients 2021, 13, 2193. [Google Scholar] [CrossRef]
- De Hoogh, I.M.; Pasman, W.J.; Boorsma, A.; van Ommen, B.; Wopereis, S. Effects of a 13-Week Personalized Lifestyle In-tervention Based on the Diabetes Subtype for People with Newly Diagnosed Type 2 Diabetes. Biomedicines 2022, 10, 643. [Google Scholar] [CrossRef] [PubMed]
- De Roos, B.; Brennan, L. Personalised interventions—A precision approach for the next generation of dietary intervention studies. Nutrients 2017, 9, 847. [Google Scholar] [CrossRef] [Green Version]
- Tabák, A.G.; Jokela, M.; Akbaraly, T.N.; Brunner, E.J.; Kivimäki, M.; Witte, D.R. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: An analysis from the Whitehall II study. Lancet 2009, 373, 2215–2221. [Google Scholar] [CrossRef]
- Coravos, A.; Doerr, M.; Goldsack, J.; Manta, C.; Shervey, M.; Woods, B.; Wood, W.A. Modernizing and designing evaluation frameworks for connected sensor technologies in medicine. NPJ Digit. Med. 2020, 3, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldsack, J.C.; Coravos, A.; Bakker, J.P.; Bent, B.; Dowling, A.V.; Fitzer-Attas, C.; Godfrey, A.; Godino, J.G.; Gujar, N.; Izmailova, E.; et al. Verification, analytical validation, and clinical validation (V3): The foundation of determining fit-for-purpose for Biometric Monitoring Technologies (BioMeTs). NPJ Digit. Med. 2020, 3, 55. [Google Scholar] [CrossRef] [Green Version]
- Coravos, A.; Khozin, S.; Mandl, K.D. Developing and adopting safe and effective digital biomarkers to improve patient outcomes. NPJ Digit. Med. 2019, 2, 14. [Google Scholar] [CrossRef] [Green Version]
- Brink, W.V.D.; Bloem, R.; Ananth, A.; Kanagasabapathi, T.; Amelink, A.; Bouwman, J.; Gelinck, G.; Van Veen, S.; Boorsma, A.; Wopereis, S. Digital Resilience Biomarkers for Personalized Health Maintenance and Disease Prevention. Front. Digit. Health 2021, 2, 614670. [Google Scholar] [CrossRef]
- Hall, H.; Perelman, D.; Breschi, A.; Limcaoco, P.; Kellogg, R.; McLaughlin, T.; Snyder, M. Glucotypes reveal new patterns of glucose dysregulation. PLoS Biol. 2018, 16, e2005143. [Google Scholar] [CrossRef] [Green Version]
- Zeevi, D.; Korem, T.; Zmora, N.; Israeli, D.; Rothschild, D.; Weinberger, A.; Ben-Yacov, O.; Lador, D.; Avnit-Sagi, T.; Lotan-Pompan, M.; et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015, 163, 1079–1094. [Google Scholar] [CrossRef] [Green Version]
- Berry, S.E.; Valdes, A.M.; Drew, D.A.; Asnicar, F.; Mazidi, M.; Wolf, J.; Capdevila, J.; Hadjigeorgiou, G.; Davies, R.; Al Khatib, H.; et al. Human postprandial responses to food and potential for precision nutrition. Nat. Med. 2020, 26, 964–973. [Google Scholar] [CrossRef]
- Bent, B.; Cho, P.J.; Henriquez, M.; Wittmann, A.; Thacker, C.; Feinglos, M.; Crowley, M.J.; Dunn, J.P. Engineering digital biomarkers of interstitial glucose from noninvasive smartwatches. NPJ Digit. Med. 2021, 4, 89. [Google Scholar] [CrossRef] [PubMed]
- Poslusna, K.; Ruprich, J.; De Vries, J.H.M.; Jakubikova, M.; Van’T Veer, P. Misreporting of energy and micronutrient intake estimated by food records and 24 hour recalls, control and adjustment methods in practice. Br. J. Nutr. 2009, 101 (Suppl. S2), S73–S85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westerterp, K.R.; Goris, A.H.C. Validity of the assessment of dietary intake: Problems of misreporting. Curr. Opin. Clin. Nutr. Metab. Care 2002, 5, 489–493. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Ni, B.; Kleinberg, S. Automated meal detection from continuous glucose monitor data through simulation and explanation. J. Am. Med. Inform. Assoc. 2019, 26, 1592–1599. [Google Scholar] [CrossRef]
- Ramkissoon, C.M.; Herrero, P.; Bondia, J.; Vehi, J. Unannounced Meals in the Artificial Pancreas: Detection Using Continuous Glucose Monitoring. Sensors 2018, 18, 884. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K.; Mitchell, R.; Cano, I.; Zhou, T.; et al. xgboost: Extreme Gradient Boosting. 2022. R Package Version 1.5.0.2. Available online: https://cran.r-project.org/web/packages/xgboost/index.html (accessed on 19 October 2022).
