Abstract
DNA methylation is an epigenetic mechanism that is crucial for mammalian development and genomic stability. Aberrant DNA methylation changes have been detected not only in malignant tumor tissues; the decrease of global DNA methylation levels is also characteristic for aging. The consumption of extra virgin olive oil (EVOO) as part of a balanced diet shows preventive effects against age-related diseases and cancer. On the other hand, consuming trans fatty acids (TFA) increases the risk of cardiovascular diseases as well as cancer. The aim of the study was to investigate the LINE-1 retrotransposon (L1-RTP) DNA methylation pattern in liver, kidney, and spleen of mice as a marker of genetic instability. For that, mice were fed with EVOO or TFA and were pretreated with environmental carcinogen 7,12-dimethylbenz[a]anthracene (DMBA)—a harmful substance known to cause L1-RTP DNA hypomethylation. Our results show that DMBA and its combination with TFA caused significant L1-RTP DNA hypomethylation compared to the control group via inhibition of DNA methyltransferase (DNMT) enzymes. EVOO had the opposite effect by significantly decreasing DMBA and DMBA + TFA-induced hypomethylation, thereby counteracting their effects.
1. Introduction
Adverse environmental effects often cause epigenetic modifications. In turn, the resulting genomic instability and abnormal methylation patterns can be observed in the background of cardiovascular and malignant diseases, obesity, type 2 diabetes mellitus, and neurodegenerative diseases [1]. A good example is the dietary intake of trans fatty acid (TFA), mainly from hydrogenated fats, which can account for 0.2–6.5% of energy intake [2]. In countries where more olive oil (OO) is consumed as an alternative to hydrogenated fats, the damage caused by TFA is lower [3].
Early epigenetic alterations may usually be reversed through chemopreventive compounds according to clinical trials [4]. Nutritional factors are the most important in chemoprevention and exert their effects mainly through antioxidation and anti-inflammatory effects. Anticancer effects of nutrition are mediated partly through gene expression ensured by genomic stability. Proapoptotic effects or antiproliferative regulation by nutritional factors can help to maintain genomic stability, as supported by numerous in vitro experiments, in vivo experiments, and clinical trials [5,6,7,8,9,10]. The frequent consumption of OO, particularly extra virgin olive oil (EVOO), has been shown to protect against cardiovascular diseases and malignancies and even to increase life expectancy [11,12]. The constituents of OO are capable to reduce the infarct size, exert strong antioxidant protection, and reduce the total cholesterol as well as triglyceride level in vivo [13]. The aging process is also accompanied by epigenetic and gene expression changes, mainly due to alterations in DNA methylation patterns toward a genome-wide more hypomethylated state [14].
1.1. Effects of Trans-Fatty Acids
TFA content of food increases the risk of cardiovascular diseases (CVD), breast cancer, prostate cancer, diabetes, and obesity [15], which also shortens life expectancy. A 16-year prospective cohort study in the United States analyzed the fat intake of 521,120 people [16]. The limits of the quintiles of amounts of daily TFA intakes were 1.41; 1.81; 2.2; and 2.73 percentages of calorie intake. Between the data from the upper and lower quintiles is a positive association with mortality based on gender- and age-normalized hazard ratio (1.03; CI 1.00–1.05; p trend = 0.0062) [16].
In a meta-analysis, 7 prospective studies of total dietary TFA intake and 5 studies of serum TFA in which participants were 26 years old or older and appeared to be healthy were analyzed. Although TFA intake does not correlate with overall cancer mortality, a positive association between dietary TFA intake and relative risk (RR) of breast cancer (1.37; 95% CI 1.04–1.81; p = 0.02) was found in postmenopausal women [17]. Another meta-analysis involving nearly 140,000 subjects demonstrated the adverse effect of TFA, namely a 2% energy intake increase in dietary intake of TFA significantly elevated the risk of cardiovascular disease (RR 1.23 95% CI 1.11–1.37; p < 0.001) [18].
