Interplay of Lymphocytes with the Intestinal Microbiota in Children with Nonalcoholic Fatty Liver Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Subject Recruitment
2.2. Data Collection
2.3. 16S rRNA Gene Sequencing
2.4. Metagenomic Sequencing
2.5. Bioinformatics and Statistical Analyses
3. Results
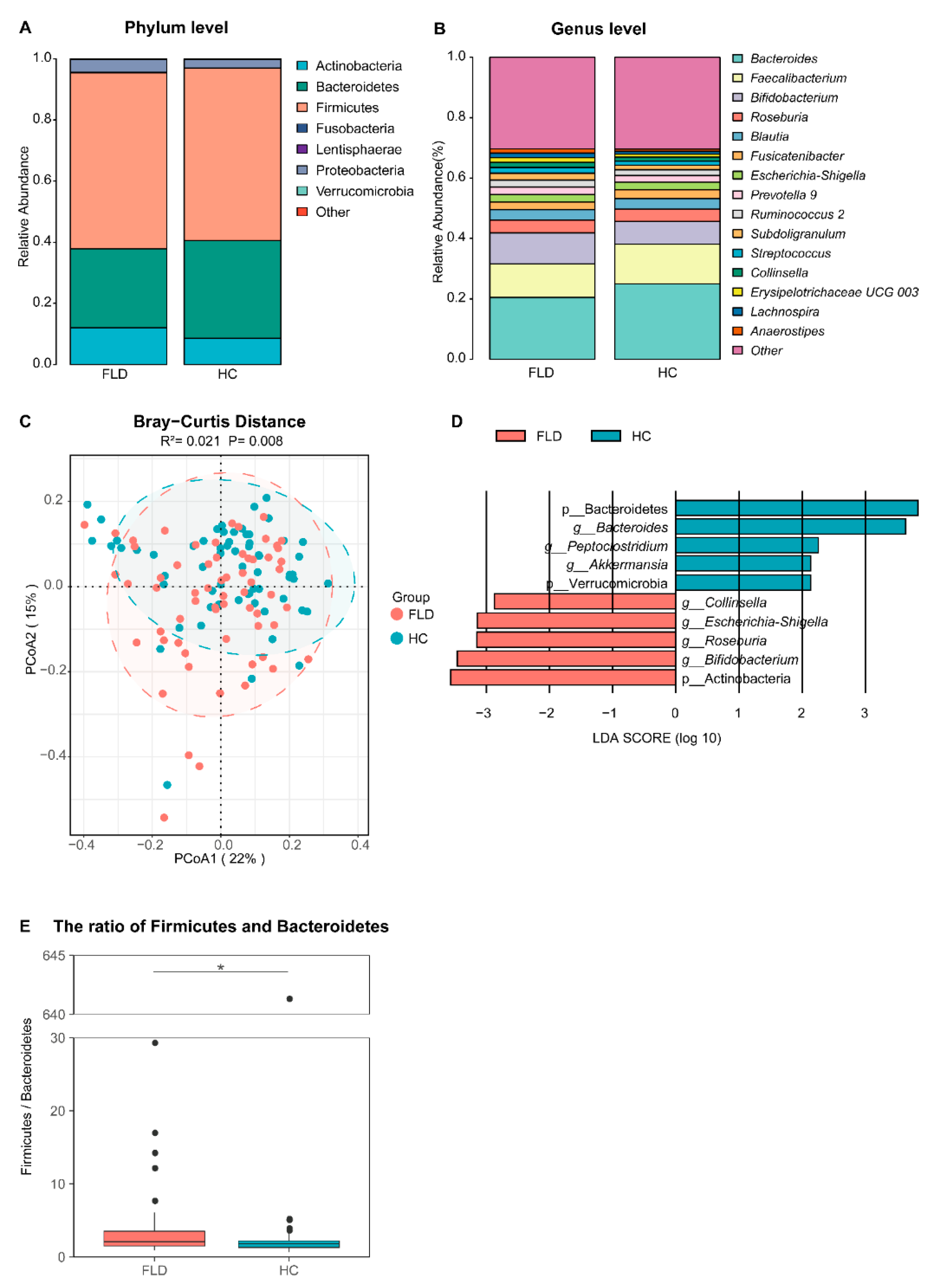
3.1. NAFLD Alters the Composition of Gut Microbiota in Children
3.2. Gut Microbiota in Children with NAFLD Interplayed with Lymphocytes
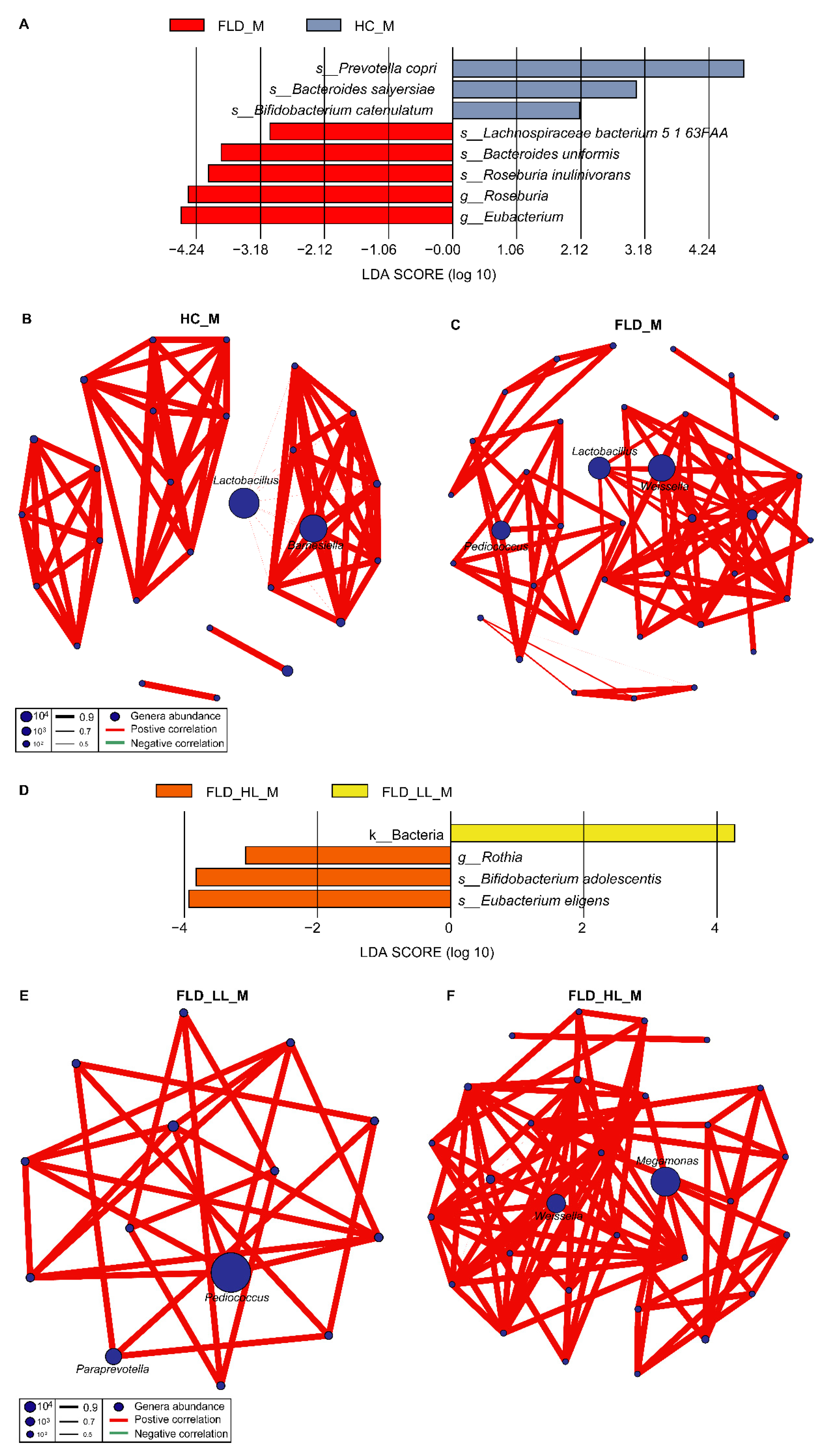
3.3. Differences in the Gut Microbiome between NAFLD and Healthy Children
3.4. Relationships between NAFLD, Gut Microbiome, and Lymphocytes
4. Discussion
4.1. Alpha and Beta Diversity of Gut Microbiota in Children Is Less Affected by NAFLD
4.2. The Disturbance of Gut Microbiota Is Associated with NAFLD and Lymphocytes
4.3. The Interplay among NAFLD, Gut Microbiome, and Lymphocytes
4.4. NAFLD Alters Gut Microbiota Microbial Networks in Children
4.5. NAFLD Alters Gut Microbiome Microbial Networks in Children
4.6. Impact of NAFLD on the Microbial Network of Clinical Indicators in Children
4.7. Influence of Lymphocytes on the Microbial Network of Clinical Indicators in Children with NAFLD
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eslam, M.; El-Serag, H.; Francque, S.; Sarin, S.; Wei, L.; Bugianesi, E.; George, J. Metabolic (dysfunction)-associated fatty liver disease in individuals of normal weight. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 638–651. [Google Scholar] [CrossRef] [PubMed]
- Gebru, Y.A.; Gupta, H.; Kim, H.S.; Eom, J.A.; Kwon, G.H.; Park, E.; Jeong, J.J.; Won, S.M.; Sharma, S.P.; Ganesan, R.; et al. T Cell Subsets and Natural Killer Cells in the Pathogenesis of Nonalcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2021, 22, 12190. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kordy, K.; Li, F.; Lee, D.J.; Kinchen, J.M.; Jew, M.H.; La Rocque, M.E.; Zabih, S.; Saavedra, M.; Woodward, C.; Cunningham, N.J.; et al. Metabolomic Predictors of Non-alcoholic Steatohepatitis and Advanced Fibrosis in Children. Front. Microbiol. 2021, 12, 713234. [Google Scholar] [CrossRef]
- Liu, J.; Mu, C.; Li, K.; Luo, H.; Liu, Y.; Li, Z. Estimating Global Prevalence of Metabolic Dysfunction-Associated Fatty Liver Disease in Overweight or Obese Children and Adolescents: Systematic Review and Meta-Analysis. Int. J. Public Health 2021, 66, 1604371. [Google Scholar] [CrossRef] [PubMed]