- Kuhn, M. caret: Classification and Regression Training. 2022. R Package Version 6.0-90. Available online: https://cran.r-project.org/web/packages/caret/index.html (accessed on 19 October 2022).
- Komisarczyk, K.; Kozminski, P.; Maksymiuk, S.; Biecek, P. treeshap: Fast SHAP values computation for tree ensemble models. 2021. R Package Version 0.1.1. Available online: https://github.com/ModelOriented/treeshap/ (accessed on 19 October 2022).
- Borchers, H.W. pracma: Practical Numerical Math Functions. 2022. R Package Version 2.3.8. Available online: https://cran.r-project.org/web/packages/pracma/index.html (accessed on 19 October 2022).
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020, 21, 56–67. [Google Scholar] [CrossRef]
- Alshurafa, N.; Lin, A.W.; Zhu, F.; Ghaffari, R.; Hester, J.; Delp, E.; Rogers, J.; Spring, B. Counting Bites with Bits: Expert Workshop Addressing Calorie and Macronutrient Intake Monitoring. J. Med. Internet Res. 2019, 21, e14904. [Google Scholar] [CrossRef]
- Samadi, S.; Turksoy, K.; Hajizadeh, I.; Feng, J.; Sevil, M.; Cinar, A. Meal Detection and Carbohydrate Estimation Using Continuous Glucose Sensor Data. IEEE J. Biomed. Health Inform. 2017, 21, 619–627. [Google Scholar] [CrossRef]
- Huo, Z.; Mortazavi, B.J.; Chaspari, T.; Deutz, N.; Ruebush, L.; Gutierrez-Osuna, R. Predicting the meal macronutrient composition from continuous glucose monitors. In Proceedings of the2019 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), Chicago, IL, USA, 19–22 May 2019. [Google Scholar] [CrossRef]
- Mazidi, M.; Valdes, A.M.; Ordovas, J.M.; Hall, W.L.; Pujol, J.C.; Wolf, J.; Hadjigeorgiou, G.; Segata, N.; Sattar, N.; Koivula, R.; et al. Meal-induced inflammation: Postprandial insights from the Personalised REsponses to DIetary Composition Trial (PREDICT) study in 1000 participants. Am. J. Clin. Nutr. 2021, 114, 1028–1038. [Google Scholar] [CrossRef]
- Adams, O.P. The impact of brief high-intensity exercise on blood glucose levels. Diabetes Metab. Syndr. Obes. Targets Ther. 2013, 6, 113–122. [Google Scholar] [CrossRef] [Green Version]