Thus, TFA-induced damages increase the risk of cardiovascular diseases (CVD), breast cancer, prostate cancer, diabetes, and obesity [15], through which TFA presumably also shortens life expectancy. In contrast, Alfin-Slater and coworkers fed rats with a TFA-enriched diet (TFA content was 0.32% of the body weight, 30 times of the human consumption/kg of body weight) and found no difference in life expectancy between rats fed with this diet and rats fed with a conventional diet [19].
However, TFA damage may be especially harmful through enhancing transforming growth factor-beta (TGF-β) production in case of solitary fibrous tumors, neoplasms (angiomyolipoma, leiomyoma, hemangioma, lymphangioma, juxtaglomerular cell tumor, renomedullary interstitial cell tumor, lipoma, and schwannoma), and malignant tumors (leiomyosarcoma, rhabdomyosarcoma, angiosarcoma, osteosarcoma, synovial sarcoma, fibrosarcoma, malignant fibrous histiocytoma) arising from renal mesangial cells [20].
1.2. Effects of Olive Oil
EVOO has 55–83% of omega-9 oleic acid, which is a monounsaturated fatty acid (MUFA), 3.5–21% of linoleic acid, which is a polyunsaturated fatty acid (PUFA), 7.5–20% of saturated palmitic acid, and 0.5–5% of stearic acid content, while triunsaturated omega-3 α-linolenic acid is present in 0–1.5% [21]. In addition, EVOO also contains protective water-soluble substances, the best-known being oleuropein and oleocanthal [22,23].
A meta-analysis by Pelucchi and coworkers found based on five case-control studies that the pooled RR of breast cancer between the lowest and the highest quartiles of the population consuming a diet containing olive oil was 0.62 (95% CI 0.44–0.88) [24]. In another study, it was found that OO consumption significantly reduced the risk of the development of lung cancer (OR: 0.65; 95% CI: 0.42–0.99; p < 0.05) [25]. A case-control study found a significant difference in the protective effect against laryngeal cancer between the highest quartile consuming 42.9 g olive oil per day and the lowest quartile consuming less than 3.2 g per day (OR: 0.4 (95% CI: 0.3–0.7; p = 0.01) [26]. In a case-control study, a statistically significant inverse dose–response relationship was also found between the risk of developing bladder cancer and the level of olive oil consumption, when comparing the data of the lower tertile and middle tertile of less than 1.6 g OO consumption per day (OR: 0.62; 95% CI: 0.39–0.99) and the data of the lower and upper tertile consuming over 3.9 g per day (OR: 0.47; 95% CI: 0.28–0.78; p-trend = 0.002) [27].
1.3. The Effect of DMBA
The environmental carcinogen 7,12-dimethylbenz[a]anthracene (DMBA) is a harmful substance that can also be found in exhaust fumes, tobacco smoke, and burnt food. DMBA increased the risk of the development of bladder cancer, skin cancer, and soft tissue malignancies in proportion to age in rodents [28]. Thus, the DMBA-induced changes in molecular epidemiological biomarkers can reliably predict both the adverse environmental effects and the protective effect of chemopreventive agents, on which animal models can be based [29,30].
According to our present knowledge, no data are available either on the annual global exposure of humans to DMBA or on its effects on reducing life expectancy, but DMBA damage causes LINE-1 retrotransposon (L1-RTP) DNA hypomethylation [31], which is a relevant biomarker of biological aging [14,32].
1.4. DNA Methylation
DNA methylation involves the substitution of the hydrogen atom of the number 5 carbon atom of the cytosine ring by a methyl group due to the action of DNA methyltransferase (DNMT) enzymes. This epigenetic regulatory mechanism silences the gene expression of the given gene by methylation at the tandem repeating CpG (cytosine preceding guanosine) islands in the promoter and/or enhancer region of genes [14,31]. The L1-RTP DNA methylation pattern is a representative biomarker of global methylation, with positive correlations between them [31,33].
Hypomethylation may be induced/caused by passive demethylation of DNA, lack of methyl-donor group containing substrates (for example methionine-deficient diet) [34], or by the altered functioning of DNMT enzymes [35]. The activity of DNMT enzymes is generally reduced in global hypomethylation, but parallelly, the activity of the DNMT1 enzyme may increase, leading to the hypermethylation of CpG islands of the tumor suppressor genes, silencing them—and thus increases the risk of carcinogenesis or malignancy [36].
