Effect of Dietary Protein and Processing on Gut Microbiota—A Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Literature Search
2.2. Study Selection Criteria
2.3. Data Extraction
- (a)
- If the most abundant population in that diet group, one asterisk was placed after classification (*);
- (b)
- If the most or least abundant population in that diet group compared to all other diet groups, two asterisks were placed after classification (**);
- (c)
- If the most abundant population in that diet group, as well as compared to other diet groups, three asterisks were placed after classification (***);
- (d)
- If least abundant compared to other diet groups but most abundant in that diet group, four asterisks were placed after classification (****);
- (e)
- If there are no differences in the number of asterisks for a particular bacterial population between diet groups, then there are no differences in the abundance of the corresponding microbial population amongst those diet groups.
3. Results
3.1. Selected Studies
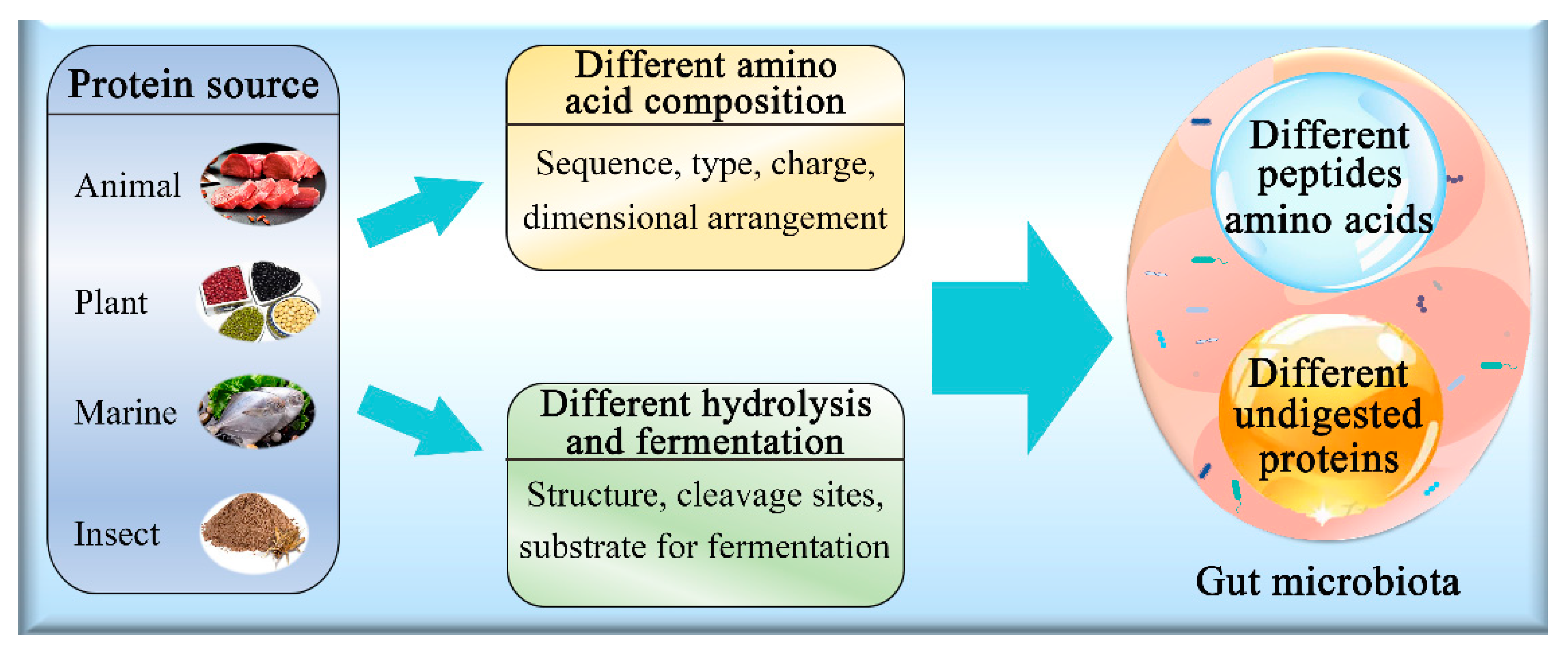
3.2. Effect of Protein Source on Gut Microbiota
3.3. Animal and Plant Proteins
3.4. Effect of Protein Content and Diet Composition on Gut Microbiota
3.4.1. Protein Content
3.4.2. Other Macronutrients
3.5. Effect of Processing Technologies on Dietary Protein to Influence Gut Microbiota
3.5.1. Protein Glycation
3.5.2. Protein Oxidation
3.6. Effect of Protein Structure on the Gut Microbiota
4. Strengths and Limitations of This Review
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Agirman, G.; Hsiao, E.Y. SnapShot: The microbiota-gut-brain axis. Cell 2021, 184, 2524. [Google Scholar] [CrossRef]
- Hill, J.H.; Round, J.L. SnapShot: Microbiota effects on host physiology. Cell 2021, 184, 2796. [Google Scholar] [CrossRef]
- Danneskiold-Samsøe, N.B.; De Freitas Queiroz Barros, H.D.; Santos, R.; Bicas, J.L.; Cazarin, C.B.B.; Madsen, L.; Kristiansen, K.; Pastore, G.M.; Brix, S.; Maróstica Júnior, M.R. Interplay between food and gut microbiota in health and disease. Food Res. Int. 2019, 115, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Wastyk, H.C.; Fragiadakis, G.K.; Perelman, D.; Dahan, D.; Merrill, B.D.; Yu, F.B.; Topf, M.; Gonzalez, C.G.; Van Treuren, W.; Han, S.; et al. Gut-microbiota-targeted diets modulate human immune status. Cell 2021, 184, 4137–4153. [Google Scholar] [CrossRef] [PubMed]
- Olson, C.A.; Iñiguez, A.J.; Yang, G.E.; Fang, P.; Pronovost, G.N.; Jameson, K.G.; Rendon, T.K.; Paramo, J.; Barlow, J.T.; Ismagilov, R.F.; et al. Alterations in the gut microbiota contribute to cognitive impairment induced by the ketogenic diet and hypoxia. Cell Host Microbe 2021, 29, 1378–1392.e6. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Duan, M.; Jia, J.; Song, S.; Ai, C. Low-molecular alginate improved diet-induced obesity and metabolic syndrome through modulating the gut microbiota in BALB/c mice. Int. J. Biol. Macromol. 2021, 187, 811–820. [Google Scholar] [CrossRef]
- Diether, N.; Willing, B. Microbial fermentation of dietary protein: An important factor in diet–microbe–host interaction. Microorganisms 2019, 7, 19. [Google Scholar] [CrossRef] [Green Version]
- Fan, P.; Li, L.; Rezaei, A.; Eslamfam, S.; Che, D.; Ma, X. Metabolites of dietary protein and peptides by intestinal microbes and their impacts on gut. Curr. Prot. Pept. Sci. 2015, 16, 646–654. [Google Scholar] [CrossRef]
- Kim, E.; Kim, D.; Park, J. Changes of mouse gut microbiota diversity and composition by modulating dietary protein and carbohydrate contents: A pilot study. Prevent. Nutr. Food Sci. 2016, 21, 57–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bekhit, A.; Giteru, S.G.; Holman, B.; Hopkins, D.L. Total volatile basic nitrogen and trimethylamine in muscle foods: Potential formation pathways and effects on human health. Comp. Rev. Food Sci. Food Saf. 2021, 20, 3620–3666. [Google Scholar] [CrossRef] [PubMed]
- Portune, K.J.; Beaumont, M.; Davila, A.; Tomé, D.; Blachier, F.; Sanz, Y. Gut microbiota role in dietary protein metabolism and health-related outcomes: The two sides of the coin. Trends Food Sci. Technol. 2016, 57, 213–232. [Google Scholar] [CrossRef] [Green Version]
- Jantchou, P.; Morois, S.; Clavel-Chapelon, F.; Boutron-Ruault, M.C.; Carbonnel, F. Animal protein intake and risk of inflammatory bowel disease: The E3N prospective study. Am. J. Gastroenterol. 2010, 105, 2195–2201. [Google Scholar] [CrossRef]
- Batterham, R.L.; Heffron, H.; Kapoor, S.; Chivers, J.E.; Chandarana, K.; Herzog, H.; Le Roux, C.W.; Thomas, E.L.; Bell, J.D.; Withers, D.J. Critical role for peptide YY in protein-mediated satiation and body-weight regulation. Cell Metab. 2006, 4, 223–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moher, D.; Shamseer, L.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst. Rev. 2015, 4, 1. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Blouin, J.; Santacruz, A.; Lan, A.; Andriamihaja, M.; Wilkanowicz, S.; Benetti, P.; Tomé, D.; Sanz, Y.; Blachier, F.; et al. High-protein diet modifies colonic microbiota and luminal environment but not colonocyte metabolism in the rat model: The increased luminal bulk connection. Am. J. Physiol.-Gastrointest. Liver Physiol. 2014, 307, G459–G470. [Google Scholar] [CrossRef] [Green Version]
- Boudry, G.; Jamin, A.; Chatelais, L.; Gras-Le, G.C.; Michel, C.; Le Huerou-Luron, I. Dietary protein excess during neonatal life alters colonic microbiota and mucosal response to inflammatory mediators later in life in female pigs. J. Nutr. 2013, 143, 1225–1232. [Google Scholar] [CrossRef] [Green Version]
- Mu, C.; Yang, Y.; Luo, Z.; Guan, L.; Zhu, W. The colonic microbiome and epithelial transcriptome are altered in rats fed a high-protein diet compared with a normal-protein diet. J. Nutr. 2016, 146, 474–483. [Google Scholar] [CrossRef] [Green Version]
- Mu, C.; Yang, Y.; Luo, Z.; Zhu, W. Temporal microbiota changes of high-protein diet intake in a rat model. Anaerobe 2017, 47, 218–225. [Google Scholar] [CrossRef]
- Kar, S.K.; Jansman, A.J.M.; Benis, N.; Ramiro-Garcia, J.; Schokker, D.; Kruijt, L.; Stolte, E.H.; Taverne-Thiele, J.J.; Smits, M.A.; Wells, J.M. Dietary protein sources differentially affect microbiota, mTOR activity and transcription of mTOR signaling pathways in the small intestine. PLoS ONE 2017, 12, e188282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Y.; Wang, C.; Zhao, D.; Zhou, G.; Li, C. Processing method altered mouse intestinal morphology and microbial composition by affecting digestion of meat proteins. Front. Microbiol. 2020, 11, 511. [Google Scholar] [CrossRef]
- Zhao, F.; Zhou, G.; Liu, X.; Song, S.; Xu, X.; Hooiveld, G.; Müller, M.; Liu, L.; Kristiansen, K.; Li, C. Dietary protein sources differentially affect the growth of Akkermansia muciniphila and maintenance of the gut mucus barrier in mice. Mol. Nutr. Food Res. 2019, 63, 1900589. [Google Scholar] [CrossRef]