- Wolever, T.M.S.; Mehling, C. Long-term effect of varying the source or amount of dietary carbohydrate on postprandial plasma glucose, insulin, triacylglycerol, and free fatty acid concentrations in subjects with impaired glucose tolerance. Am. J. Clin. Nutr. 2003, 77, 612–621. [Google Scholar] [CrossRef] [Green Version]
- Frank, S.; Jbaily, A.; Hinshaw, L.; Basu, R.; Basu, A.; Szeri, A.J. Modeling the acute effects of exercise on glucose dynamics in healthy nondiabetic subjects. J. Pharmacokinet. Pharmacodyn. 2021, 9, 225–239. [Google Scholar] [CrossRef] [PubMed]
- MacLeod, S.F.; Terada, T.; Chahal, B.S.; Boulé, N.G. Exercise lowers postprandial glucose but not fasting glucose in type 2 diabetes: A meta-analysis of studies using continuous glucose monitoring. Diabetes/Metab. Res. Rev. 2013, 29, 593–603. [Google Scholar] [CrossRef] [PubMed]
- DiMenna, F.J.; Arad, A.D. The acute vs. chronic effect of exercise on insulin sensitivity: Nothing lasts forever. Cardiovasc. Endocrinol. Metab. 2020, 10, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, E.; Vogt, J.A.; Wolever, T.M.S. The Effects of Fat and Protein on Glycemic Responses in Nondiabetic Humans Vary with Waist Circumference, Fasting Plasma Insulin, and Dietary Fiber Intake. J. Nutr. 2006, 136, 2506–2511. [Google Scholar] [CrossRef] [Green Version]
- Spiegel, K.; Tasali, E.; Leproult, R.; Van Cauter, E. Effects of poor and short sleep on glucose metabolism and obesity risk. Nat. Rev. Endocrinol. 2009, 5, 253–261. [Google Scholar] [CrossRef]
- Dempsey, P.C.; Larsen, R.N.; Sethi, P.; Sacre, J.W.; Straznicky, N.E.; Cohen, N.D.; Cerin, E.; Lambert, G.W.; Owen, N.; Kingwell, B.A.; et al. Benefits for Type 2 Diabetes of Interrupting Prolonged Sitting with Brief Bouts of Light Walking or Simple Resistance Activities. Diabetes Care 2016, 39, 964–972. [Google Scholar] [CrossRef] [Green Version]
- Russell, W.R.; Baka, A.; Björck, I.; Delzenne, N.; Gao, D.; Griffiths, H.R.; Hadjilucas, E.; Juvonen, K.; Lahtinen, S.; Lansink, M.; et al. Impact of Diet Composition on Blood Glucose Regulation. Crit. Rev. Food Sci. Nutr. 2016, 56, 541–590. [Google Scholar] [CrossRef]