Different organs in vivo and tumor cells in vitro show various correlation patterns between their aging and the possibility of the occurrence of mutations in them [37,38,39,40]. Mahmood and coworkers have found a positive association between L1-RTP hypomethylation measured in the DNA content of the cell-free fraction of blood and aging and the increased likelihood of malignant tumorous diseases [41,42]. Interestingly, however, the correlation between aging and the probability of development of somatic mutations in the kidney renal cell carcinoma (KIRC) and the kidney renal papillary cell carcinoma (KIRP) cell lines is inverse (Horvath, 2013)—which provides a basis for studying the methylation pattern of renal DNA.
1.4.1. DNA Methylation and Malignant Tumors
Global DNA hypomethylation occurs in malignant tumor tissues, but this is not a permanent process but a sudden one, usually preceding malignant transformation (Sheaffer, 2016). For example, there is a significant (p < 0.001) correlation between the incidence of hepatocellular carcinoma and the hypomethylation of serum L1-RTP DNA [43]. According to published results C-MYC gene expression—which is also important in carcinogenesis—increased with aging due to the hypomethylation of the promoter region in both the spleen and the liver of mice—and this may also cause downregulation of P53, which protects against aging through treating DNA damage [44,45,46].
1.4.2. DNA Methylation and Aging
With aging global DNA methylation levels tend to decline continuously—this phenomenon is known as “epigenetic drift” [14,28,35], which is also strongly influenced by environmental factors [38,47]. On the other hand, the “epigenetic clock” represents with respect to specific DNA segments and organs, how methylation of CpG regions changes with aging [Horvath, 2013; Jones, 2015; Lim, 2018]. For example, aging correlates with the hypomethylation of the liver tissue DNA both in human and rodent liver [48].
Obesity, smoking, and the lack of exercise are also positively associated with L1-RTP DNA hypomethylation in white blood cells [49] and with reduced life expectancy [47]. In Europe, smoking shortened life expectancy on average by 19.8% in men and by 18.9% in women, and overweight and obesity by 7.7% in men and by 11.7% in women [50].
1.5. Objective
Our study aimed to examine the L1-RTP DNA methylation pattern in the liver, spleen, and kidneys of DMBA-treated TFA—and EVOO-fed mice to determine whether the change in the quantitative values compared to the DMBA-treated and the controls free of DMBA reflects the expected harmful effect of TFA and the protective effect of EVOO, as reported in the literature. Furthermore, we also examined whether the change in the L1-RTP DNA methylation pattern was associated with the predictors of life expectancy of dietary TFA and EVOO consumption, and with DMBA exposure, based upon literature data.
A further aim of the experiment was to determine whether the effects of these carcinogenic/chemopreventive agents could be examined with the L1-RTP DNA methylation pattern as potentially relevant biomarker.
Our present study aims to investigate the extent of L1-RTP DNA methylation on the effects of DMBA exposure combined with TFA or EVOO consumption in the liver, spleen, and kidneys of mice in vivo.
2. Materials and Methods
We used eight groups of 12-week-old female CBA/Ca mice (n = 6) in our study. Untreated control and DMBA-treated control groups received no prefeeding, while one group of animals received 300 mg/day/animal of olive oil (Agraria Riva Del Garda SCA) and 300 mg/day/animal of TFA (trans-3-hexadecenoic acid) (Sigma Aldrich), respectively, in addition to their usual diet for 2 weeks before DMBA treatment. Table 1. contains the exposure details for DMBA, TFA and olive oil.

Table 1.
Treatment and feeding of the study groups.
Apart from the untreated (negative control) control group, the other seven groups received 20 mg/kg bodyweight DMBA intraperitoneally (Sigma-Aldrich) dissolved in 0.1 mL of corn oil. The negative control group was also injected with 0.1 mL corn oil. (Although the corn oil contains chemopreventive linoleic acid in 58–62% in earlier experiments, the effect of DMBA was proper, or even due to n-6 essential fatty acid content it could enhance the effect of DMBA [51,52,53]. After 24 h of DMBA exposure, the organs to be tested (liver, kidneys, and spleen) were removed after cervical dislocation.
