Bifidobacterium longum Subspecies infantis Strain EVC001 Decreases Neonatal Murine Necrotizing Enterocolitis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. EVC001 Dose Optimization
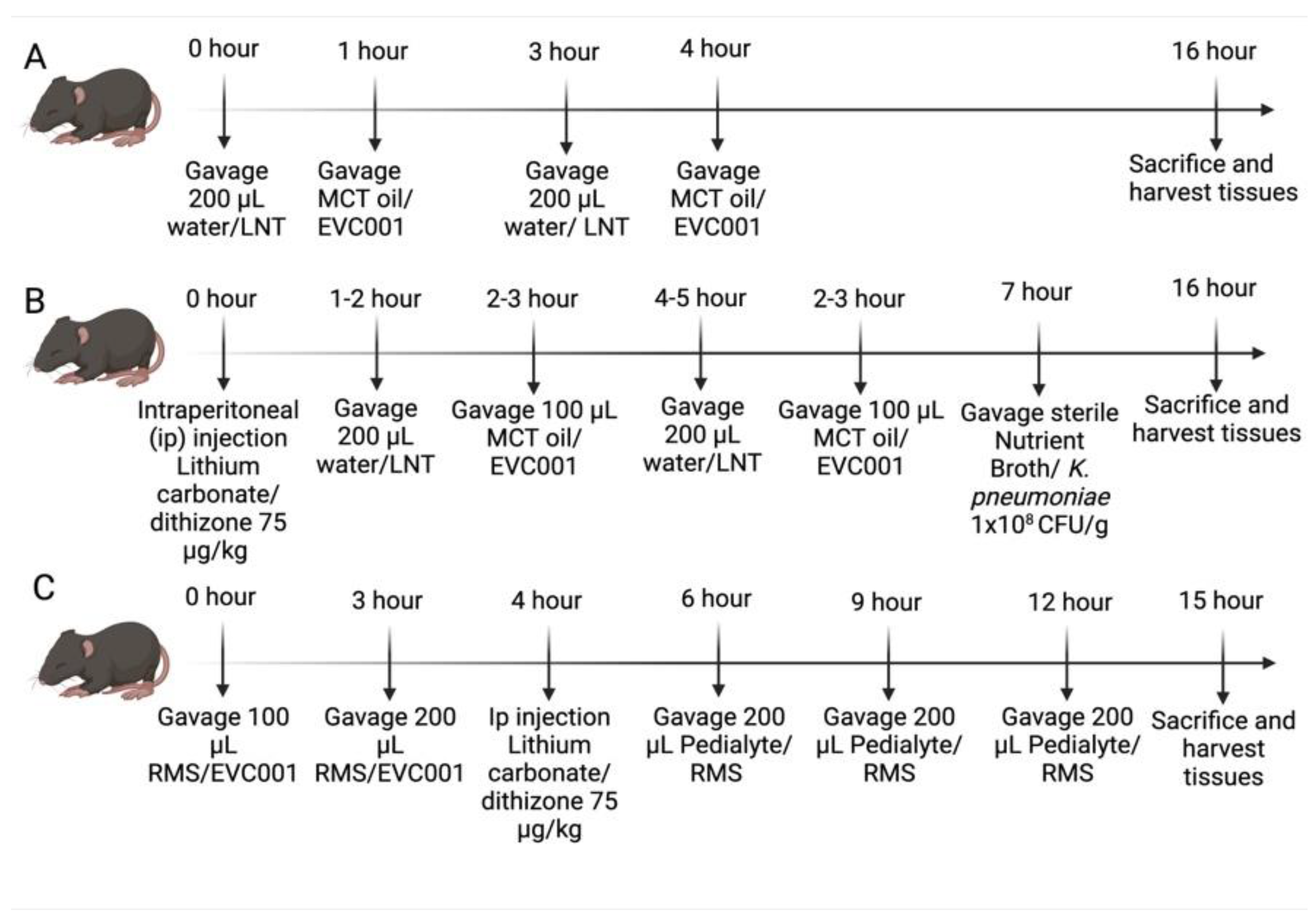
2.3. NEC Models
2.4. Injury Scoring
2.5. Serum Cytokine Measurements
2.6. Microbiome Analysis
2.7. Quantitative PCR
2.8. Wound Healing Assay
2.9. Statistics
3. Results
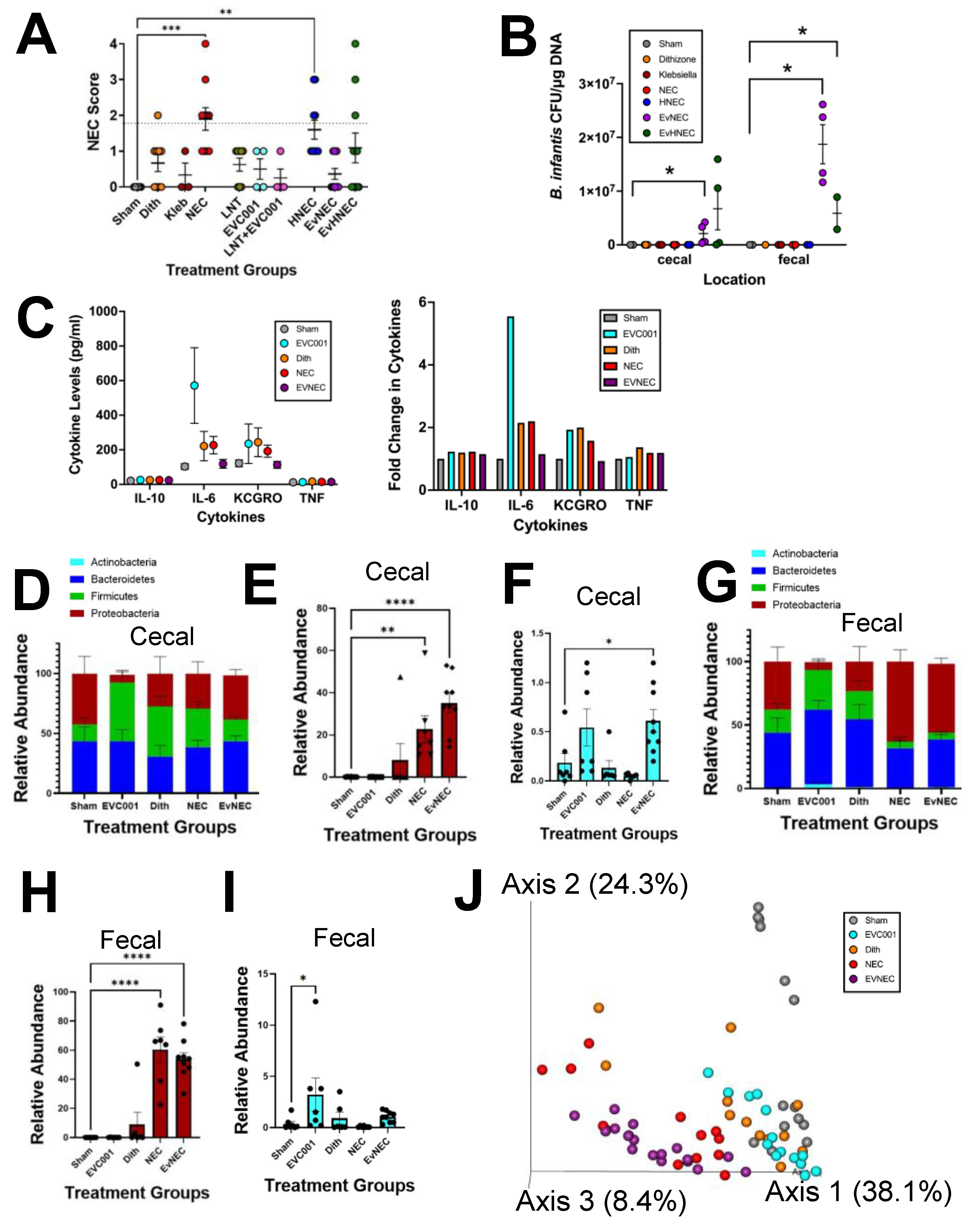
3.1. EVC001 Delivery to Mice at Human Relevant Concentrations Causes Intestinal Injury and Inflammation Due to MCT Oil Toxicity
3.2. EVC001 Supplementation Decreases NEC Injury without LNT Supplementation
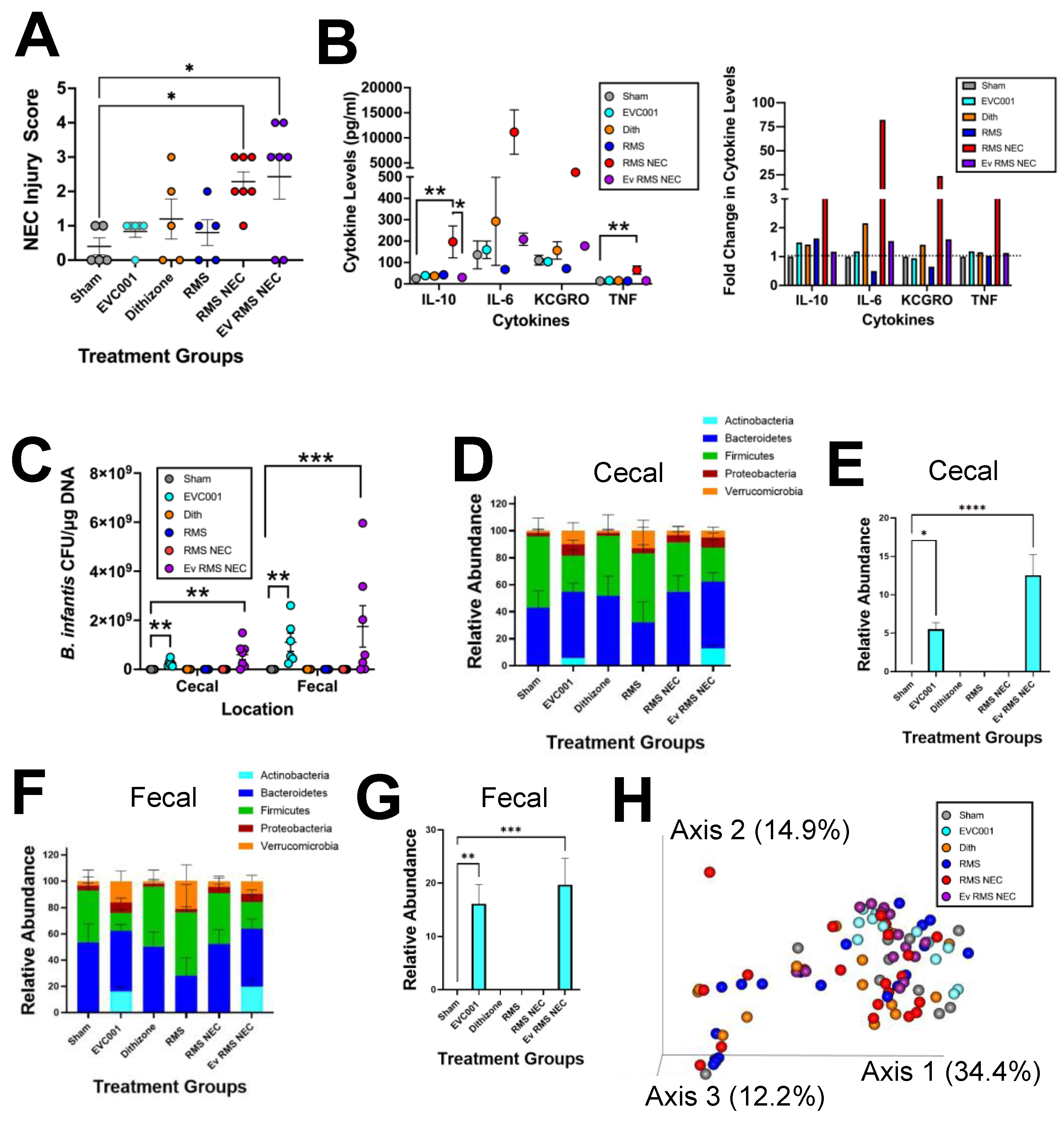
3.3. Effects of EVC001 Are Model Dependent
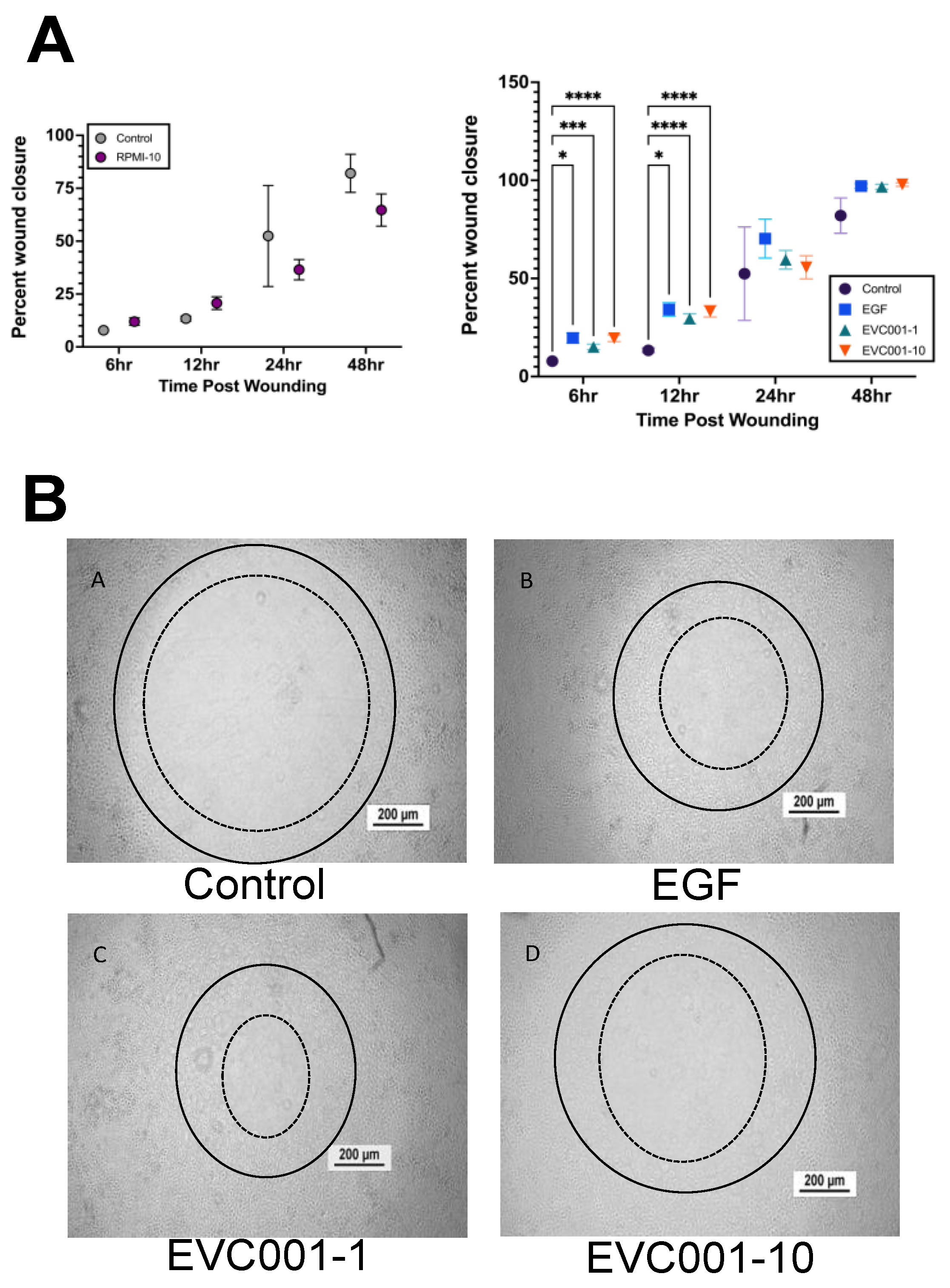
3.4. EVC001 Improves Intestinal Epithelial Wound Healing
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- March of Dimes; The Partnership for Maternal, Newborn & Child Health; Save the Children; WHO. Born Too Soon: The Global Action Report on Preterm Birth, 1st ed.; World Health Organization: Geneva, Switzerland, 2012; pp. 21–45. [Google Scholar]
- Claud, E.C.; Walker, W.A. Hypothesis: Inappropriate colonization of the premature intestine can cause neonatal necrotizing enterocolitis. FASEB J. 2001, 15, 1398–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, P.V.; Christensen, R.; Weitkamp, J.H.; Maheshwari, A. Mapping the new world of necrotizing enterocolitis (NEC): Review and opinion. EJ Neonatol. Res. 2012, 2, 145–172. [Google Scholar] [PubMed]
- Mizrahi, A.; Barlow, O.; Berdon, W.; Blanc, W.A.; Silverman, W.A. Necrotizing enterocolitis in premature infants. J. Pediatr. 1965, 66, 697–706. [Google Scholar] [CrossRef]
- Neu, J.; Walker, W.A. Necrotizing enterocolitis. N. Engl. J. Med. 2011, 364, 255–264. [Google Scholar] [CrossRef] [Green Version]
- Tanner, S.M.; Berryhill, T.F.; Ellenburg, J.L.; Jilling, T.; Cleveland, D.S.; Lorenz, R.G.; Martin, C.A. Pathogenesis of necrotizing enterocolitis: Modeling the innate immune response. Am. J. Pathol. 2015, 185, 4–16. [Google Scholar] [CrossRef] [Green Version]