- Qi, H.; Xiang, Z.; Han, G.; Yu, B.; Huang, Z.; Chen, D. Effects of different dietary protein sources on cecal microflora in rats. Afr. J. Biotechnol. 2011, 10, 3704–3708. [Google Scholar]
- An, C.; Kuda, T.; Yazaki, T.; Takahashi, H.; Kimura, B. Caecal fermentation, putrefaction and microbiotas in rats fed milk casein, soy protein or fish meal. Appl. Microbiol. Biotechnol. 2014, 98, 2779–2787. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Song, S.; Ma, Y.; Xu, X.; Zhou, G.; Li, C. A short-term feeding of dietary casein increases abundance of Lactococcus lactis and upregulates gene expression involving obesity prevention in cecum of young rats compared with dietary chicken protein. Front. Microbiol. 2019, 10, 511. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Lin, X.; Zhao, F.; Shi, X.; Li, H.; Li, Y.; Zhu, W.; Xu, X.; Li, C.; Zhou, G. Meat, dairy and plant proteins alter bacterial composition of rat gut bacteria. Sci. Rep. 2015, 5, 15220. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Lin, X.; Li, H.; Li, Y.; Shi, X.; Zhao, F.; Xu, X.; Li, C.; Zhou, G. Intake of meat proteins substantially increased the relative abundance of genus Lactobacillus in rat feces. PLoS ONE 2016, 11, e152678. [Google Scholar] [CrossRef] [Green Version]
- Zhao, F.; Huang, Z.; Zhou, G.; Li, H.; Xu, X.; Li, C. Dietary proteins rapidly altered the microbial composition in rat caecum. Curr. Microbiol. 2017, 74, 1447–1452. [Google Scholar] [CrossRef]
- Zhu, Y.; Shi, X.; Lin, X.; Ye, K.; Xu, X.; Li, C.; Zhou, G. Beef, chicken, and soy proteins in diets induce different gut microbiota and metabolites in rats. Front. Microbiol. 2017, 8, 1395. [Google Scholar] [CrossRef]
- Yu, H.; Qiu, N.; Meng, Y.; Keast, R. A comparative study of the modulation of the gut microbiota in rats by dietary intervention with different sources of egg-white proteins. J. Sci. Food Agric. 2020, 100, 3622–3629. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Jin, W.; Mao, Z.; Dong, S.; Zhang, Q.; Yang, Y.; Chen, B.; Wu, H.; Zeng, M. Microbiome and butyrate production are altered in the gut of rats fed a glycated fish protein diet. J. Funct. Foods 2018, 47, 423–433. [Google Scholar] [CrossRef]
- Oberli, M.; Douard, V.; Beaumont, M.; Jaoui, D.; Devime, F.; Laurent, S.; Chaumontet, C.; Mat, D.; Le Feunteun, S.; Michon, C.; et al. Lipo-protein emulsion structure in the diet affects protein digestion kinetics, intestinal mucosa parameters and microbiota composition. Mol. Nutr. Food Res. 2018, 62, 1700570. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, M.; Jaoui, D.; Douard, V.; Mat, D.; Koeth, F.; Goustard, B.; Mayeur, C.; Mondot, S.; Hovaghimian, A.; Le Feunteun, S.; et al. Structure of protein emulsion in food impacts intestinal microbiota, caecal luminal content composition and distal intestine characteristics in rats. Mol. Nutr. Food Res. 2017, 61, 1700078. [Google Scholar] [CrossRef]
- Ortman, J.; Sinn, S.M.; Gibbons, W.R.; Brown, M.L.; DeRouchey, J.M.; St-Pierre, B.; Saqui-Salces, M.; Levesque, C.L. Comparative analysis of the ileal bacterial composition of post-weaned pigs fed different high-quality protein sources. Animal 2020, 14, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Opheim, M.; Strube, M.L.; Sterten, H.; Overland, M.; Kjos, N.P. Atlantic salmon (Salmo salar) protein hydrolysate in diets for weaning piglets horizontal line effect on growth performance, intestinal morphometry and microbiota composition. Arch. Anim. Nutr. 2016, 70, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Rist, V.T.S.; Weiss, E.; Sauer, N.; Mosenthin, R.; Eklund, M. Effect of dietary protein supply originating from soybean meal or casein on the intestinal microbiota of piglets. Anaerobe 2014, 25, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Chang, L.; Hou, G.; Song, Z.; Fan, Z.; He, X.; Hou, D. Colonic microbiota and metabolites response to different dietary protein sources in a Piglet model. Front. Nutr. 2019, 6, 151. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Li, Z.; Chen, W.; Rong, T.; Wang, G.; Ma, X. Hermetia illucens larvae as a potential dietary protein source altered the microbiota and modulated mucosal immune status in the colon of finishing pigs. J. Anim. Sci. Biotechnol. 2019, 10, 50. [Google Scholar] [CrossRef]
- Schutkowski, A.; König, B.; Kluge, H.; Hirche, F.; Henze, A.; Schwerdtle, T.; Lorkowski, S.; Dawczynski, C.; Gabel, A.; Große, I.; et al. Metabolic footprint and intestinal microbial changes in response to dietary proteins in a pig model. J. Nutr. Biochem. 2019, 67, 149–160. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, H.; Dong, S.; Jin, W.; Han, K.; Ren, Y.; Zeng, M. Glycation of fish protein impacts its fermentation metabolites and gut microbiota during in vitro human colonic fermentation. Food Res. Int. 2018, 113, 189–196. [Google Scholar] [CrossRef]
- Ge, Y.; Lin, S.; Li, B.; Yang, Y.; Tang, X.; Shi, Y.; Sun, J.; Le, G. Oxidized pork induces oxidative stress and inflammation by altering gut microbiota in mice. Mol. Nutr. Food Res. 2020, 64, 1901012. [Google Scholar] [CrossRef]
- Van Hecke, T.; Vossen, E.; Goethals, S.; Boon, N.; De Vrieze, J.; De Smet, S. In vitro and in vivo digestion of red cured cooked meat: Oxidation, intestinal microbiota and fecal metabolites. Food Res. Int. 2021, 142, 110203. [Google Scholar] [CrossRef]
- Snelson, M.; Clarke, R.E.; Nguyen, T.V.; Penfold, S.A.; Forbes, J.M.; Tan, S.M.; Coughlan, M.T. Long term high protein diet feeding alters the microbiome and increases intestinal permeability, systemic inflammation and kidney injury in mice. Mol. Nutr. Food Res. 2021, 65, 2000851. [Google Scholar] [CrossRef]
- Nasrollahzadeh, M.; Nezafat, Z.; Shafiei, N. Biopolymer-Based Metal Nanoparticle Chemistry for Sustainable Applications. In Proteins in Food Industry; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Hicks, L.M.; Verbeek, C.J.R. Meat Industry Protein by-Products: Sources and Characteristics. In Protein Byproducts; Gurpreet, S.D., Ed.; Academic Press: Edmonton, AB, Canada, 2016. [Google Scholar]
- Feher, J. Protein Structure. In Quantitative Human Physiology; Joseph, F., Ed.; Academic Press: Richmond, VA, USA, 2012; pp. 100–109. [Google Scholar]
- Wu, G. Dietary protein intake and human health. Food Funct. 2016, 7, 1251–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.; Bekhit, A.E.A.; Wu, Q.; Chen, M.; Liao, X.; Wang, J.; Ding, Y. Bioactive peptides and gut microbiota: Candidates for a novel strategy for reduction and control of neurodegenerative diseases. Trends Food Sci. Technol. 2021, 108, 164–176. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, X.; Liu, H.; Brown, M.A.; Qiao, S. Dietary protein and gut microbiota composition and function. Curr. Prot. Pept. Sci. 2019, 20, 145. [Google Scholar] [CrossRef]