Mice were housed according to the principles and guidelines of animal experimentation. Every effort was made to minimize their suffering. The experiment was conducted by following the ethical standards in force (University of Pécs, Animal Welfare Committee; Ethical approval number: BA02/2000-79/2017).
2.1. Isolation of DNA
DNA was isolated using the High Pure PCR Template Preparation Kit (Roche, Madison, WI, USA) according to the manufacturer’s instructions.
2.2. LINE-1 DNA Methylation
We used EpiTect Bisulfite kit (Qiagen, Hilden, Germany) for bisulfite conversion according to the manufacturer’s instructions. This process resulted in the conversion of unmethylated cytosines into uracil. High-resolution melting (HRM) analysis was then performed, which, based on the difference in melting point, was able to distinguish between uracil and methylated cytosine bases. If the DNA contains highly methylated regions, bisulfite conversion and subsequent amplification will result in a higher melting point because the retention of more cytosine will result in a higher GC content of the amplified fragment (there are three hydrogen bonds between guanine and cytosine). In less methylated regions, unmethylated cytosines are converted to adenine resulting in a lower melting temperature.
For the HRM analysis, primers targeting the CpG-rich region of LINE-1 were used [Newman, 2012], and the sequences were as follows: forward: 5′-GGT TGA GGT AGT ATT TTG TGT G-3′, reverse: 5′- TCC AAA AAC TAT CAA ATT CTC TAA C-3′. Amplification was performed in 96-well plates in a Roche LightCycler480 qPCR instrument (Roche, Madison, WI, USA). The reaction mix contained 20 ng of bisulfite-treated DNA, 0.75-0.75 μM forward and reverse primers, 1xLightCycler 480 High Resolution Melting Master (Roche, Madison, WI, USA) in 20 μL final volume [Bray, 2018)]. The parameters of PCR were the following: heating to 95 °C for 5 min, followed by 35 cycles: 1. 95 °C for 20 s, 2. 60 °C for 30 s, 3. 72 °C for 20 s. Then melting point/melting curve analysis was performed between 73 °C and 84 °C with temperature steps of 0.1 °C/2 s.
We used mouse high methylated genomic DNA (EpigenDx, Hopkinton, MA, USA) and mouse low methylated genomic DNA (EpigenDx, Hopkinton, MA, USA) for positive and negative controls, respectively, and their mixtures in different proportions to allow quantification of the methylation level of our samples.
2.3. Calculation and Statistical Analysis
We calculated and compared the relative L1-RTP DNA methylation levels of L1-RTP DNA expression levels using the 2-ΔΔCT method. The Kolmogorov–Smirnov test was used to examine the distribution of the results and Levene’s F-test and T-test were used to compare means. Calculations and analyses were performed using IBM SPSS 21 statistical software and the level of statistical significance was set at a p-value of <0.05.
Average DNA methylation levels were expressed as the percentage of untreated animals (negative controls).