- Wejryd, E.; Martí, M.; Marchini, G.; Werme, A.; Jonsson, B.; Landberg, E.; Abrahamsson, T.R. Low diversity of human milk oligosaccharides is associated with necrotising enterocolitis in extremely low birth weight infants. Nutrients 2018, 10, 1556. [Google Scholar] [CrossRef] [Green Version]
- Underwood, M.A. Impact of probiotics on necrotizing enterocolitis. Semin. Perinatol. 2018, 41, 41–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Athalye-Jape, G.; Rao, S.; Patole, S. Effects of probiotics on experimental necrotizing enterocolitis: A systematic review and meta-analysis. Pediatr. Res. 2018, 83, 16–22. [Google Scholar] [CrossRef] [Green Version]
- Patel, R.M.; Underwood, M.A. Probiotics and necrotizing enterocolitis. Semin. Pediatr. Surg. 2018, 27, 39–46. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Lucas, A.; Cole, T.J. Breast milk and neonatal necrotising enterocolitis. Lancet 1990, 336, 1519–1523. [Google Scholar] [CrossRef]
- Maffei, D.; Schanler, R.J. Human milk is the feeding strategy to prevent necrotizing enterocolitis! Semin. Perinatol. 2017, 41, 36–40. [Google Scholar] [CrossRef] [Green Version]
- Gephart, S.M.; McGrath, J.M.; Effken, J.A.; Halpern, M.D. Necrotizing enterocolitis risk; State of the science. Adv. Neonatal. 2012, 12, 77–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, S.; Schanler, R.J.; Kim, J.H.; Patel, A.L.; Trawöger, R.; Kiechl-Kohlendorfer, U.; Chan, G.M.; Blanco, C.L.; Abrams, S.; Cotton, C.M.; et al. An exclusively human milk-based diet is associated with a lower rate of necrotizing enterocolitis than a diet of human milk and bovine milk-based products. J. Pediatr. 2010, 156, 562–567. [Google Scholar] [CrossRef] [Green Version]
- Nolan, L.S.; Parks, O.B.; Good, M. A review of the immunomodulation components of maternal breast milk and protection against necrotizing enterocolitis. Nutrients 2020, 12, nu12010014. [Google Scholar]