- Lund, M.N.; Heinonen, M.; Baron, C.P.; Estévez, M. Protein oxidation in muscle foods: A review. Mol. Nutr. Food Res. 2011, 55, 83–95. [Google Scholar] [CrossRef]
- Vlieg, J.E.V.H.; Veiga, P.; Zhang, C.; Derrien, M.; Zhao, L. Impact of microbial transformation of food on health-from fermented foods to fermentation in the gastro-intestinal tract. Curr. Opin. Biotechnol. 2011, 22, 211–219. [Google Scholar] [CrossRef]
- Wu, L.; Tang, Z.; Chen, H.; Ren, Z.; Ding, Q.; Liang, K.; Sun, Z. Mutual interaction between gut microbiota and protein/amino acid metabolism for host mucosal immunity and health. Anim. Nutr. 2021, 7, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Li, Y.; Han, H.; Liu, Z.; Zeng, X.; Li, T.; Yin, Y. Long-term effects of lysine concentration on growth performance, intestinal microbiome, and metabolic profiles in a pig model. Food Funct. 2018, 9, 4153–4163. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, Y.; Xu, Y.; Luo, T.; Ge, Y.; Jiang, Y.; Shi, Y.; Sun, J.; Le, G. Dietary methionine restriction improves the gut microbiota and reduces intestinal permeability and inflammation in high-fat-fed mice. Food Funct. 2019, 10, 5952–5968. [Google Scholar] [CrossRef]
- Windey, K.; De Preter, V.; Verbeke, K. Relevance of protein fermentation to gut health. Mol. Nutr. Food Res. 2012, 56, 184–196. [Google Scholar] [CrossRef]
- Li, R.; Hou, G.F.; Song, Z.H.; Zhao, J.F.; Fan, Z.Y.; Hou, D.; He, X. Nutritional value of enzyme-treated soybean meal, concentrated degossypolized cottonseed protein, dried porcine solubles and fish meal for 10- to −20 kg pigs. Anim. Feed Sci. Technol. 2019, 252, 23–33. [Google Scholar] [CrossRef]
- Dangin, M.; Boirie, Y.; Garcia-Rodenas, C.; Gachon, P.; Fauquant, J.; Callier, P.; Ballevre, O.; Beaufrere, B. The digestion rate of protein is an independent regulating factor of postprandial protein retention. Am. J. Physiol. Endocrinol. Metab. 2001, 280, E340–E348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhe, A.M.; Collier, G.R.; O’Dea, K. A comparison of the effects of beef, chicken and fish protein on satiety and amino acid profiles in lean male subjects. J. Nutr. 1992, 122, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Fouillet, H.; Juillet, B.; Gaudichon, C.; Mariotti, F.; Tomé, D.; Bos, C. Absorption kinetics are a key factor regulating postprandial protein metabolism in response to qualitative and quantitative variations in protein intake. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 297, R1691–R1705. [Google Scholar] [CrossRef] [Green Version]
- Kostic, A.D.; Gevers, D.; Pedamallu, C.S.; Michaud, M.; Duke, F.; Earl, A.M.; Ojesina, A.I.; Jung, J.; Bass, A.J.; Tabernero, J.; et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012, 22, 292–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Higuchi, Y.; Hosojima, M.; Kabasawa, H.; Kuwahara, S.; Goto, S.; Toba, K.; Kaseda, R.; Tanaka, T.; Kitamura, N.; Takihara, H.; et al. Rice endosperm protein administration to juvenile mice regulates gut microbiota and suppresses the development of high-fat diet-induced obesity and related disorders in adulthood. Nutrients 2019, 11, 2919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashaolu, T.J. Soy bioactive peptides and the gut microbiota modulation. Appl. Microbiol. Biotechnol. 2020, 104, 9009–9017. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Krishnan, H.B.; Pham, Q.; Yu, L.L.; Wang, T.T.Y. Soy and gut microbiota: Interaction and implication for human health. J. Agric. Food Chem. 2016, 64, 8695–8709. [Google Scholar] [CrossRef]
- Han, W.; Zhuang, X.; Liu, Q.; Sun, B.; Miao, H.; Zhang, X. Fermented soy whey induced changes on intestinal microbiota and metabolic influence in mice. Food Sci. Hum. Wellness 2022, 11, 41–48. [Google Scholar] [CrossRef]
- Kumar, S.; Bhat, Z.F.; Kumar, P. Functional meat and meat products. In Animal Products Technology; Mandal, P.K., Biswas, A.K., Eds.; Studium Press: New Delhi, India, 2013; pp. 404–455. [Google Scholar]
- Bhat, Z.F.; Bhat, H. Fibre-based functional meat products. Asian J. Food Agro-Ind. 2011, 4, 261–273. [Google Scholar]
- Mariotti, F. Vegetarian and plant-based diets in health and disease prevention. In Plant Protein, Animal Protein, and Protein Quality; Academic Press: Cambridge, MA, USA, 2017; pp. 621–642. [Google Scholar]
- Pereira, P.M.D.C.; Vicente, A.F.D.R. Meat nutritional composition and nutritive role in the human diet. Meat Sci. 2013, 93, 586–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, J.A.; Bendsen, N.T.; Tremblay, A.; Astrup, A. Effect of proteins from different sources on body composition. Nutr. Metab. Cardiovasc. Dis. 2011, 21, B16–B31. [Google Scholar] [CrossRef]
- Rodriguez, N.R.; Miller, S.L. Effective translation of current dietary guidance: Understanding and communicating the concepts of minimal and optimal levels of dietary protein. Am. J. Clinic. Nutr. 2015, 101, 1353S–1358S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguirre, M.; Eck, A.; Koenen, M.E.; Savelkoul, P.H.M.; Budding, A.E.; Venema, K. Diet drives quick changes in the metabolic activity and composition of human gut microbiota in a validated in vitro gut model. Res. Microbiol. 2016, 167, 114–125. [Google Scholar] [CrossRef]
- Kim, E.; Coelho, D.; Blachier, F. Review of the association between meat consumption and risk of colorectal cancer. Nutr. Res. 2013, 33, 983–994. [Google Scholar] [CrossRef]
- Stecher, B.; Hardt, W. Mechanisms controlling pathogen colonization of the gut. Curr. Opin. Microbiol. 2010, 14, 82–91. [Google Scholar] [CrossRef]
- Amaretti, A.; Gozzoli, C.; Simone, M.; Raimondi, S.; Righini, L.; Pérez-Brocal, V.; García-López, R.; Moya, A.; Rossi, M. Profiling of protein degraders in cultures of human gut microbiota. Front. Microbiol. 2019, 10, 2614. [Google Scholar] [CrossRef] [PubMed]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef] [Green Version]
- Smith, E.A.; Macfarlane, G.T. Enumeration of amino acid fermenting bacteria in the human large intestine: Effects of pH and starch on peptide metabolism and dissimilation of amino acids. Fem. Microbiol. Ecol. 1998, 25, 355–368. [Google Scholar] [CrossRef]
- Zackular, J.P.; Baxter, N.T.; Iverson, K.D.; Sadler, W.D.; Petrosino, J.F.; Chen, G.Y.; Schloss, P.D.; Blaser, M.J. The gut microbiome modulates colon tumorigenesis. Mbio 2013, 4, e692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Q.; Jin, Z.; Wu, W.; Gao, R.; Guo, B.; Gao, Z.; Yang, Y.; Qin, H.; Hold, G.L. Analysis of the intestinal lumen microbiota in an animal model of colorectal cancer. PLoS ONE 2014, 9, e90849. [Google Scholar] [CrossRef] [PubMed]