- Hariri, N.; Thibault, L. High-fat diet-induced obesity in animal models. Nutr. Res. Rev. 2010, 23, 270–299. [Google Scholar] [CrossRef] [Green Version]
- Tokuhara, D. Role of the Gut Microbiota in Regulating Non-alcoholic Fatty Liver Disease in Children and Adolescents. Front. Nutr. 2021, 8, 700058. [Google Scholar] [CrossRef]
- Han, H.; Jiang, Y.; Wang, M.; Melaku, M.; Liu, L.; Zhao, Y.; Everaert, N.; Yi, B.; Zhang, H. Intestinal dysbiosis in nonalcoholic fatty liver disease (NAFLD): Focusing on the gut-liver axis. Crit. Rev. Food Sci. Nutr. 2021, 8, 1–18. [Google Scholar] [CrossRef]
- Bruneau, A.; Hundertmark, J.; Guillot, A.; Tacke, F. Molecular and Cellular Mediators of the Gut-Liver Axis in the Progression of Liver Diseases. Front. Med. 2021, 8, 725390. [Google Scholar] [CrossRef]
- Pan, X.; Kaminga, A.C.; Liu, A.; Wen, S.W.; Luo, M.; Luo, J. Gut Microbiota, Glucose, Lipid, and Water-Electrolyte Metabolism in Children With Nonalcoholic Fatty Liver Disease. Front. Cell Infect. Microbiol. 2021, 11, 683743. [Google Scholar] [CrossRef]
- Sahuri-Arisoylu, M.; Brody, L.P.; Parkinson, J.R.; Parkes, H.; Navaratnam, N.; Miller, A.D.; Thomas, E.L.; Frost, G.; Bell, J.D. Reprogramming of hepatic fat accumulation and ‘browning’ of adipose tissue by the short-chain fatty acid acetate. Int. J. Obes. 2016, 40, 955–963. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; He, J.; Gao, N.; Lu, X.; Li, M.; Wu, X.; Liu, Z.; Jin, Y.; Liu, J.; Xu, J.; et al. Probiotics may delay the progression of nonalcoholic fatty liver disease by restoring the gut microbiota structure and improving intestinal endotoxemia. Sci. Rep. 2017, 7, 45176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, N.; Baker, S.S.; Chapa-Rodriguez, A.; Liu, W.; Nugent, C.A.; Tsompana, M.; Mastrandrea, L.; Buck, M.J.; Baker, R.D.; Genco, R.J.; et al. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut 2018, 67, 1881–1891. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Manchester, J.K.; Semenkovich, C.F.; Gordon, J.I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA 2007, 104, 979–984. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Chantar, M.L.; Delgado, T.C.; Beraza, N. Revisiting the Role of Natural Killer Cells in Non-Alcoholic Fatty Liver Disease. Front. Immunol. 2021, 12, 640869. [Google Scholar] [CrossRef]
- Dao, M.C.; Everard, A.; Aron-Wisnewsky, J.; Sokolovska, N.; Prifti, E.; Verger, E.O.; Kayser, B.D.; Levenez, F.; Chilloux, J.; Hoyles, L.; et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: Relationship with gut microbiome richness and ecology. Gut 2016, 65, 426–436. [Google Scholar] [CrossRef] [Green Version]