- Fong, B.; Ma, K.; McJarrow, P. Quantification of bovine milk oligosaccharides using liquid chromatography selected reaction monitoring mass spectrometry. J. Agric. Food Chem. 2011, 59, 9788–9795. [Google Scholar] [CrossRef] [PubMed]
- Bode, L. Human milk oligosaccharides; every baby needs a sugar mama. Glycobiology 2012, 22, 1147–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aldredge, D.L.; Geronimo, M.R.; Hua, S.; Nwosu, C.C.; Lebrilla, C.B.; Barile, D. Annotation and structural elucidation of bovine milk oligosaccharides and determination of novel fucosylated structures. Glycobiology 2013, 23, 644–676. [Google Scholar] [CrossRef]
- Bode, L. Human milk oligosaccharides in prevention of necrotizing enterocolitis: A journey from in vitro and in vivo models to mother-infant cohort studies. Front. Pediatr. 2018, 6, 385. [Google Scholar] [CrossRef]
- Underwood, M.A. Human milk for the premature infant. Pediatr. Clin. N. Am. 2013, 60, 189–207. [Google Scholar] [CrossRef] [Green Version]
- Frese, S.; Shafizadeh, T. The influence of human milk on the preterm infant gut microbiome. Intensive Care Med. 2017, 30, 25–26. [Google Scholar]
- Sela, D.A.; Chapman, J.; Adeuya, A.; Kim, J.H.; Chen, F.; Whitehead, T.R.; Lapidus, A.; Rokhsar, D.S.; Lebrilla, C.B.; German, J.B.; et al. The genome sequence of Bifidiboacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 18964–18969. [Google Scholar] [PubMed] [Green Version]
- LoCascio, R.G.; Desai, P.; Sela, D.A.; Weimer, B.; Mills, D.A. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis revealed by comparative genomic hybridization. Appl. Environ. Microbiol. 2010, 76, 7373–7381. [Google Scholar]
- Underwood, M.A.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Bifidobacterium longum subspecies infantis: Champion colonizer of the infant gut. Pediatr. Res. 2015, 77, 229–235. [Google Scholar] [CrossRef] [Green Version]
- György, P.N.R.; Rose, C.S. Bifidus factor I. A variant of Lactobacillus bifidus requiring a special growth factor. Arch. Biochem. Biophys. 1954, 48, 193–201. [Google Scholar] [CrossRef]