- Varsha, K.K.; Nampoothiri, K.M. Appraisal of lactic acid bacteria as protective cultures. Food Cont. 2016, 69, 61–64. [Google Scholar] [CrossRef]
- Durica-Mitic, S.; Gopel, Y.; Gorke, B. Carbohydrate utilization in bacteria: Making the most out of sugars with the help of small regulatory RNAs. Microbiol. Spectr. 2018, 6, 2. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Q.; Cheng, L.; Buch, H.; Zhang, F. Akkermansia muciniphila is a promising probiotic. Microb. Biotechnol. 2019, 12, 1109–1125. [Google Scholar] [CrossRef] [Green Version]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.; Rodriguez-Palacios, A. The genus Alistipes: Gut bacteria with emerging implications to inflammation, cancer, and mental health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef]
- Drewes, J.L.; Domingue, J.C.; Housseau, F. Microbiota, Mucosal Immunity, and Colon Cancer; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 157–209. [Google Scholar]
- Nakamura, S.; Kuda, T.; Midorikawa, Y.; Takahashi, H.; Kimura, B. Typical gut indigenous bacteria in ICR mice fed a normal or soy protein-based low-protein diet. Curr. Res. Food Sci. 2021, 4, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.; Martín, M.Á. Impact of diet on gut microbiota. Curr. Opin. Food Sci. 2021, 37, 83–90. [Google Scholar] [CrossRef]
- Mokkala, K.; Houttu, N.; Cansev, T.; Laitinen, K. Interactions of dietary fat with the gut microbiota: Evaluation of mechanisms and metabolic consequences. Clin. Nutr. 2020, 39, 994–1018. [Google Scholar] [CrossRef]
- Payling, L.; Fraser, K.; Loveday, S.M.; Sims, I.; Roy, N.; McNabb, W. The effects of carbohydrate structure on the composition and functionality of the human gut microbiota. Trends Food Sci. Technol. 2020, 97, 233–248. [Google Scholar] [CrossRef]
- Mithul Aravind, S.; Wichienchot, S.; Tsao, R.; Ramakrishnan, S.; Chakkaravarthi, S. Role of dietary polyphenols on gut microbiota, their metabolites and health benefits. Food Res. Int. 2021, 142, 110189. [Google Scholar] [CrossRef]
- Ma, N.; Tian, Y.; Wu, Y.; Ma, X. Contributions of the interaction between dietary protein and gut microbiota to intestinal health. Curr. Prot. Pept. Sci. 2017, 18, 795–808. [Google Scholar] [CrossRef]
- Nakata, T.; Kyoui, D.; Takahashi, H.; Kimura, B.; Kuda, T. Inhibitory effects of soybean oligosaccharides and water-soluble soybean fibre on formation of putrefactive compounds from soy protein by gut microbiota. Int. J. Biol. Macromol. 2017, 97, 173–180. [Google Scholar] [CrossRef]
- Biddle, A.; Stewart, L.; Blanchard, J.; Leschine, S. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity 2013, 5, 627–640. [Google Scholar] [CrossRef]
- Russell, W.R.; Gratz, S.W.; Duncan, S.H.; Holtrop, G.; Ince, J.; Scobbie, L.; Duncan, G.; Johnstone, A.M.; Lobley, G.E.; Wallace, R.J.; et al. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am. J. Clin. Nutr. 2011, 93, 1062–1072. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Niu, F.; Li, J.; Su, Y.; Liu, Y.; Yang, Y. Interactions between tea polyphenol and two kinds of typical egg white proteins—ovalbumin and lysozyme: Effect on the gastrointestinal digestion of both proteins in vitro. Food Res. Int. 2014, 59, 100–107. [Google Scholar] [CrossRef]
- Mirmoghtadaie, L.; Aliabadi, S.S.; Hosseini, S.M. Recent approaches in physical modification of protein functionality. Food Chem. 2016, 199, 619–627. [Google Scholar] [CrossRef]
- Mitra, B.; Rinnan, Å.; Ruiz-Carrascal, J. Tracking hydrophobicity state, aggregation behaviour and structural modifications of pork proteins under the influence of assorted heat treatments. Food Res. Int. 2017, 101, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Neis, E.P.J.G.; Dejong, C.H.; Rensen, S.S. The role of microbial amino acid metabolism in host metabolism. Nutrients 2015, 7, 2930–2946. [Google Scholar] [CrossRef] [Green Version]
- Davila, A.M.; Blachier, F.; Gotteland, M.; Andriamihaja, M.; Benetti, P.H.; Sanz, Y.; Tome, D. Intestinal luminal nitrogen metabolism: Role of the gut microbiota and consequences for the host. Pharmacol. Res. 2013, 69, 114–126. [Google Scholar] [CrossRef]
- Blachier, F.; Mariotti, F.; Huneau, J.F.; Tome, D. Effects of amino acid-derived luminal metabolites on the colonic epithelium and physiopathological consequences. Amino Acids 2007, 33, 547–562. [Google Scholar] [CrossRef]
- Bhat, Z.F.; Morton, J.D.; Bekhit, A.E.A.; Kumar, S.; Bhat, H.F. Emerging processing technologies for improved digestibility of muscle proteins. Trends Food Sci. Technol. 2021, 110, 226–239. [Google Scholar] [CrossRef]
- Bhat, Z.F.; Morton, J.D.; Bekhit, A.E.D.A.; Kumar, S.; Bhat, H.F. Processing technologies for improved digestibility of milk proteins. Trends Food Sci. Technol. 2021, 118, 1–16. [Google Scholar] [CrossRef]
- Bhat, Z.F.; Morton, J.D.; Bekhit, A.E.D.A.; Kumar, S.; Bhat, H.F. Processing technologies for improved digestibility of egg proteins. Comp. Rev. Food Sci. 2021, 20, 1–36. [Google Scholar]
- Zhang, Z.; Zhang, X.; Chen, W.; Zhou, P. Conformation stability, in vitro digestibility and allergenicity of tropomyosin from shrimp (Exopalaemon modestus) as affected by high intensity ultrasound. Food Chem. 2018, 245, 997–1009. [Google Scholar] [CrossRef] [PubMed]
- Rysman, T.; Van Hecke, T.; Van Poucke, C.; De Smet, S.; Van Royen, G. Protein oxidation and proteolysis during storage and in vitro digestion of pork and beef patties. Food Chem. 2016, 209, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Bhat, Z.F.; Morton, J.D.; Bekhit, A.E.A.; Kumar, S.; Bhat, H.F. Non-thermal processing has an impact on the digestibility of the muscle proteins. Crit. Rev. Food Sci. 2021, 1–28. [Google Scholar] [CrossRef]
- Zhao, D.; He, J.; Zou, X.; Nian, Y.; Xu, X.; Zhou, G.; Li, C. Influence of salting process on the structure and in vitro digestibility of actomyosin. J. Food Sci. Technol. 2019, 57, 1763–1773. [Google Scholar] [CrossRef] [PubMed]
- Bhat, Z.F.; Morton, J.D.; Bekhit, A.E.D.A.; Kumar, S.; Bhat, H.F. Thermal processing implications on the digestibility of meat, fish and seafood proteins. Comp. Rev. Food Sci. 2021, 20, 4511–4548. [Google Scholar] [CrossRef]
- Bhat, Z.F.; Morton, J.D.; Zhang, X.; Mason, S.L.; Bekhit, A. Sous-vide cooking improves the quality and in-vitro digestibility of Semitendinosus from culled dairy cows. Food Res. Int. 2020, 127, 108708. [Google Scholar] [CrossRef]
- Canfora, E.E.; Meex, R.; Venema, K.; Blaak, E.E. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat. Rev. Endocrinol. 2019, 15, 261–273. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes ratio: A relevant marker of gut dysbiosis in obese patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef] [PubMed]
