A Systems Analysis of Phenotype Heterogeneity in APOE*3Leiden.CETP Mice Induced by Long-Term High-Fat High-Cholesterol Diet Feeding
Abstract
:1. Introduction
2. Methods
2.1. Animals, Diet and Housing
2.2. Experimental Setup
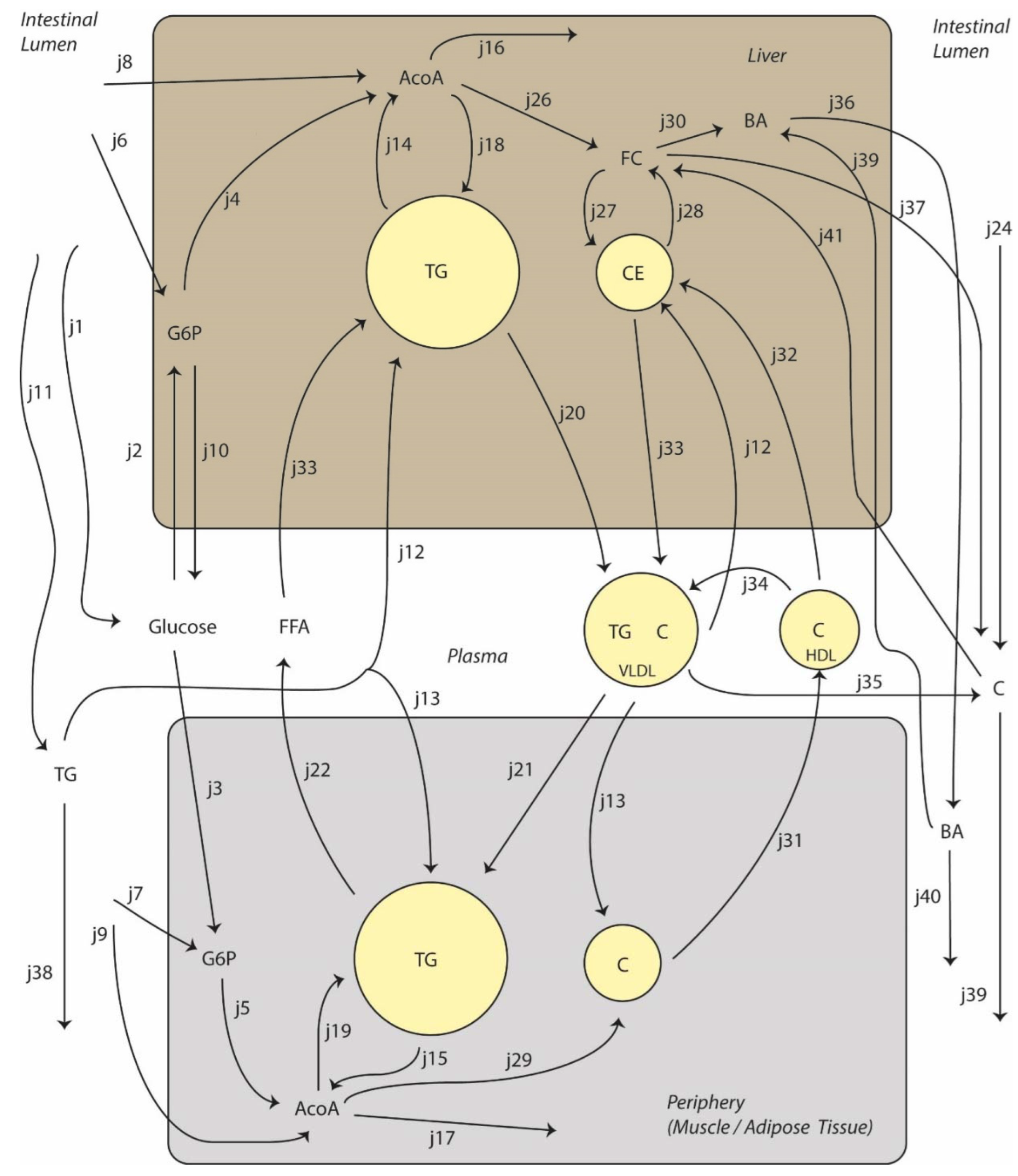
2.3. The ADAPT Method
2.4. Experimental Data and Modeling Constraints
2.5. Choices Concerning Model Design
2.6. Validation Experiment
3. Results
3.1. Stratification to Responder and Non-Responder Phenotypes
3.2. Application of ADAPT
3.3. ADAPT Predicts Decreased Cholesterol Absorption for Non-Responders
3.4. Validation Experiment of Decreased Cholesterol Absorption
3.5. ADAPT Predicts Higher Glucose Oxidation Rates in Non-responders
3.6. ADAPT Predicts Lower Fat Absorption in Non-responders
3.7. Decreased Fat Absorption Is Associated with a Lower Hydrophobicity Index of Fecal Bile Acids
4. Discussion
4.1. Hybrid Modeling Using ADAPT
4.2. Fat Absorption and Energy Expenditure
4.3. Bile Acid Metabolism
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Grundy, S.M. Hypertriglyceridemia, Atherogenic Dyslipidemia, and the Metabolic Syndrome. Am. J. Cardiol. 1998, 81, 18B–25B. [Google Scholar] [CrossRef]
- Locke, A.E.; Kahali, B.; Berndt, S.I.; Justice, A.E.; Pers, T.H.; Day, F.R.; Powell, C.; Vedantam, S.; Buchkovich, M.L.; Yang, J.; et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paalvast, Y.; Gerding, A.; Wang, Y.; Bloks, V.W.; van Dijk, T.H.; Havinga, R.; Willems van Dijk, K.; Rensen, P.C.; Bakker, B.M.; Kuivenhoven, J.A.; et al. Male apoE*3-Leiden.CETP mice on high-fat high-cholesterol diet exhibit a biphasic dyslipidemic response, mimicking the changes in plasma lipids observed through life in men. Physiol. Rep. 2017, 5, e13376. [Google Scholar] [CrossRef]
- Westerterp, M.; van der Hoogt, C.C.; de Haan, W.; Offerman, E.H.; Dallinga-Thie, G.M.; Jukema, J.W.; Havekes, L.M.; Rensen, P.C. Cholesteryl ester transfer protein decreases high-density lipoprotein and severely aggravates atherosclero-sis in APOE*3-Leiden mice. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 2552–2559. [Google Scholar] [CrossRef] [Green Version]
- Tiemann, C.A.; Vanlier, J.; Hilbers, P.A.; van Riel, N.A. Parameter adaptations during phenotype transitions in progressive diseases. BMC Syst. Biol. 2011, 5, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozendaal, Y.J.W.; Wang, Y.; Paalvast, Y.; Tambyrajah, L.L.; Li, Z.; Van Dijk, K.W.; Rensen, P.C.N.; Kuivenhoven, J.A.; Groen, A.K.; Hilbers, P.A.J.; et al. In vivo and in silico dynamics of the development of Metabolic Syndrome. PLOS Comput. Biol. 2018, 14, e1006145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiemann, C.A.; Vanlier, J.; Oosterveer, M.H.; Groen, A.K.; Hilbers, P.A.J.; van Riel, N.A.W. Parameter Trajectory Analysis to Identify Treatment Effects of Pharmacological Interventions. PLoS Comput. Biol. 2013, 9, e1003166. [Google Scholar] [CrossRef] [Green Version]
- Rye, K.-A.; Barter, P.J. Regulation of High-Density Lipoprotein Metabolism. Circ. Res. 2014, 114, 143–156. [Google Scholar] [CrossRef] [Green Version]