- Henrick, B.M.; Hutton, A.A.; Palumbo, M.C.; Casaburi, G.; Mitchell, R.D.; Underwood, M.A.; Smilowitz, J.T.; Frese, S.A. Elevated fecal pH indicates a profound change in the breastfed infant gut microbiome due to reduction of Bifidobacterium over the past century. mSphere 2018, 3, e00041-18. [Google Scholar] [CrossRef] [Green Version]
- Underwood, M.A.; Umberger, E.; Patel, R.M. Safety and efficacy of probiotic administration to preterm infants: Ten common questions. Pediatr. Res. 2020, 88, 48–55. [Google Scholar] [CrossRef]
- Poindexter, B.; Committee on Fetus and Newborn. Use of probiotics in preterm infants. Pediatrics 2021, 147, e2021051485. [Google Scholar] [CrossRef] [PubMed]
- Lewis, Z.T.; Shani, G.; Masarweh, C.F.; Popovic, M.; Frese, S.A.; Sela, D.A.; Underwood, M.A.; Mills, D.A. Validating bifidobacterial species and subspecies identity in commercial probiotic products. Peds. Res. 2015, 79, 445–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frese, S.A.; Hutton, A.A.; Contreras, L.N.; Shaw, C.A.; Palumbo, M.C.; Casaburi, G.; Xu, G.; Davis, J.C.C.; Lebrilla, C.; Henrick, B.M.; et al. Persistence of supplemented Bifidobacterium longum subsp. infantis EVC001 in breastfed infants. mSphere 2017, 2, e00501-17. [Google Scholar]
- Smilowitz, J.T.; Moya, J.; Breck, M.A.; Cook, C.; Fineberg, A.; Angkustrisiri, K.; Underwood, M.A. Safety and tolerability of Bifidobacteirum longum subspecies infantis EVC001 supplementation in healthy term breastfed infants: A phase 1 clinical trial. BMC Pediatr. 2017, 17, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Casaburi, G.; Frese, S.A. Colonization of breastfed infants by Bifidobacteirum longum subsp. infantis EVC001 reduces virulence gene abundance. Hum. Microbiome J. 2018, 9, 7–10. [Google Scholar]