- Kaur, U.S.; Shet, A.; Rajnala, N.; Gopalan, B.P.; Moar, P.; Himanshu, D.; Singh, B.P.; Chaturvedi, R.; Tandon, R. High abundance of genus Prevotella in the gut of perinatally HIV-infected children is associated with IP-10 levels despite therapy. Sci. Rep. 2018, 8, 17679. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.; Liu, P.; Song, P.; Chen, X.; Ma, X. Moderate dietary protein restriction alters the composition of gut microbiota and improves ileal barrier function in adult pig model. Sci. Rep. 2017, 7, 43412. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhou, G.; Bai, Y.; Wang, C.; Zhu, S.; Xu, X.; Li, C. The effect of meat processing methods on changes in disulfide bonding and alteration of protein structures: Impact on protein digestion products. RSC Adv. 2018, 8, 17595–17605. [Google Scholar] [CrossRef] [Green Version]
- Collado, L.; Figueras, M.J. Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin. Microbiol. Rev. 2011, 24, 174–192. [Google Scholar] [CrossRef] [Green Version]
- Polansky, O.; Sekelova, Z.; Faldynova, M.; Sebkova, A.; Sisak, F.; Rychlik, I. Important metabolic pathways and biological processes expressed by chicken cecal microbiota. Appl. Environ. Microbiol. 2015, 82, 1569–1576. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Bayjanov, J.R.; Renckens, B.; Nauta, A.; Siezen, R.J. The proteolytic system of lactic acid bacteria revisited: A genomic comparison. BMC Genom. 2010, 11, 36. [Google Scholar] [CrossRef] [Green Version]
- Binda, C.; Lopetuso, L.R.; Rizzatti, G.; Gibiino, G.; Cennamo, V.; Gasbarrini, A. Actinobacteria: A relevant minority for the maintenance of gut homeostasis. Digest. Liver Dis. 2018, 50, 421–428. [Google Scholar] [CrossRef]
- Naito, Y.; Uchiyama, K.; Takagi, T. A next-generation beneficial microbe: Akkermansia muciniphila. J. Clin. Biochem. Nutr. 2018, 63, 33–35. [Google Scholar] [CrossRef] [Green Version]
- Geerlings, S.Y.; Kostopoulos, I.; de Vos, W.M.; Belzer, C. Akkermansia muciniphila in the Human Gastrointestinal Tract: When, Where, and How? Microorganisms 2018, 6, 75. [Google Scholar] [CrossRef] [Green Version]
- Berardo, A.; Devreese, B.; De Maere, H.; Stavropoulou, D.A.; Van Royen, G.; Leroy, F.; De Smet, S. Actin proteolysis during ripening of dry fermented sausages at different pH values. Food Chem. 2017, 221, 1322–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traore, S.; Aubry, L.; Gatellier, P.; Przybylski, W.; Jaworska, D.; Kajak-Siemaszko, K.; Sante-Lhoutellier, V. Higher drip loss is associated with protein oxidation. Meat Sci. 2012, 90, 917–924. [Google Scholar] [CrossRef]
- Sante-Lhoutellier, V.; Aubry, L.; Gatellier, P. Effect of oxidation on in vitro digestibility of skeletal muscle myofibrillar proteins. J Agric. Food Chem. 2007, 55, 5343–5348. [Google Scholar] [CrossRef] [PubMed]
- Cabanillas, B.; Novak, N. Effects of daily food processing on allergenicity. Crit. Rev. Food Sci. 2019, 59, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Vanga, S.K.; Wang, J.; Raghavan, V. Effects of ultrasonic and microwave processing on Avidin assay and secondary structures of egg white protein. Food Bioprocess Technol. 2018, 11, 1974–1984. [Google Scholar] [CrossRef]
- Wang, X.B.N.A.; Chi, Y.J.N.A. Microwave-assisted phosphorylation of soybean protein isolates and their physicochemical properties. Czech J. Food Sci. 2012, 30, 99–107. [Google Scholar] [CrossRef] [Green Version]
- Morzel, M.; Gatellier, P.; Sayd, T.; Renerre, M.; Laville, E. Chemical oxidation decreases proteolytic susceptibility of skeletal muscle myofibrillar proteins. Meat Sci. 2006, 73, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Gratz, M.; Schottroff, F.; Gall, L.; Zejma, B.; Simon, F.; Jaeger, H. Advantages of ohmic cooking in the kilohertz-range-part I: Impact of conductivity and frequency on the heating uniformity of potatoes. Innov. Food Sci. Emerg. 2021, 67, 102595. [Google Scholar] [CrossRef]
- Rodrigues, R.M.; Pereira, R.N.; Vicente, A.A.; Cavaco-Paulo, A.; Ribeiro, A. Ohmic heating as a new tool for protein scaffold engineering. Mat. Sci. Eng. C 2021, 120, 111784. [Google Scholar] [CrossRef]
- Moreira, T.; Pereira, R.N.; Vicente, A.A.; Da, C.R. Effect of Ohmic heating on functionality of sodium caseinate—A relationship with protein gelation. Food Res. Int. 2019, 116, 628–636. [Google Scholar] [CrossRef] [Green Version]
- Alizadeh, O.; Aliakbarlu, J. Effects of ultrasound and ohmic heating pretreatments on hydrolysis, antioxidant and antibacterial activities of whey protein concentrate and its fractions. Food Sci. Technol. 2020, 131, 109913. [Google Scholar] [CrossRef]
- Jaeger, H.; Janositz, A.; Knorr, D. The Maillard reaction and its control during food processing. The potential of emerging technologies. Pathol. Biol. 2010, 58, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Nooshkam, M.; Varidi, M.; Verma, D.K. Functional and biological properties of Maillard conjugates and their potential application in medical and food: A review. Food Res. Int. 2020, 131, 109003. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Yao, Y.; Dong, S.; Jin, S.; Xiao, H.; Wu, H.; Zeng, M. Chemical characterization of the glycated myofibrillar proteins from grass carp (Ctenopharyngodon idella) and their impacts on the human gut microbiota in vitro fermentation. Food Funct. 2017, 8, 1184–1194. [Google Scholar] [CrossRef]
- Yang, M.; Liu, J.; Yang, X.; Li, S.; Li, C.; Liu, B.; Ma, S.; Liu, X.; Du, Z.; Zhang, T.; et al. Effect of glycation degree on the in vitro simulated gastrointestinal digestion: A promising formulation for egg white gel with controlled digestibility. Food Chem. 2021, 349, 129096. [Google Scholar] [CrossRef]
- Jiménez-Saiz, R.; Belloque, J.; Molina, E.; López-Fandinño, R. Human immunoglobulin E (IgE) binding to heated and glycated ovalbumin and ovomucoid before and after in vitro digestion. J. Agric. Food Chem. 2011, 59, 10044–10051. [Google Scholar] [CrossRef] [PubMed]
- Hellwig, M. Analysis of protein oxidation in food and feed products. J. Agric. Food Chem. 2020, 68, 12870–12885. [Google Scholar] [CrossRef]
- Soladoye, O.P.; Juárez, M.L.; Aalhus, J.L.; Shand, P.; Estévez, M. Protein oxidation in processed meat: Mechanisms and potential implications on human health. Comp. Rev. Food Sci. 2015, 14, 106–122. [Google Scholar] [CrossRef] [PubMed]
- Bax, M.; Buffière, C.; Hafnaoui, N.; Gaudichon, C.; Savary-Auzeloux, I.; Dardevet, D.; Santé-Lhoutellier, V.; Rémond, D.; Blachier, F. Effects of meat cooking, and of ingested amount, on protein digestion speed and entry of residual proteins into the colon: A study in minipigs. PLoS ONE 2013, 8, e61252. [Google Scholar] [CrossRef]
- Gong, X.; Morton, J.D.; Bhat, Z.F.; Mason, S.L.; Bekhit, A.E.D.A. Comparative efficacy of actinidin from green and gold kiwi fruit extract on in vitro simulated protein digestion of beef Semitendinosus and its myofibrillar protein fraction. Int. J. Food Sci. Technol. 2020, 55, 742–750. [Google Scholar] [CrossRef]