- Van Herck, M.A.; Weyler, J.; Kwanten, W.J.; Dirinck, E.L.; De Winter, B.Y.; Francque, S.M.; Vonghia, L. The Differential Roles of T Cells in Non-alcoholic Fatty Liver Disease and Obesity. Front. Immunol. 2019, 10, 82. [Google Scholar] [CrossRef] [Green Version]
- Shi, Z.; Lei, H.; Chen, G.; Yuan, P.; Cao, Z.; Ser, H.; Zhu, X.; Wu, F.; Liu, C.; Dong, M.; et al. Impaired Intestinal Akkermansia muciniphila and Aryl Hydrocarbon Receptor Ligands Contribute to Nonalcoholic Fatty Liver Disease in Mice. Msystems 2021, 6, e00985-20. [Google Scholar] [CrossRef]
- Krishnan, S.; Ding, Y.; Saedi, N.; Choi, M.; Sridharan, G.V.; Sherr, D.H.; Yarmush, M.L.; Alaniz, R.C.; Jayaraman, A.; Lee, K. Gut Microbiota-Derived Tryptophan Metabolites Modulate Inflammatory Response in Hepatocytes and Macrophages. Msystems Cell Rep. 2018, 23, 1099–1111. [Google Scholar] [CrossRef]
- Li, M.; Shu, W.; Zunong, J.; Amaerjiang, N.; Xiao, H.; Li, D.; Vermund, S.H.; Hu, Y. Predictors of non-alcoholic fatty liver disease in children. Pediatr. Res. 2021, 92, 322–330. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.; Costello, E.; Fierer, N.; Peña, A.; Goodrich, J.; Gordon, J.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truong, D.; Franzosa, E.; Tickle, T.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 2015, 12, 902–903. [Google Scholar] [CrossRef]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 18. [Google Scholar] [CrossRef]
- Zhu, W.; Lomsadze, A.; Borodovsky, M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res 2010, 38, e132. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [Green Version]
- Kelly, B.J.; Gross, R.; Bittinger, K.; Sherrill-Mix, S.; Lewis, J.D.; Collman, R.G.; Bushman, F.D.; Li, H. Power and sample-size estimation for microbiome studies using pairwise distances and PERMANOVA. Bioinformatics 2015, 31, 2461–2468. [Google Scholar] [CrossRef] [Green Version]
- Feehley, T.; Plunkett, C.H.; Bao, R.; Choi Hong, S.M.; Culleen, E.; Belda-Ferre, P.; Campbell, E.; Aitoro, R.; Nocerino, R.; Paparo, L.; et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat. Med. 2019, 25, 448–453. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome. Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, T.; Liu, F.; Liu, L.; Zhang, Z.; Dong, W.; Bai, S.; Ma, L.; Kang, L. Effects of Helicobacter pylori Infection on the Oral Microbiota of Reflux Esophagitis Patients. Front. Cell Infect. Microbiol. 2021, 11, 732613. [Google Scholar] [CrossRef] [PubMed]