- Henrick, B.M.; Chew, S.; Casaburi, G.; Brown, H.K.; Frese, S.A.; Zhou, Y.; Underwood, M.A.; Smilowitz, J.T. Colonization by B. infantis EVC001 modulates enteric inflammation in exclusively breastfed infants. Pediatr. Res. 2019, 86, 749–757. [Google Scholar] [CrossRef]
- Duar, R.M.; Casaburi, G.; Mitchell, R.D.; Scofield, L.N.C.; Ortega Ramirez, C.A.; Barile, D.; Henrick, B.M.; Frese, S.A. Comparative genome analysis of Bifidobacterium longum subsp. infantis strains reveals variation in human milk oligosaccharide utilization genes among commercial probiotics. Nutrients 2020, 12, 3247. [Google Scholar]
- Nguyen, M.; Holdbrooks, H.; Mishra, P.; Abrantes, M.A.; Eskew, S.; Garma, M.; Oca, C.G.; McGuckin, C.; Hein, C.B.; Mitchell, R.D.; et al. Impact of probiotic B. infantis EVC001 feeding in premature infants on the gut microbiome, noscomially acquired antibiotic resistance and enteric inflammation. Front. Pediatr. 2021, 9, 618009. [Google Scholar] [CrossRef] [PubMed]
- Stanford, A.H.; Gong, H.; Noonan, M.; Lewis, A.N.; Gong, Q.; Lanik, W.E.; Hsieh, J.J.; Lueschow, S.R.; Frey, M.R.; Good, M.; et al. A direct comparison of mouse and human intestinal development using epithelial gene expression patterns. Pediatr. Res. 2019, 88, 66–76. [Google Scholar] [CrossRef]
- White, J.R.; Gong, H.; Pope, B.; Schlievert, P.; McElroy, S.J. Paneth-cell-disruption-induced necrotizing enterocolitis in mice requires live bacteria and occurs independently of TLR4 signaling. Dis. Model Mech. 2017, 10, 727–736. [Google Scholar] [PubMed] [Green Version]
- Lueschow, S.R.; Stumphy, J.; Gong, H.; Kern, S.L.; Elgin, T.G.; Underwood, M.A.; Kalanetra, K.M.; Mills, D.A.; Wong, M.H.; Meyerholz, D.K.; et al. Loss of murine Paneth cell function alters the immature intestinal microbiome and mimics changes seen in neonatal necrotizing enterocolitis. PLoS ONE 2018, 13, e0204967. [Google Scholar] [CrossRef]
- Lueschow, S.R.; Kern, S.L.; Gong, H.; Grobe, J.L.; Segar, J.L.; Carlson, S.J.; McElroy, S.J. Feeding formula eliminates the necessity of bacterial dysbiosis and induces inflammation and injury in the Paneth cell disruption murine NEC model in an osmolality-dependent manner. Nutrients 2020, 12, 900. [Google Scholar] [CrossRef] [Green Version]