- Du, X.; Sun, Y.; Pan, D.; Wang, Y.; Ou, C.; Cao, J. Change of the structure and the digestibility of myofibrillar proteins in Nanjing dry-cured duck during processing. J. Sci. Food Agric. 2018, 98, 3140–3147. [Google Scholar] [CrossRef]
- Kaur, L.; Maudens, E.; Haisman, D.R.; Boland, M.J.; Singh, H. Microstructure and protein digestibility of beef: The effect of cooking conditions as used in stews and curries. Food Sci. Technol. 2014, 55, 612–620. [Google Scholar] [CrossRef]
- Wei, T.; Dang, Y.; Cao, J.; Wu, Z.; He, J.; Sun, Y.; Pan, D.; Tian, Z. Different duck products protein on rat physiology and gut microbiota. J. Proteom. 2019, 206, 103436. [Google Scholar] [CrossRef]
- Cao, C.; Tang, M.; Zhao, N.; Dong, S.; Wu, H. Effects of fish protein with glycation extent on gut microbiota and colonic barrier function in mice fed a high-fat diet. J. Funct. Foods 2021, 85, 104636. [Google Scholar] [CrossRef]
- Kanauchi, O.; Fujiyama, Y.; Mitsuyama, K.; Araki, Y.; Ishii, T.; Nakamura, T.; Hitomi, Y.; Agata, K.; Saiki, T.; Andoh, A.; et al. Increased growth of Bifidobacterium and Eubacterium by germinated barley foodstuff, accompanied by enhanced butyrate production in healthy volunteers. Int. J. Mol. Med. 1999, 3, 175. [Google Scholar] [CrossRef]
- Santiago-López, L.; Hernández-Mendoza, A.; Vallejo-Cordoba, B.; Wall-Medrano, A.; González-Córdova, A.F. Th17 immune response in inflammatory bowel disease: Future roles and opportunities for lactic acid bacteria and bioactive compounds released in fermented milk. Trends Food Sci. Technol. 2021, 112, 109–117. [Google Scholar] [CrossRef]
- Bermingham, E.N.; Maclean, P.; Thomas, D.G.; Cave, N.J.; Young, W. Key bacterial families (Clostridiaceae, Erysipelotrichaceae and Bacteroidaceae) are related to the digestion of protein and energy in dogs. PeerJ 2017, 5, e3019. [Google Scholar] [CrossRef] [PubMed]
- Muniz, P.D.; Chen, J.; Hillmann, B.; Jeraldo, P.; Al-Ghalith, G.; Taneja, V.; Davis, J.M.; Knights, D.; Nelson, H.; Faubion, W.A.; et al. An increased abundance of Clostridiaceae characterizes Arthritis in inflammatory bowel disease and rheumatoid arthritis: A Cross-sectional study. Inflamm. Bowel Dis. 2019, 25, 902–913. [Google Scholar] [CrossRef]
- Krzyściak, W.; Pluskwa, K.K.; Jurczak, A.; Kościelniak, D. The pathogenicity of the Streptococcus genus. Eur. J. Clin. Microbiol. 2013, 32, 1361–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goertz, S.; de Menezes, A.B.; Birtles, R.J.; Fenn, J.; Lowe, A.E.; MacColl, A.D.C.; Poulin, B.; Young, S.; Bradley, J.E.; Taylor, C.H. Geographical location influences the composition of the gut microbiota in wild house mice (Mus musculus domesticus) at a fine spatial scale. PLoS ONE 2019, 14, e222501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Model (Type; Gender; Age of Animals; Number of Animals Examined; Acclimation Period; Feeding Period) | Sample and Analytical Method | Protein Source (Dose) | Microbial Populations Increased: Phylum (P), Family (F), Genus (G), Species (S) | Microbial Populations Decreased: Phylum (P), Family (F), Genus (G), Species (S) | References |
|---|---|---|---|---|---|
| Mouse (Balb/c; female; 6-week-old; 4; 1 week; 2 weeks) | Feces; pyrosequencing | NP: Casein (20%) HP: Casein (30%) | Bacteroidetes (P) (HP *) Bacteroides (G) (HP *) Parabacteroides (G) (HP) | Firmicutes (P) (HP) Lachnospiraceae (F) (HP *) Ruminococcaceae (F) (HP) Oscillibacter (G) (HP) Enterococcus (G) (HP) | [9] |
| Rat (Wistar; male; NA 1; 16; 6 days; 15 days) | Cecal contents and Colonic contents; qPCR 2 and DGGE 2 | NP: Whole milk proteins (14%) HP: Whole milk proteins (53%) | Cecal contents Fusobacterium (G) (HP) | Cecal contents and Colonic contents Clostridium (G) (HP) Cecal contents Escherichia coli (S) (HP) Colonic contents Bifidobacterium (G) (HP) Fusobacterium (G) (HP) | [15] |
| Piglets (Pietrain; Large White × Landrace; male and female; new-born; 48; 1 week; 2 weeks) | Colonic contents; qPCR 2 | NP: Whey (15%) + Potassium caseinate (8%) HP: Whey (20%) + Potassium caseinate (15%) | No difference was observed in composition of the major gut microbiota. | No difference was observed in composition of the major gut microbiota. | [16] |
| Rat (Wistar; male; NA 1; 20; 1 week; 6 weeks) | Colonic contents; qPCR 2 | NP: Casein (20%) HP: Casein (54%) | Bacteroidetes (P) (HP *) Lawsonia (G) (HP) Bacteroides (G) (HP) Parabacteroides (G) (HP) Escherichia/Shigella (G) (HP) Enterococcus (G) (HP) Streptococcus (G) (HP) Lactobacillus (G) (HP) Lactococcus (G) (HP) Alistipes (G) (HP) Eubacterium (G) (HP) | Firmicutes (P) (HP) Actinobacteria (P) (HP) Acidobacteria (P) (HP) Sporobacter (G) (HP) Bifidobacterium (G) (HP) Ruminococcus (G) (HP) Akkermansia (G) (HP) Prevotella (G) (HP) Barnesiella (G) (HP *) Blautia (G) (HP) Roseburia (G) (HP) Allobaculum (G) (HP) Coprococcus (G) (HP) | [17] |
| Rat (Wistar; male; NA 1; 20; 1 week; 6 weeks) | Feces; qPCR 2 | NP: Casein (20%) HP: Casein (54%) | Week 1 Lactobacillus (G) (HP) Bifidobacterium (G) (HP) | Week 2 Prevotella (G) (HP) Lactobacillus (G) (HP) Bifidobacterium (G) (HP) Week 4 Prevotella (G) (HP) Bifidobacterium (G) (HP) Week 6 Bifidobacterium (G) (HP) Prevotella (G) (HP) | [18] |
| Mouse (C57BL/6J; male; 3-week-old; 36; 1 week; 4 weeks) | Ileal contents; Illumina sequencing technology 3 | Sm: Soybean meal (30%) Ca: Casein (30%) Dw: Delactosed whey (30%) Sdp: Spray dried plasma (30%) Wgm: Wheat gluten meal (30%) Ymw: Yellow meal worm (30%) | Bacteroidetes (P) (Sm ***) Firmicutes (P) (Ca *, Dw *, Wgm *, Sdp ***, Ymw ***) Actinobacteria (P) (Ca, Sdp, Dw **) Proteobacteria (P) (Ca **) Deferribacteres (P) (Sdp **) | Firmicutes (P) (Sm **) Bacteroidetes (P) (Ca, Dw, Sdp, Wgm, Ymw **) Deferribacteres (P) (Dw **) Proteobacteria (P) (Ymw **) | [19] |
| Mouse (C57BL/6J; male; 4-week-old; 60; 2 weeks; 8 months) | Cecal contents; Illumina sequencing technology 3 | Ca: Casein (20%) So: Soy (20%) Esp: Emulsion-type sausage protein (20%) Dpp: Dry-cured pork protein (20%) Spp: Stewed pork protein (20%) Cpp: Steam-cooked pork protein (20%) | Firmicutes (P) (Ca *, Dpp *, Spp *, Cpp *) Bacteroidetes (P) (So, Esp, Dpp, Spp, Cpp) Proteobacteria (P) (So, Spp, Cpp) Actinobacteria (P) (So **, Esp **) Muribaculaceae-Norank (G) (So, Esp, Dpp, Spp, Cpp) Lactobacillus (G) (So **) Faecalibaculum (G) (So *, Esp *, Dpp *, Cpp *, Spp ***) Lachnospiraceae-Uncultured (G) (Dpp **) | Bacteroidetes (P) (Ca **) Firmicutes (P) (So ****, Esp ****) Verrucomicrobia (P) (Cpp, Spp **) Blautia (G) (Esp, Dpp, Spp, Cpp, So **) Akkermansia (G) (Cpp, Spp **) Lachnospiraceae-Uncultured (G) (So **) Lachnospiraceae Nk4a136 (G) (So **) Lachnoclostridium (G) (So **) Ruminiclostridium 9 (G) (So **) | [20] |