- Umpierre, D.; Ribeiro, P.A.B.; Kramer, C.K.; Leitão, C.B.; Zucatti, A.T.N.; Azevedo, M.J.; Gross, J.L.; Ribeiro, J.P.; Schaan, B.D. Physical Activity Advice Only or Structured Exercise Training and Association with HbA1c Levels in Type 2 Diabetes. JAMA 2011, 305, 1790–1799. [Google Scholar] [CrossRef] [Green Version]
- Dunstan, D.W.; Kingwell, B.A.; Larsen, R.; Healy, G.N.; Cerin, E.; Hamilton, M.T.; Shaw, J.E.; Bertovic, D.A.; Zimmet, P.Z.; Salmon, J.; et al. Breaking Up Prolonged Sitting Reduces Postprandial Glucose and Insulin Responses. Diabetes Care 2012, 35, 976–983. [Google Scholar] [CrossRef] [Green Version]
- Paing, A.C.; McMillan, K.A.; Kirk, A.F.; Collier, A.; Hewitt, A.; Chastin, S.F.M. Impact of free-living pattern of sedentary behaviour on intra-day glucose regulation in type 2 diabetes. Eur. J. Appl. Physiol. 2020, 120, 171–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bent, B.; Cho, P.J.; Wittmann, A.; Thacker, C.; Muppidi, S.; Snyder, M.; Crowley, M.J.; Feinglos, M.; Dunn, J.P. Non-invasive wearables for remote monitoring of HbA1c and glucose variability: Proof of concept. BMJ Open Diabetes Res. Care 2021, 9, e002027. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, R.; Tam, D.N.H.; Shah, J.; Moriyama, M.; Varney, J.; Huy, N.T. Effects of sleep intervention on glucose control: A narrative review of clinical evidence. Prim. Care Diabetes 2021, 15, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Mendes-Soares, H.; Raveh-Sadka, T.; Azulay, S.; Edens, K.; Ben-Shlomo, Y.; Cohen, Y.; Ofek, T.; Bachrach, D.; Stevens, J.; Colibaseanu, D.; et al. Assessment of a Personalized Approach to Predicting Postprandial Glycemic Responses to Food Among Individuals Without Diabetes. JAMA Netw. Open 2019, 2, e188102. [Google Scholar] [CrossRef] [Green Version]
- Gillum, R.F.; Makuc, D.M.; Feldman, J.J. Pulse rate, coronary heart disease, and death: The NHANES I Epidemiologic Fol-low-up Study. Am. Heart J. 1991, 121, 172–177. [Google Scholar] [CrossRef]
- Weijs, P.J.M. Validity of predictive equations for resting energy expenditure in US and Dutch overweight and obese class I and II adults aged 18–65 y. Am. J. Clin. Nutr. 2008, 88, 959–970. [Google Scholar] [CrossRef] [Green Version]