- Alamooti, A.A.; Ardalan, F.A.; Abdollahi, A.; Zeidi, M.; Firouzjaie, F. Determination of lymphocyte subsets reference values in healthy Iranian men by a single platform flow cytometric method. Cytom. A 2010, 77, 890–894. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Qiu, Z.; Xie, J.; Li, D.; Li, T. Reference ranges and age-related changes of peripheral blood lymphocyte subsets in Chinese healthy adults. Sci. China C Life Sci. 2009, 52, 643–650. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, Z.; Cui, B.; Jiang, A.; Tao, H.; Cheng, S.; Liu, Y. Combination of Chronic Alcohol Consumption and High-Salt Intake Elicits Gut Microbial Alterations and Liver Steatosis in Mice. J. Agric. Food Chem. 2020, 68, 1750–1759. [Google Scholar] [CrossRef]
- Yoshida, K.; Yokota, K.; Kutsuwada, Y.; Nakayama, K.; Watanabe, K.; Matsumoto, A.; Miyashita, H.; Khor, S.S.; Tokunaga, K.; Kawai, Y.; et al. Genome-Wide Association Study of Lean Nonalcoholic Fatty Liver Disease Suggests Human Leukocyte Antigen as a Novel Candidate Locus. Hepatol. Commun. 2020, 4, 1124–1135. [Google Scholar] [CrossRef]
- Tsai, H.J.; Tsai, Y.C.; Hung, W.W.; Hung, W.C.; Chang, C.C.; Dai, C.Y. Gut Microbiota and Non-Alcoholic Fatty Liver Disease Severity in Type 2 Diabetes Patients. J. Pers. Med. 2021, 11, 238. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Li, J.; Zhang, Y.; Guo, Y.; Chang, Q.; Chen, L.; Wang, Y.; Wang, S.; Song, Y.; et al. Sini Decoction Ameliorates Colorectal Cancer and Modulates the Composition of Gut Microbiota in Mice. Front. Pharmacol. 2021, 12, 609992. [Google Scholar] [CrossRef]
- Rodloff, A.C.; Widera, P.; Ehlers, S.; Montag, T.; Lucas, M.; Schmidt, G.; Hahn, H. Suppression of Blastogenic Transformation of Lymphocytes by Bacteroides fragilis in vitro and in vivo. Zent. Für Bakteriol. 1990, 274, 406–416. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhou, J.; Liu, J.; Wang, Z.; Chen, M.; Zhou, S. Metagenome of Gut Microbiota of Children With Nonalcoholic Fatty Liver Disease. Front. Pediatr. 2019, 7, 518. [Google Scholar] [CrossRef] [PubMed]
- Wahlang, B.; Alexander, N.C., 2nd; Li, X.; Rouchka, E.C.; Kirpich, I.A.; Cave, M.C. Polychlorinated biphenyls altered gut microbiome in CAR and PXR knockout mice exhibiting toxicant-associated steatohepatitis. Toxicol. Rep. 2021, 8, 536–547. [Google Scholar] [CrossRef]