| Mouse (C57BL/6J; male; 5-week-old; 18; 1 week; 4 weeks) | Cecal contents and Colonic contents; Illumina sequencing technology 3 and qPCR 2 | So: Soy (20%) Ch: Chicken (20%) | Firmicutes (P) (So *, Ch *) Proteobacteria (P) (So, Ch) Actinobacteria (P) (So, Ch) Verrucomicrobia (P) (Ch) Lactobacillus (G) (So, Ch) | Bacteroidetes (P) (So, Ch) Verrucomicrobia (P) (So) Deferribacteres (P) (So, Ch) | [21] |
| Rat (Sprague-Dawley; male; NA 1; 30; NA 1; 2 weeks) | Cecal; qPCR 2 | Ca: Casein (21%) So: Soy (20%) Ze: Zein (24%) | Lactobacillus (G) (Ca **) Bifidobacterium (G) (Ca **) Escherichia (G) (Ze, So **) | Escherichia (G) * (Ca **) Lactobacillus (G) (So, Ze **) Bifidobacterium (G) (So, Ze **) | [22] |
| Rat (Wistar; male; 4-week-old; 18; 7 days; 16 days) | Cecal contents; pyrosequencing and DGGE 2 | Ca: Casein (20%) So: Soy (20%) Fm: Fish meal (20%) | Firmicutes (P) (Ca *, So *, Fm *) Turibacter (G) (Ca **) Oscillibacter (G) (So **) Lactobacillus (G) (Fm ***) | Ruminococcaceae (F) (Ca **, Fm **) Lactobacillaceae (F) (So **, Fm **) | [23] |
| Rat (Sprague-Dawley; male; 3-week-old; 20; 1 week; 1 week) | Cecal; Illumina sequencing technology 3 | Ca: Casein (20%) Ch: Chicken (20%) | Bacteroidetes (P) (Ca *) Firmicutes (P) (Ch *) Verrucomicrobia (P) (Ch) Bacteroides (G) * (Ca *, Ch *) | Bacteroidetes (P) (Ch *) Proteobacteria (P) (Ch) Actinobacteria (P) (Ch) Mycobacterium (G) (Ch) Tetragenococcus (G) (Ch) Lactococcus (G) (Ch) | [24] |
| Rat (Sprague-Dawley; male; NA 1; 66; 7 days; 90 days) | Cecal contents; Illumina sequencing technology 3 | Ca: Casein (20%) Ch: Chicken (20%) Fi: Fish (20%) Po: Pork (20%) Be: Beef (20%) So: Soy (20%) | Firmicutes (P) (Ca *, So *, Be *, Po *, Ch ***, Fi ***) Fusobacteria (P) (Ca **) Bacteroidetes (P) (So **) Roseburia (G) (Ca **, So **) Bacteroides (G) (Ca, Be, Po, So*) Alloprevotella (G) (Ca, So) Proteobacteria (P) (Be **) Tenericutes (P) (Be **, Po **) Oscillibacter (G) (Be, Po) Actinobacteria (P) (Ch **) Lactobacillus (G) (Fi, Ch **) | Fusobacteria (P) (So, Be, Po, Ch, Fi) Bacteroidetes (P) (Ch **, Fi **) Lactobacillus (G) (Ca **) | [25] |
| Rat (Sprague-Dawley; male; 4-week-old; 55; 7 days; 14 days) | Feces; Illumina sequencing technology 3 | Ca: Casein (19%) Fi: Fish (19%) Po: Pork (19%) Be: Beef (19%) So: Soy (19%) | Bacteroidetes (P) (Ca ***, So ***) Spirochaetae (P) (Ca **) Proteobacteria (P) (So **) Firmicutes (P) (Po *, Fi *, Be ***) Bacteroides (G) (Ca *, So ***) Alloprevotella (G) (Po, Be, S **) Blautia (G) (So **, Be **, Fi **) Lactobacillus (G) (Be *, Fi *, Po ***) | Spirochaetae (P) (So, Po, Be, Fi) Bacteroidetes (P) (Po, Fi, Be **) Fusobacteria (P) (Po, Be, Fi) Lactobacillus (G) (Ca **) Treponema (G) (Po, Be, Fi, So **) Bacteroides (G) (Be, Fi, Po **) Fusobacterium (G) (Po, Be, Fi) Alloprevotella (G) (Po **) | [26] |
| Rat (Sprague-Dawley; male; 3-week-old; 190; 7 days; 14 days) | Cecal contents; DGGE 2 | Ca: Casein (20%) Ch: Chicken (20%) Fi: Fish (20%) Po: Pork (20%) Be: Beef (20%) So: Soy (20%) | Day 7 Robinsoniella peoriensis (S) (So, Be, Ch, Fi, Ca **, Po **) Clostridium hathewayi (S) (Ca **, Be **) Blautia wexlerae (S) (So **) Day 14 Blautia wexlerae (S) (So **) | Day 14 Robinsoniella peoriensis (S) (Ca, So, Be, Po, Ch, Fi,) Clostridium hathewayi (S) (Ca, So, Be, Po, Ch, Fi,) Akkermansia muciniphila (S) (So **) | [27] |
| Rat (Sprague-Dawley; male; 4-week-old, 32; 7 days; 90 days) | Colonic contents; Illumina sequencing technology 3 | Ca: Casein (20%) Ch: Chicken (20%) Be: Beef (20%) So: Soy (20%) | Firmicutes (P) (Ca ***) Bacteroidetes (P) (So, Be, Ch **) Spirochaeta (P) (Ch, So **) Proteobacteria (P) (Be **) Tenericutes (P) (Ch **) Lactobacillus (G) (Ch **) | Bacteroidetes (P) (Ca **) Firmicutes (P) (So*, Be*, Ch ****) | [28] |
| Rat (Sprague-Dawley; male; NA 1; 40; 1 week; 8 weeks) | Cecal contents; Illumina sequencing technology 3 | Ca: Casein (20%) Hew: Hen egg white (20%) Dew: Duck egg white (20%) Pew: Preserved egg white (20%) | Firmicutes (P) (Ca ***) Actinobacteria (P) (Ca **) Bacteroidetes (P) (Dew, Pew, Ca, Hew **) Verrucomicrobia (P) (Pew, Ca, Hew **) Proteobacteria (P) (Dew **) Akkermansia (G) (Ca, Hew **) | Bacteroidetes (P) (Ca **) Firmicutes (P) (Dew, Pew, Hew ****) Actinobacteria (P) (Dew, Pew, Hew) | [29] |
| Rat (Sprague-Dawley; male; NA 1; 32; 1 week; 2 weeks) | Cecal contents; Illumina sequencing technology 3 | Ca: Casein (18%) FiC: Fish (12%) + Casein (6%) HfC: Heated fish (12%) + Casein (6%) GfC: Glycated fish (12%) + Casein (6%) | Firmicutes (P) (FiC *) Actinobacteria (P) (HfC, GfC **) Lactobacillus (G) (FiC, GfC, HfC **) Collinsella (G) (HfC) Ruminococcaceae_UCG-014 (G) (GfC **) Turicibacter (G) (GfC **) | Bacteroidetes (P) (FiC, GfC, HfC **) Fusobacteria (P) (FiC, HfC) Proteobacteria (P) (HiC, GfC) Allobaculum (G) (FiC) Fusobacterium (G) (FiC, HfC, GfC **) Bacteroides (G) (HfC**, GfC **) Subdoligranulum (HfC**, GfC **) Erysipelatoclostridium (G) (GfC **) Escherichia-Shigella (G) (GfC **) Ruminococcaceae_UCG-009 (G) GfC **) | [30] |
| Rat (Wistar Han; male; 7-week-old; 40; 1 week; 3 weeks) | Feces; Illumina sequencing technology 3 | Lfe: Lipid-protein liquid-fine emulsions (17%) Gce: Gelled-coarse emulsions (17%) | Firmicutes (P) (Lfe *, Gfe *) Bacteroidetes (P) (Lfe, Gfe) Parabacteroides (G) (Lfe, Gfe) Clostridium Cluster Xiv (G) (Gfe) Bifidobacterium (G) (Gfe) Sutterella (G) (Gfe) | Proteobacteria (P) (Lfe, Gfe) Actinobacteria (P) (Lfe, Gfe) Deferribacteres (P) (Lfe, Gfe) Tenericutes (P) (Lfe, Gfe) Parasutterella (G) (Lfe, Gfe) Clostridium Cluster Xiv (G) (Lfe)Bifidobacterium (G) (Lfe) Sutterella (G) (Lfe) Parasutterella (G) (Lfe) | [31] |
| Rat (Wistar Han; male; 7-week-old; 16; NA 1; 3 weeks) | Ileal contents, Cecal contents, and Cecal mucus; Illumina sequencing technology 3 | Lfe: Lipid-protein liquid-fine emulsions (21%) Gce: Gelled-coarse emulsions (21) | Ileal Firmicutes (P) (Lfe *, Gfe *) Proteobacteria (P) (Lfe, Gfe) Lactobacillus (G) (Lfe) Bifidobacterium (G) (Gce) Cecal Bacteroidetes (P) (Lfe *, Gfe *) Firmicutes (P) (Lfe, Gfe) Lactobacillus (G) (Lfe) Coprococcus (G) (Lfe) Verrucomicrobia (P) (Gce) Bifidobacterium (G) (Gce) Cecal Mucus Coprococcus (G) (Lfe, Gce) Lactobacillus (G) (Lfe) Bifidobacterium (G) (Gce) | Ileal Bacteroidetes (P) (Lfe, Gfe) Actinobacteria (P) (Lfe, Gfe) Bifidobacterium (G) (Lfe) Lactobacillus (G) (Gce) Cecal Proteobacteria (P) (Lfe, Gfe) Deferribacteres (P) (Lfe, Gfe) Cyanobacteria (P) (Lfe, Gfe) Verrucomicrobia (P) (Lfe) Bifidobacterium (G) (Lfe) Lactobacillus (G) (Gce) Coprococcus (G) (Gce) Cecal Mucus Bifidobacterium (G) (Lfe) Oscillospira (G) (Lfe)Lactobacillus (G) (Gce) Coprococcus (G) (Gce) | [32] |