- Dvorak, B.; McWilliam, D.L.; Williams, C.S.; Dominguez, J.A.; Machen, N.W.; McCuskey, R.S.; Philipps, A.F. Artificial formula induces precocious maturation of the small intestine of artificially reared suckling rats. J. Pediatr. Gastroenterol. Nutr. 2000, 31, 162–169. [Google Scholar] [CrossRef] [PubMed]
- McElroy, S.J.; Castle, S.L.; Bernard, J.K.; Almohazey, D.; Hunter, C.J.; Bell, B.A.; Al Alam, D.; Wang, L.; Ford, H.R.; Frey, M.R. The ErbB4 ligand neuregulin-4 protects against experimental necrotizing enterocolitis. Am. J. Pathol. 2014, 184, 2768–2778. [Google Scholar] [CrossRef]
- Zhang, C.; Sherman, M.P.; Prince, L.S.; Bader, D.; Weitkamp, J.H.; Slaughter, J.C.; McElroy, S.J. Paneth cell ablation in the presence of Klebsiella pneumoniae induces necrotizing enterocolitis (NEC)-like injury in the small intestine of immature mice. Dis. Models Mech. 2012, 5, 522–532. [Google Scholar] [CrossRef] [Green Version]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable, and extensible microbiome data science using Qiime 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2011, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. Gigascience 2013, 2, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawley, B.; Munro, K.; Hughes, A.; Hodgkinson, A.J.; Prosser, C.G.; Lowry, D.; Zhou, S.J.; Makrides, M.; Gibson, R.A.; Lay, C.; et al. Differentiation of Bifidobacterium longum subspecies longum and infantis by quantitative PCR using functional gene targets. PeerJ 2017, 5, e3375. [Google Scholar] [CrossRef] [Green Version]
- Frey, M.R.; Dise, R.S.; Edelblum, K.L.; Polk, D.B. p38 kinase regulates epidermal growth factor receptor downregulation and cellular migration. EMBO J. 2006, 25, 5683–5692. [Google Scholar] [CrossRef] [Green Version]