- Rattigan, R.; Sweeney, T.; Vigors, S.; Rajauria, G.; O’Doherty, J.V. Effects of reducing dietary crude protein concentration and supplementation with laminarin or zinc oxide on the faecal scores and colonic microbiota in newly weaned pigs. J. Anim. Physiol. Anim. Nutr. 2020, 104, 1471–1483. [Google Scholar] [CrossRef] [PubMed]
- Doden, H.L.; Wolf, P.G.; Gaskins, H.R.; Anantharaman, K.; Alves, J.M.P.; Ridlon, J.M. Completion of the gut microbial epi-bile acid pathway. Gut Microbes 2021, 13, 1907271. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, Y.; Guo, X.; Jin, X.; Yan, W.; Lin, B.; Cai, T.; Wei, Y. Activation of p38 mitogen-activated protein kinase pathway by lipopolysaccharide aggravates postoperative ileus in colorectal cancer patients. J. Gastroenterol. Hepatol. 2022, 37, 518–530. [Google Scholar] [CrossRef] [PubMed]
- Matsui, K.; Wirotesangthong, M.; Nishikawa, A. Peptidoglycan from Staphylococcus aureus induces IL-4 production from murine spleen cells via an IL-18-dependent mechanism. Int. Arch. Allergy Immunol. 2008, 146, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.Y.; Lu, H.C.; Chou, Y.H.; Liu, P.Y.; Chen, H.Y.; Huang, M.C.; Lin, C.H.; Tsai, C.N. Gut Microbial Signatures for Glycemic Responses of GLP-1 Receptor Agonists in Type 2 Diabetic Patients: A Pilot Study. Front. Endocrinol. 2021, 12, 814770. [Google Scholar] [CrossRef]
- Hu, X.; Fan, Y.; Li, H.; Zhou, R.; Zhao, X.; Sun, Y.; Zhang, S. Impacts of Cigarette Smoking Status on Metabolomic and Gut Microbiota Profile in Male Patients With Coronary Artery Disease: A Multi-Omics Study. Front. Cardiovasc. Med. 2021, 8, 766739. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N.; Luo, C.; Yajnik, V.; Khalili, H.; Garber, J.J.; Stevens, B.W.; Cleland, T.; Xavier, R.J. Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases. Cell Host. Microbe 2017, 21, 603–610. [Google Scholar] [CrossRef] [Green Version]
- Nagayama, M.; Yano, T.; Atarashi, K.; Tanoue, T.; Sekiya, M.; Kobayashi, Y.; Sakamoto, H.; Miura, K.; Sunada, K.; Kawaguchi, T.; et al. TH1 cell-inducing Escherichia coli strain identified from the small intestinal mucosa of patients with Crohn’s disease. Gut Microbes 2020, 12, 1788898. [Google Scholar] [CrossRef]