| Piglets (Crossbred; 184 male and 152 female; 21-day-old; 336; NA 1; 21 days) | Ileal contents; Illumina sequencing technology 3 | SW: Soybean meal + Whey FSW: Fish meal + Soybean meal + Whey MSSW: Microbially enhanced soybean meal + Soybean meal + Whey | Firmicutes (P) (SW ***) Proteobacteria (P) (MSSW, FSW **) Actinobacteria (P) (MSSW **) | Firmicutes (P) (FSW *, MSSW ****) Actinobacteria (P) (SMW **, FSW **) Proteobacteria (P) (SMW **) | [33] |
| Piglets (Crossbred; male and female; 34 days; 96; NA 1; 21 days) | Ileal contents; Illumina sequencing technology 3 | SSm: 12% Soy + 9% Soybean meal FSS: 4% Fish meal + 7% Soy + 9% Soybean SHSS: Salmon protein hydrolysate (10%) + Soy (7%) + Soybean meal (9%) | Firmicutes (P) (SSm *, FSS *, SHSS *) Lactobacillus (G) (SSm *, FSS *, SHSS *) Turicibacter (G) (SSm *, FSS *, SHSS *) | Enterobacteriaceae (F) (SSm, FSS, SHSS) | [34] |
| Piglets (German Landrace × Piétrain; NA 1; 48; NA 1; 16 days) | Feces and Ileal contents; qPCR 2 | Ca: Casein (10%, 16%, 22%, 27%, 33%, 39%) Sm: Soybean meal (21%, 34%, 46%, 59%, 72%, 84%) | Effect of increasing protein level: Ileal Lactobacillus (G) (Ca, Sm) Bifidobacteria (G) (Sm) Fecal Bacteroides (G) (Sm) Bifidobacteria (G) (Sm) | Effect of increasing protein level: Ileal Clostridium Cluster XIVa (G) (Sm) Bacteroides (G) (Sm) Fecal Clostridium Cluster IV (G) (Sm) | [35] |
| Piglets (German Landrace × Piétrain; NA 1; 48; NA1; 16 days) | Feces and Ileal contents; qPCR 2 | Ca: Casein Sm: Soybean meal | Ileal Bacteroides (G) (Ca) Clostridium Cluster XIVa (G) (Ca) Fecal Bifidobacteria (G) (Sm) Lactobacillus (G) (Sm) Bacteroides (G) (Sm) Clostridium Cluster IV (G) (Sm) Clostridium Cluster XIVa (G) (Sm) | Ileal Bacteroides (G) (Sm) Clostridium Cluster XIVa (G) (Sm) Fecal Bifidobacteria (G) (Ca) Lactobacillus (G) (Ca) Bacteroides (G) (Ca) Clostridium Cluster IV (G) (Ca) Clostridium Cluster XIVa (G) (Ca) | [35] |
| Piglets (Crossbred; male; NA1; 24; NA 1; 10 days) | Colonic contents; Illumina sequencing technology 3 | Dpmt: Dried porcine mucosal tissue (34%) ESm: Enzyme-treated soybean meal (35%) Cdcp: Concentrated degossypolized cottonseed protein (29%) Sdfm: Steam dried fish meal (26%) | Firmicutes (P) (Dpmt *, ESm *, Cdcp ***) Bacteroidetes (P) (Dpmt *, ESm *, Sdfm ***) Proteobacteria (P) (Dpmt **) Spirochaetes (P) (ESm, Sdfm) Escherichia (G) (ESm, Sdfm, Dpmt **) Clostridium (G) (Dpmt, Sdfm) Campylobacter (G) (Dpmt **) Faecalibacterium (G) (Dpmt **, ESm **) Prevotella (G) (ESm ***, FM ***) Roseburia (G) (Dpmt **) Turibacter (G) (Dpmt **) Gemmiger (G) (ESm) Oscillospira (G) (ESm **) Lactobacillus (G) (Cdcp **) Megasphaera (G) (Cdcp **) Bacteroides (G) (Sdfm **) Parabacteroides (G) (Sdfm **) Prevotella (G) (Sdfm ***) Ruminococcus (G) (Sdfm **) | Spirochaetes (P) (Dpmt **, Cdcp **) Proteobacteria (P) (Sdfm, ESm, Cdcp **) Bacteroidetes (P) (Cdcp **) Firmicutes (P) (Sdfm **) Ruminococcus (G) (Dpmt **, SBM **) Prevotella (G) (ESm ****, CDCP ****) Roseburia (G) (Cdcp **) Phascolarctobacterium (G) (Cdcp **) Roseburia (G) (Cdcp **) | [36] |
| Piglets (Duroc × Landrace × Large White; female; NA 1; 72; NA 1; 46 days) | Colonic contents; Illumina sequencing technology 3 and qPCR 2 | Sm: Soybean meal (17%) H1Sm: Hermetia illucens larvae (4%) + Soybean meal (14%) H2Sm: Hermetia illucens larvae (8%) + Soybean meal (11%) | Firmicutes (P) (Sm *, H1Sm *, H2Sm ***) Bacteroidetes (P) (Sm **, H1Sm **) Actinobacteria (P) (H1Sm **, H2Sm **) Proteobacteria (P) (H2Sm **) Bacteroides (G) (SM ** Pseudobutyrivibrio (G) (H1Sm) Oribacterium (G) (H1Sm) Lactobacillus (G) (H2Sm, H1Sm **) Roseburia (G) (H2Sm, H1Sm **) Faecalibacterium (G) (H2Sm, H1Sm **) Clostridium cluster IV (G) (H2Sm, H1Sm) | Bacteroidetes (P) (H2Sm **) Streptococcus (G) (H2Sm, H1Sm **) Treponema (G) (H2Sm, H1Sm **) Bacteroides (G) (H2Sm, H1Sm **) Eubacterium (G) (H1Sm **) Barnesiella (G) (H1Sm) Oscillibacter (G) (H1Sm) | [37] |
| Piglets (Crossbred; female; 13-week-old; 45; NA 1; 4 weeks) | Feces; qPCR 2 | Ca: Casein (13–15%) Lu: Lupin (13–15%) Be: Beef (13–15%) | Proteobacteria (P) (Ca *, Be ***) Actinobacteria (P) (Ca **, Lu **) Firmicutes (P) (Lu ***) Bacteroidetes (P) (Lu **) | Bacteroidetes (P) (Ca *, Be ***) Firmicutes (P) (Ca *, Be ***) Proteobacteria (P) (Lu *) Actinobacteria (P) (Be **) | [38] |
| In vitro batch fermentation model of human distal colon | - | Fp: Fish protein HFp24: Heated (24 h) fish protein HFp48: Heated (48 h) fish protein GFp24: Glycated (24 h) fish protein GFp48: Glycated (48 h) fish protein | Fusobacteria (P) (FP *, HFp24 *, HFp48 *, GFp48 *) Bacteroidetes (P) (Fp **, HFp24 **) Proteobacteria (P) (Fp, HFp24, HFp48, GFp24, GFp48) Firmicutes (P) (HFp48) Clostridium_sensu_stricto_1 (G) (HFp48 **) Streptococcus (G) (HFp48 **) Arcobacter (G) (HFP48 **) Holdemania (G) (GFP48 **) | Proteobacteria (P) (Fp **) Firmicutes (P) (Fp **, HFP **) Bacteroidetes (P) (HFp24) | [39] |
| Mouse (Balb/c; male; 4-week-old; 30; 1 week; 12 weeks) | Cecal contents; Illumina sequencing technology 3 | HOP: Low-oxidative damage pork MOP: Medium-oxidative damage pork LOP: High-oxidative damage pork | Escherichia-Shigella (G) (HOP) Mucispirillum (G) (HOP) | Lactobacillus (G) (HOP) Bifidobacterium (G) (HOP) Desulfovibrio (G) (HOP) | [40] |
| Rat (Sprague-Dawley; male; NA 1; 40; 5 days; 21 days) | Colonic contents; Illumina sequencing technology 3 | FC: Fresh chicken CC: Cured chicken FB: Fresh beef CB: Cured beef | Ruminococcaceae (P) (CC, CB) Oscillibacter (G) (CB) | Marvinbryantia (G) (CC) | [41] |
| Mouse (C57BL/6J; male; 6 or 8-week-old; 20; NA 1; 24 weeks) | Cecal contents; Illumina sequencing technology 3 | NP: Casein (20%) HP: Casein (52%) | Actinobacteria (P) (HP) Bifidobacterium (G) (HP) Bacteroides (G) (HP) Parabacteroides (G) (HP) Oscillospira (G) (HP) | Saccharibacteria (P) (HP) | [42] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, S.; Bhat, Z.F.; Gounder, R.S.; Mohamed Ahmed, I.A.; Al-Juhaimi, F.Y.; Ding, Y.; Bekhit, A.E.-D.A. Effect of Dietary Protein and Processing on Gut Microbiota—A Systematic Review. Nutrients 2022, 14, 453. https://doi.org/10.3390/nu14030453
Wu S, Bhat ZF, Gounder RS, Mohamed Ahmed IA, Al-Juhaimi FY, Ding Y, Bekhit AE-DA. Effect of Dietary Protein and Processing on Gut Microbiota—A Systematic Review. Nutrients. 2022; 14(3):453. https://doi.org/10.3390/nu14030453
Chicago/Turabian StyleWu, Shujian, Zuhaib F. Bhat, Rochelle S. Gounder, Isam A. Mohamed Ahmed, Fahad Y. Al-Juhaimi, Yu Ding, and Alaa E. -D. A. Bekhit. 2022. "Effect of Dietary Protein and Processing on Gut Microbiota—A Systematic Review" Nutrients 14, no. 3: 453. https://doi.org/10.3390/nu14030453
APA StyleWu, S., Bhat, Z. F., Gounder, R. S., Mohamed Ahmed, I. A., Al-Juhaimi, F. Y., Ding, Y., & Bekhit, A. E.-D. A. (2022). Effect of Dietary Protein and Processing on Gut Microbiota—A Systematic Review. Nutrients, 14(3), 453. https://doi.org/10.3390/nu14030453