- Hower, I.M.; Harper, S.A.; Buford, T.W. Circadian rhythms, exercise, and cardiovascular health. J. Circadian Rhythm. 2018, 16, 7. [Google Scholar] [CrossRef]
- Khozin, S.; Coravos, A. Decentralized Trials in the Age of Real-World Evidence and Inclusivity in Clinical Investigations. Clin. Pharmacol. Ther. 2019, 106, 25–27. [Google Scholar] [CrossRef]
- Nielsen, J. Usability 101: Introduction to Usability. 2012. Available online: https://www.nngroup.com/arti-cles/usability-101-introduction-to-usability/ (accessed on 7 September 2022).

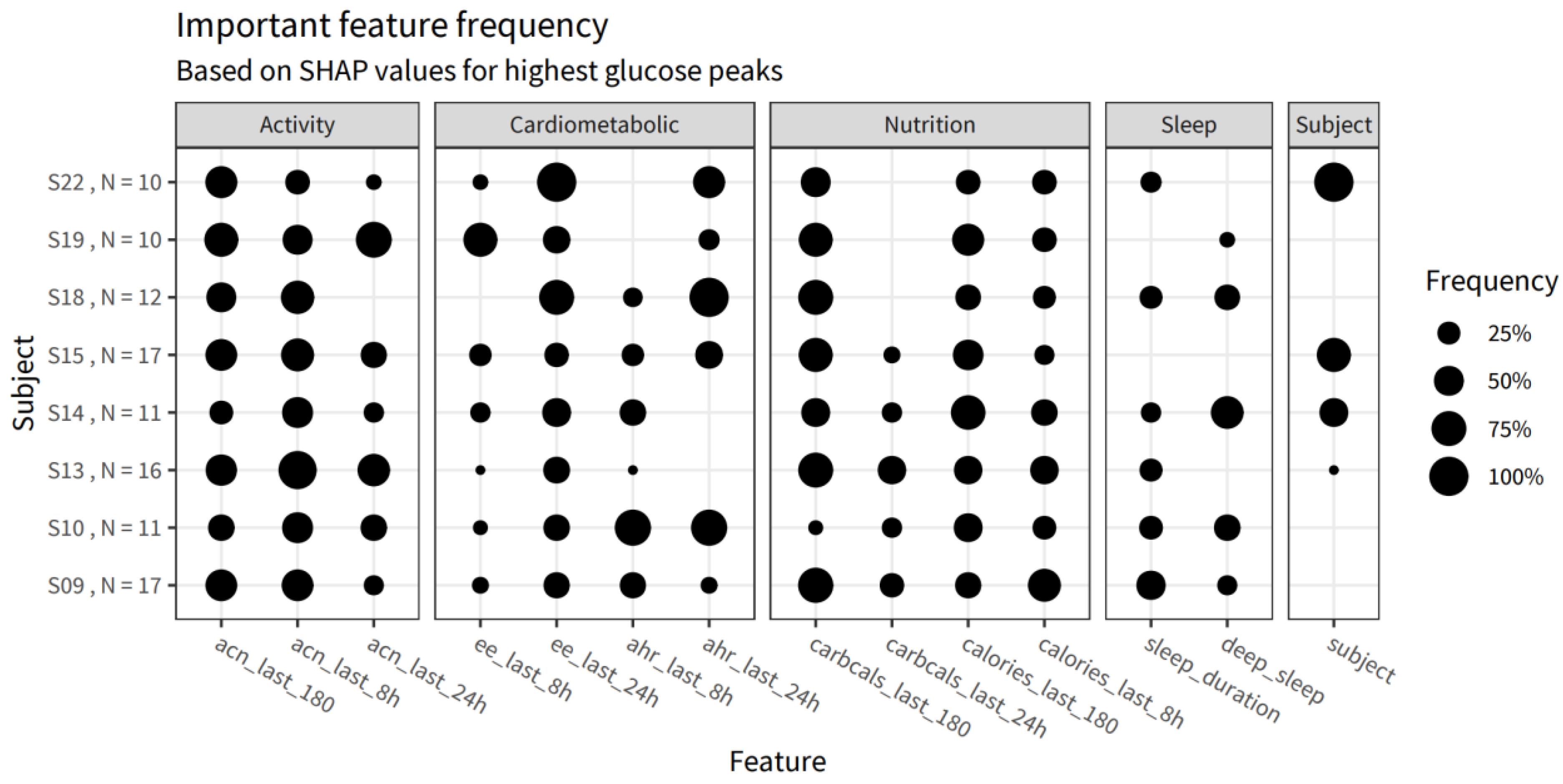
| Group | Weight |
|---|---|
| Cardiometabolic factors | 26.7% |
| Subject | 24.1% |
| Activity—Long term | 12.5% |
| Nutrition—Short term | 10.9% |
| Sleep | 10.7% |
| Nutrition—Long term | 8.7% |
| Activity—Short term | 6.5% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van den Brink, W.J.; van den Broek, T.J.; Palmisano, S.; Wopereis, S.; de Hoogh, I.M. Digital Biomarkers for Personalized Nutrition: Predicting Meal Moments and Interstitial Glucose with Non-Invasive, Wearable Technologies. Nutrients 2022, 14, 4465. https://doi.org/10.3390/nu14214465
van den Brink WJ, van den Broek TJ, Palmisano S, Wopereis S, de Hoogh IM. Digital Biomarkers for Personalized Nutrition: Predicting Meal Moments and Interstitial Glucose with Non-Invasive, Wearable Technologies. Nutrients. 2022; 14(21):4465. https://doi.org/10.3390/nu14214465
Chicago/Turabian Stylevan den Brink, Willem J., Tim J. van den Broek, Salvator Palmisano, Suzan Wopereis, and Iris M. de Hoogh. 2022. "Digital Biomarkers for Personalized Nutrition: Predicting Meal Moments and Interstitial Glucose with Non-Invasive, Wearable Technologies" Nutrients 14, no. 21: 4465. https://doi.org/10.3390/nu14214465
APA Stylevan den Brink, W. J., van den Broek, T. J., Palmisano, S., Wopereis, S., & de Hoogh, I. M. (2022). Digital Biomarkers for Personalized Nutrition: Predicting Meal Moments and Interstitial Glucose with Non-Invasive, Wearable Technologies. Nutrients, 14(21), 4465. https://doi.org/10.3390/nu14214465