- Vrins, C.L.; Ottenhoff, R.; van den Oever, K.; de Waart, D.R.; Kruyt, J.K.; Zhao, Y.; van Berkel, T.J.; Havekes, L.M.; Aerts, J.M.; van Eck, M.; et al. Trans-intestinal cholesterol efflux is not mediated through high density lipo-protein. J. Lipid Res. 2012, 53, 2017–2023. [Google Scholar] [CrossRef] [Green Version]
- van der Veen, J.N.; van Dijk, T.H.; Vrins, C.L.; van Meer, H.; Havinga, R.; Bijsterveld, K.; Tietge, U.J.; Groen, A.K.; Kuipers, F. Faculty Opinions recommendation of Activation of the liver X receptor stimulates trans-intestinal excretion of plasma cholesterol. J. Biol. Chem. 2009, 284, 19211–19219. [Google Scholar] [CrossRef]
- Van den Kamer, J.H.; Huinink, B.O.; Weyers, H.A. Method for the determination of fat in feces. J. Biol. Chem. 1949, 177, 347–355. [Google Scholar] [CrossRef]
- Siperstein, M.; Guest, M. Studies on the site of the feedback control of cholesterol synthesis. J. Biol. Chem. 1960, 39, 642–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heuman, D.M. Quantitative estimation of the hydrophilic-hydrophobic balance of mixed bile salt solutions. J. Lipid Res. 1989, 30, 719–730. [Google Scholar] [CrossRef]
- Wang, D.Q.H.; Tazuma, S.; Cohen, D.E.; Carey, M.C. Feeding natural hydrophilic bile acids inhibits intestinal choles-terol absorption: Studies in the gallstone-susceptible mouse. Am. J. Physiol. Gastrointest. Liver Physiol. 2003, 285, G494–G502. [Google Scholar] [CrossRef] [Green Version]
- Chevre, R.; Trigueros-Motos, L.; Castaño, D.; Chua, T.; Corlianò, M.; Patankar, J.V.; Sng, L.; Sim, L.; Juin, T.L.; Carissimo, G.; et al. Therapeutic modulation of the bile acid pool by Cyp8b1 knockdown protects against nonalcoholic fatty liver disease in mice. FASEB J. 2018, 32, 3792–3802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higuchi, S.; Ahmad, T.R.; Argueta, D.A.; Perez, P.A.; Zhao, C.; Schwartz, G.J.; DiPatrizio, N.V.; Haeusler, R.A. Bile acid composition regulates GPR119-dependent intestinal lipid sensing and food intake regulation in mice. Gut 2020, 69, 1620–1628. [Google Scholar] [CrossRef] [Green Version]
- Regazzoni, F.; Chapelle, D.; Moireau, P. Combining data assimilation and machine learning to build data-driven models for unknown long time dynamics—Applications in cardiovascular modeling. Int. J. Numer. Methods Biomed. Eng. 2021, 37, e3471. [Google Scholar] [CrossRef]
- van Riel, N.A.W.; Tiemann, C.A.; Hilbers, P.A.J.; Groen, A.K. Metabolic Modeling Combined with Machine Learning Integrates Longitudinal Data and Identifies the Origin of LXR-Induced Hepatic Steatosis. Front. Bioeng. Biotechnol. 2021, 8, 536957. [Google Scholar] [CrossRef]
- Tarasco, E.; Pellegrini, G.; Whiting, L.; Lutz, T.A. Phenotypical heterogeneity in responder and nonresponder male ApoE*3Leiden.CETP mice. Am. J. Physiol. Liver Physiol. 2018, 315, G602–G617. [Google Scholar] [CrossRef]
- Butler, A.A.; Kozak, L.P. A recurring problem with the analysis of energy expenditure in genetic models express-ing lean and obese phenotypes. Diabetes 2010, 59, 323–329. [Google Scholar] [CrossRef]