- Plaza-Diaz, J.; Solis-Urra, P.; Rodriguez-Rodriguez, F.; Olivares-Arancibia, J.; Navarro-Oliveros, M.; Abadia-Molina, F.; Alvarez-Mercado, A.I. The Gut Barrier, Intestinal Microbiota, and Liver Disease: Molecular Mechanisms and Strategies to Manage. Int. J. Mol. Sci. 2020, 21, 8351. [Google Scholar] [CrossRef]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zheng, J.; Shi, W.; Du, N.; Xu, X.; Zhang, Y.; Ji, P.; Zhang, F.; Jia, Z.; Wang, Y.; et al. Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut 2018, 67, 1614–1625. [Google Scholar] [CrossRef] [PubMed]
- Caparros, E.; Wiest, R.; Scharl, M.; Rogler, G.; Gutierrez Casbas, A.; Yilmaz, B.; Wawrzyniak, M.; Frances, R. Dysbiotic microbiota interactions in Crohn’s disease. Gut Microbes 2021, 13, 1949096. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Torralba, M.G.; Moncera, K.J.; DiLello, L.; Petrini, J.; Nelson, K.E.; Pieper, R. Gastro-intestinal and oral microbiome signatures associated with healthy aging. Geroscience 2019, 41, 907–921. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Bai, Y.; Zhou, J.; Huang, W.; Yan, J.; Tao, J.; Fan, Q.; Liu, Y.; Mei, D.; Yan, Q.; et al. Core Fucosylation of Maternal Milk N-Glycan Evokes B Cell Activation by Selectively Promoting the l-Fucose Metabolism of Gut Bifidobacterium spp. and Lactobacillus spp. mBio 2019, 10, e00128-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, D.; Pan, Q.; Xin, F.Z.; Zhang, R.N.; He, C.X.; Chen, G.Y.; Liu, C.; Chen, Y.W.; Fan, J.G. Sodium butyrate attenuates high-fat diet-induced steatohepatitis in mice by improving gut microbiota and gastrointestinal barrier. World J. Gastroenterol. 2017, 23, 60–75. [Google Scholar] [CrossRef]
- Jiang, W.; Wu, N.; Wang, X.; Chi, Y.; Zhang, Y.; Qiu, X.; Hu, Y.; Li, J.; Liu, Y. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci. Rep. 2015, 5, 8096. [Google Scholar] [CrossRef] [Green Version]
- Zhuge, A.; Li, S.; Lou, P.; Wu, W.; Wang, K.; Yuan, Y.; Xia, J.; Li, B.; Li, L. Longitudinal 16S rRNA Sequencing Reveals Relationships among Alterations of Gut Microbiota and Nonalcoholic Fatty Liver Disease Progression in Mice. J. Microbiol. Spectr. 2022, 10, e0004722. [Google Scholar] [CrossRef]
- Kang, H.; You, H.J.; Lee, G.; Lee, S.H.; Yoo, T.; Choi, M.; Joo, S.K.; Park, J.H.; Chang, M.S.; Lee, D.H.; et al. Interaction effect between NAFLD severity and high carbohydrate diet on gut microbiome alteration and hepatic de novo lipogenesis. Gut Microbes 2022, 14, 2078612. [Google Scholar] [CrossRef]
- Zhuang, P.; Li, H.; Jia, W.; Shou, Q.; Zhu, Y.; Mao, L.; Wang, W.; Wu, F.; Chen, X.; Wan, X.; et al. Eicosapentaenoic and docosahexaenoic acids attenuate hyperglycemia through the microbiome-gut-organs axis in db/db mice. Microbiome 2021, 9, 185. [Google Scholar] [CrossRef]