- Jantscher-Krenn, E.; Zherebtsov, M.; Nissan, C.; Goth, K.; Guner, Y.S.; Naidu, N.; Choudhury, B.; Grishin, A.V.; Ford, H.R.; Bode, L. The human milk oligosaccharide disialyllacto-N-tetraose prevents necrotising enterocolitis in neonatal rats. Gut 2012, 61, 1417–1425. [Google Scholar] [CrossRef]
- Ewaschuk, J.B.; Diaz, H.; Meddings, L.; Diederichs, B.; Dmytrash, A.; Backer, J.; Looijer-van Langen, M.; Madsen, K.L. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 295, G1025–G1034. [Google Scholar] [CrossRef] [Green Version]
- Henrick, B.M.; Rodriguez, L.; Lakshmikanth, T.; Pou, C.; Henckel, E.; Arzoomand, A.; Olin, A.; Wang, J.; Mikes, J.; Tan, Z.; et al. Bifidobacteria-mediated immune system imprinting early in life. Cell 2021, 184, 3884–3898. [Google Scholar] [CrossRef] [PubMed]
- Laursen, M.F.; Sakanaka, M.; von Burg, N.; Mörbe, U.; Andersen, D.; Moll, J.M.; Pekmez, C.T.; Rivollier, A.; Michaelsen, K.F.; Mølgaard, C.; et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat. Microbiol. 2021, 6, 1367–1382. [Google Scholar] [CrossRef]
- Underwood, M.A.; Arriola, J.; Gerber, C.W.; Kaveti, A.; Kalanetra, K.M.; Kananurak, A.; Bevins, C.L.; Mills, D.A.; Dvorak, B. Bifidobacterium longum subsp. infantis in experimental necrotizing enterocolitis: Alteration in inflammation, innate immune response, and the microbiota. Pediatr. Res. 2014, 76, 326–333. [Google Scholar]
- Good, M.; Sodhi, C.P.; Yamaguchi, Y.; Jia, H.; Lu, P.; Fulton, W.B.; Martin, L.Y.; Prindle, T.; Niño, D.F.; Zhou, Q.; et al. The human milk oligosaccharide 2’-fucosyllactose attenuates the severity of experimental necrotizing enterocolitis by enhancing mesenteric perfusion in the neonatal intestine. Br. J. Nutr. 2016, 116, 1175–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sodhi, C.P.; Wipf, P.; Yamaguchi, Y.; Fulton, W.B.; Kovler, M.; Niño, D.F.; Zhou, Q.; Banfield, E.; Werts, A.D.; Ladd, M.R.; et al. The human milk oligosaccharides 2’-fucosyllactose and 6’-fucosyllactose protect against the development of necrotizing enterocolitis by inhibiting toll-like receptor 4 signalling. Pediatr. Res. 2021, 89, 91–101. [Google Scholar] [CrossRef]