- Even, P.C.; Nadkarni, N.A. Indirect calorimetry in laboratory mice and rats: Principles, practical considerations, interpretation and perspectives. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 303, R459–R476. [Google Scholar] [CrossRef] [PubMed]
- Buhman, K.K.; Smith, S.J.; Stone, S.J.; Repa, J.J.; Wong, J.S.; Knapp, F.; Burri, B.J.; Hamilton, R.L.; Abumrad, N.A.; Farese, R.V., Jr. DGAT1 Is Not Essential for Intestinal Triacylglycerol Absorption or Chylomicron Synthesis. J. Biol. Chem. 2002, 277, 25474–25479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carswell, K.A.; Vincent, R.P.; Belgaumkar, A.P.; Sherwood, R.A.; Amiel, S.A.; Patel, A.G.; Le Roux, C.W. The effect of bari-atric surgery on intestinal absorption and transit time. Obes. Surg. 2014, 24, 796–805. [Google Scholar] [CrossRef]
- Gribble, F.M.; Reimann, F. Enteroendocrine Cells: Chemosensors in the Intestinal Epithelium. Annu. Rev. Physiol. 2016, 78, 277–299. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Alves, J.M.; Hylemon, P.B.; Bajaj, J.S. Cirrhosis, bile acids and gut microbiota: Unraveling a complex relationship. Gut Microbes 2013, 4, 382–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridlon, J.M.; Kang, D.-J.; Hylemon, P.B.; Bajaj, J.S. Gut Microbiota, Cirrhosis, and Alcohol Regulate Bile Acid Metabolism in the Gut. Dig. Dis. 2015, 33, 338–345. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paalvast, Y.; Zhou, E.; Rozendaal, Y.J.W.; Wang, Y.; Gerding, A.; van Dijk, T.H.; de Boer, J.F.; Rensen, P.C.N.; van Dijk, K.W.; Kuivenhoven, J.A.; et al. A Systems Analysis of Phenotype Heterogeneity in APOE*3Leiden.CETP Mice Induced by Long-Term High-Fat High-Cholesterol Diet Feeding. Nutrients 2022, 14, 4936. https://doi.org/10.3390/nu14224936
Paalvast Y, Zhou E, Rozendaal YJW, Wang Y, Gerding A, van Dijk TH, de Boer JF, Rensen PCN, van Dijk KW, Kuivenhoven JA, et al. A Systems Analysis of Phenotype Heterogeneity in APOE*3Leiden.CETP Mice Induced by Long-Term High-Fat High-Cholesterol Diet Feeding. Nutrients. 2022; 14(22):4936. https://doi.org/10.3390/nu14224936
Chicago/Turabian StylePaalvast, Yared, Enchen Zhou, Yvonne J. W. Rozendaal, Yanan Wang, Albert Gerding, Theo H. van Dijk, Jan Freark de Boer, Patrick C. N. Rensen, Ko Willems van Dijk, Jan A. Kuivenhoven, and et al. 2022. "A Systems Analysis of Phenotype Heterogeneity in APOE*3Leiden.CETP Mice Induced by Long-Term High-Fat High-Cholesterol Diet Feeding" Nutrients 14, no. 22: 4936. https://doi.org/10.3390/nu14224936
APA StylePaalvast, Y., Zhou, E., Rozendaal, Y. J. W., Wang, Y., Gerding, A., van Dijk, T. H., de Boer, J. F., Rensen, P. C. N., van Dijk, K. W., Kuivenhoven, J. A., Bakker, B. M., van Riel, N. A. W., & Groen, A. K. (2022). A Systems Analysis of Phenotype Heterogeneity in APOE*3Leiden.CETP Mice Induced by Long-Term High-Fat High-Cholesterol Diet Feeding. Nutrients, 14(22), 4936. https://doi.org/10.3390/nu14224936