- Elshaghabee, F.M.F.; Ghadimi, D.; Habermann, D.; de Vrese, M.; Bockelmann, W.; Kaatsch, H.J.; Heller, K.J.; Schrezenmeir, J. Effect of Oral Administration of Weissella confusa on Fecal and Plasma Ethanol Concentrations, Lipids and Glucose Metabolism in Wistar Rats Fed High Fructose and Fat Diet. Hepat. Med. 2020, 12, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.Y.; Yoon, S.J.; Han, D.H.; Gupta, H.; Youn, G.S.; Shin, M.J.; Ham, Y.L.; Kwak, M.J.; Kim, B.Y.; Yu, J.S.; et al. Lactobacillus and Pediococcus ameliorate progression of non-alcoholic fatty liver disease through modulation of the gut microbiome. Gut Microbes 2020, 11, 882–899. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wang, J.; Ren, W.; Bian, Y.; Wang, Y.; Wang, L.; Guo, L.; Lei, J.; Jia, J.; Miao, J. Effect of Jiangan-Jiangzhi Pill on Gut Microbiota and Chronic Inflammatory Response in Rats with Non-Alcoholic Fatty Liver. Chem. Biodivers 2022, 19, e202100987. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, K.; Nishida, T.; Fukushima, A. Synergistic induction of eotaxin and VCAM-1 expression in human corneal fibroblasts by staphylococcal peptidoglycan and either IL-4 or IL-13. Allergol. Int. 2011, 60, 355–363. [Google Scholar] [CrossRef] [Green Version]
- Rong, Q.; Ling, C.; Lu, D.; Zhang, C.; Zhao, H.; Zhong, K.; Nong, X.; Qin, X. Sb(III) resistance mechanism and oxidation characteristics of Klebsiella aerogenes X. Chemosphere 2022, 293, 133453. [Google Scholar] [CrossRef]


| Parameters | HC (n = 63) | FLD (n = 63) | t/χ2/Z | p-Value | ||
|---|---|---|---|---|---|---|
| ± SD | M (q1, q3) | ± SD | M (q1, q3) | |||
| Boy (n, %) 1 | 47 (74.60) | 47 (74.60) | <0.01 | 1 | ||
| Age (years) 3 | 6.72 ± 0.32 | 6.73 (6.44, 7.02) | 6.75 ± 0.31 | 6.71 (6.55, 6.93) | −0.32 | 0.75 |
| Height (cm) 2 | 122.90 ± 4.81 | 122.45 (119.78, 125.75) | 127.72 ± 4.45 | 127.6 (124.4, 131) | −5.84 | p < 0.001 |
| Waist (cm) 2 | 55.79 ± 5.13 | 55.3 (52.31, 58.35) | 71.18 ± 8.35 | 70.5 (65.2, 77.3) | −12.44 | p < 0.001 |
| Hip (cm) 2 | 67.16 ± 5.32 | 67.95 (62.64, 70.51) | 79.41 ± 5.84 | 78.75 (75.38, 82.88) | −12.27 | p < 0.001 |
| Weight (kg) 2 | 24.39 ± 3.64 | 24.5 (21.75, 26.8) | 36.03 ± 6.40 | 34.7 (32.15, 39.5) | −12.56 | p < 0.001 |
| BMI (kg/m2) 3 | 16.07 ± 1.62 | 16.46 (14.73, 17.42) | 21.98 ± 3.03 | 21.54 (20.19, 23.91) | −9.13 | p < 0.001 |