- Autran, C.A.; Kellman, B.P.; Kim, J.H.; Asztalos, E.; Blood, A.B.; Hamilton Spence, E.C.; Patel, A.L.; Hou, J.; Lewis, N.E.; Bode, L. Human milk oligasccharide composition predicts risk of necrotising enterocolitis in preterm infants. Gut 2018, 67, 1064–1070. [Google Scholar] [CrossRef] [PubMed]
- Perretta, L.; Ouldibbat, L.; Hagadorn, J.; Brumberg, H.L. High versus low medium chain triglyceride content of formula for promoting short-term growth of preterm infants. Cochrane Database Syst. Rev. 2021, 2, CD002777. [Google Scholar]
- Costeloe, K.; Hardy, P.; Juszczak, E.; Wilks, M.; Millar, M.R.; The Probiotics in Preterm Infants Study Collaborative Group. Bifidobacterium breve BBG-001 in very preterm infants: A randomized controlled phase 3 trial. Lancet 2016, 387, 649–660. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lueschow, S.R.; Boly, T.J.; Frese, S.A.; Casaburi, G.; Mitchell, R.D.; Henrick, B.M.; McElroy, S.J. Bifidobacterium longum Subspecies infantis Strain EVC001 Decreases Neonatal Murine Necrotizing Enterocolitis. Nutrients 2022, 14, 495. https://doi.org/10.3390/nu14030495
Lueschow SR, Boly TJ, Frese SA, Casaburi G, Mitchell RD, Henrick BM, McElroy SJ. Bifidobacterium longum Subspecies infantis Strain EVC001 Decreases Neonatal Murine Necrotizing Enterocolitis. Nutrients. 2022; 14(3):495. https://doi.org/10.3390/nu14030495
Chicago/Turabian StyleLueschow, Shiloh R., Timothy J. Boly, Steven A. Frese, Giorgio Casaburi, Ryan D. Mitchell, Bethany M. Henrick, and Steven J. McElroy. 2022. "Bifidobacterium longum Subspecies infantis Strain EVC001 Decreases Neonatal Murine Necrotizing Enterocolitis" Nutrients 14, no. 3: 495. https://doi.org/10.3390/nu14030495
APA StyleLueschow, S. R., Boly, T. J., Frese, S. A., Casaburi, G., Mitchell, R. D., Henrick, B. M., & McElroy, S. J. (2022). Bifidobacterium longum Subspecies infantis Strain EVC001 Decreases Neonatal Murine Necrotizing Enterocolitis. Nutrients, 14(3), 495. https://doi.org/10.3390/nu14030495