| PBF (%) 2 | 20.13 ± 6.02 | 19.5 (15.25, 23.8) | 36.05 ± 6.23 | 36.7 (32.9, 39) | −14.58 | p < 0.001 |
| VFA (cm2) 3 | 20.81 ± 8.44 | 18 (15.65, 24.75) | 66.06 ± 30.13 | 64.9 (46.2, 82.6) | −8.66 | p < 0.001 |
| Parameters | FLD_HL (n = 41) | FLD_LL (n = 22) | t/χ2/Z | p-Value | ||
|---|---|---|---|---|---|---|
| ± SD | M (q1, q3) | ± SD | M (q1, q3) | |||
| Boy (n, %) 1 | 33 (80.49) | 14 (63.64) | 2.15 | 0.143 | ||
| Age (years) 2 | 6.72 ± 0.30 | 6.71 (6.46, 6.9) | 6.79 ± 0.32 | 6.73 (6.6, 6.96) | −0.74 | 0.462 |
| Height (cm) 2 | 127.57 ± 4.53 | 127.3 (124.35, 129.25) | 127.99 ± 4.39 | 127.68 (124.85, 131.11) | −0.35 | 0.725 |
| Waist (cm) 2 | 71.27 ± 8.01 | 70.25 (66.75, 77.35) | 71.01 ± 9.14 | 70.88 (63.98, 74.21) | 0.11 | 0.909 |
| Hip (cm) 2 | 78.92 ± 5.79 | 78.75 (74.7, 82.75) | 80.32 ± 5.94 | 79.9 (77.06, 82.41) | −0.9 | 0.371 |
| Weight (kg) 2 | 35.84 ± 6.05 | 34.6 (32.7, 39.8) | 36.40 ± 7.13 | 35.6 (31.28, 38.98) | −0.31 | 0.755 |
| BMI (kg/m2) 3 | 21.89 ± 2.63 | 21.57 (20.41, 24.18) | 22.14 ± 3.73 | 21.13 (19.92, 23.36) | −0.45 | 0.65 |
| PBF (%) 2 | 35.73 ± 6.27 | 36.8 (33.1, 38.4) | 36.65 ± 6.27 | 36.35 (32.85, 40.38) | −0.55 | 0.583 |
| VFA (cm2) 3 | 64.52 ± 28.72 | 65.8 (46.1, 82.4) | 68.92 ± 33.10 | 61.85 (48.5, 82.4) | −0.08 | 0.937 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, T.; Li, D.; Zunong, J.; Li, M.; Amaerjiang, N.; Xiao, H.; Khattab, N.M.; Vermund, S.H.; Hu, Y. Interplay of Lymphocytes with the Intestinal Microbiota in Children with Nonalcoholic Fatty Liver Disease. Nutrients 2022, 14, 4641. https://doi.org/10.3390/nu14214641
Liang T, Li D, Zunong J, Li M, Amaerjiang N, Xiao H, Khattab NM, Vermund SH, Hu Y. Interplay of Lymphocytes with the Intestinal Microbiota in Children with Nonalcoholic Fatty Liver Disease. Nutrients. 2022; 14(21):4641. https://doi.org/10.3390/nu14214641
Chicago/Turabian StyleLiang, Tian, Dan Li, Jiawulan Zunong, Menglong Li, Nubiya Amaerjiang, Huidi Xiao, Nourhan M. Khattab, Sten H. Vermund, and Yifei Hu. 2022. "Interplay of Lymphocytes with the Intestinal Microbiota in Children with Nonalcoholic Fatty Liver Disease" Nutrients 14, no. 21: 4641. https://doi.org/10.3390/nu14214641
APA StyleLiang, T., Li, D., Zunong, J., Li, M., Amaerjiang, N., Xiao, H., Khattab, N. M., Vermund, S. H., & Hu, Y. (2022). Interplay of Lymphocytes with the Intestinal Microbiota in Children with Nonalcoholic Fatty Liver Disease. Nutrients, 14(21), 4641. https://doi.org/10.3390/nu14214641