3. Results
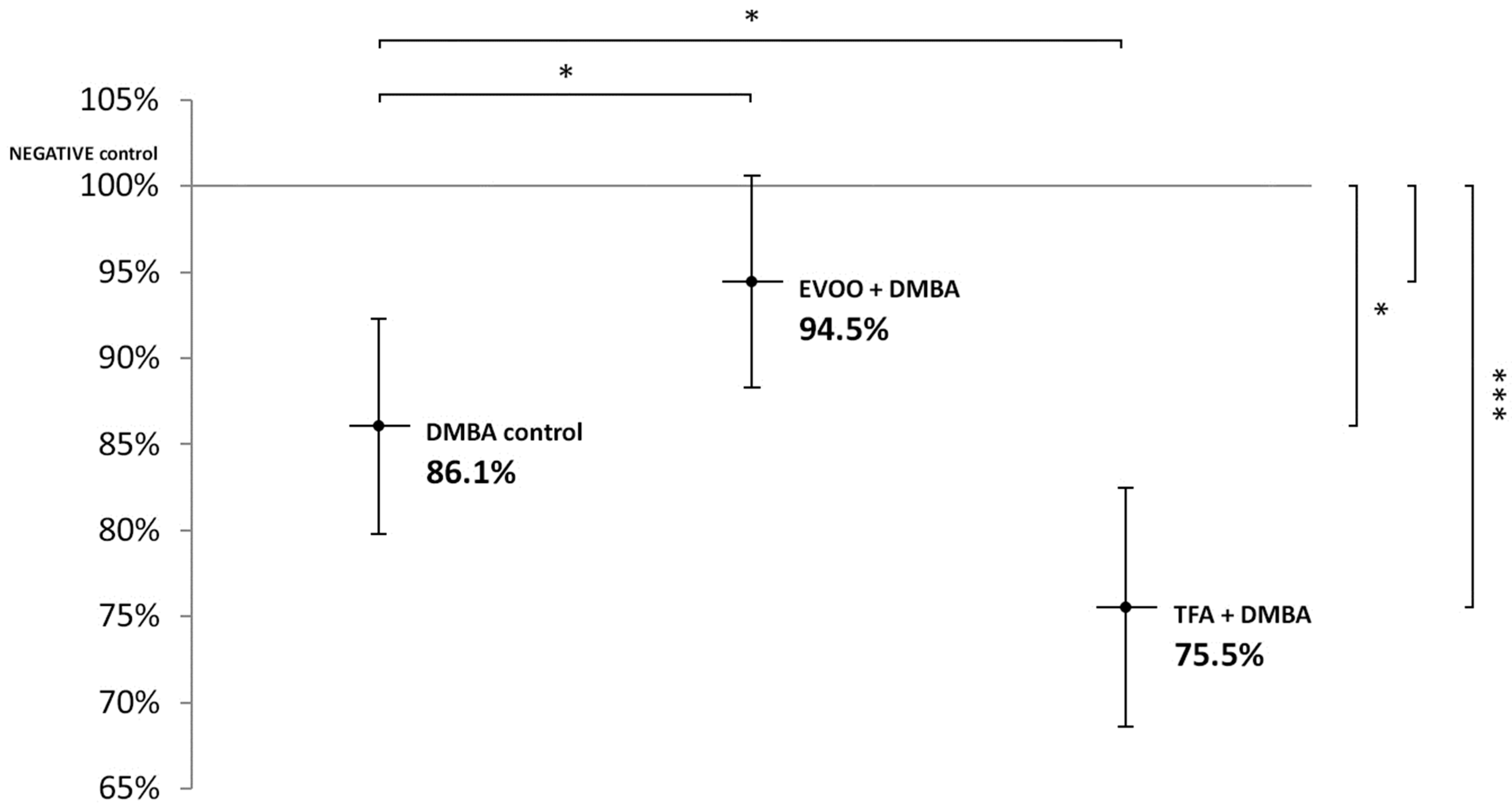
Compared to untreated control, EVOO coadministered with DMBA could partly ameliorate the hypomethylating effect of DMBA. DMBA alone and DMBA + TFA-induced significant L1-RTP DNA hypomethylation in the spleen (Figure 1).
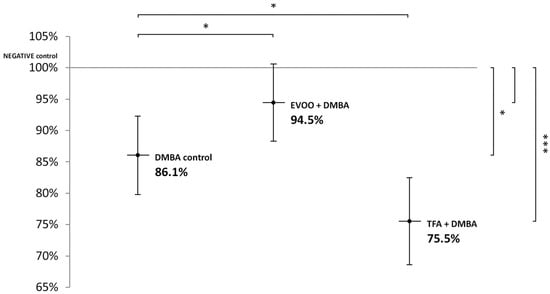
Figure 1.
L1-RTP DNA methylation pattern in the spleen of CBA/Ca female mice (n = 6) exposed to the effect of DMBA, and the effect of EVOO or TFA coadministered with DMBA, expressed as the percentage of untreated control (* p < 0.05; *** p < 0.001). L1-RTP DNA: LINE-1 retrotransposon deoxyribonucleic acid, EVOO: extra virgin olive oil, TFA: trans-fatty acid.
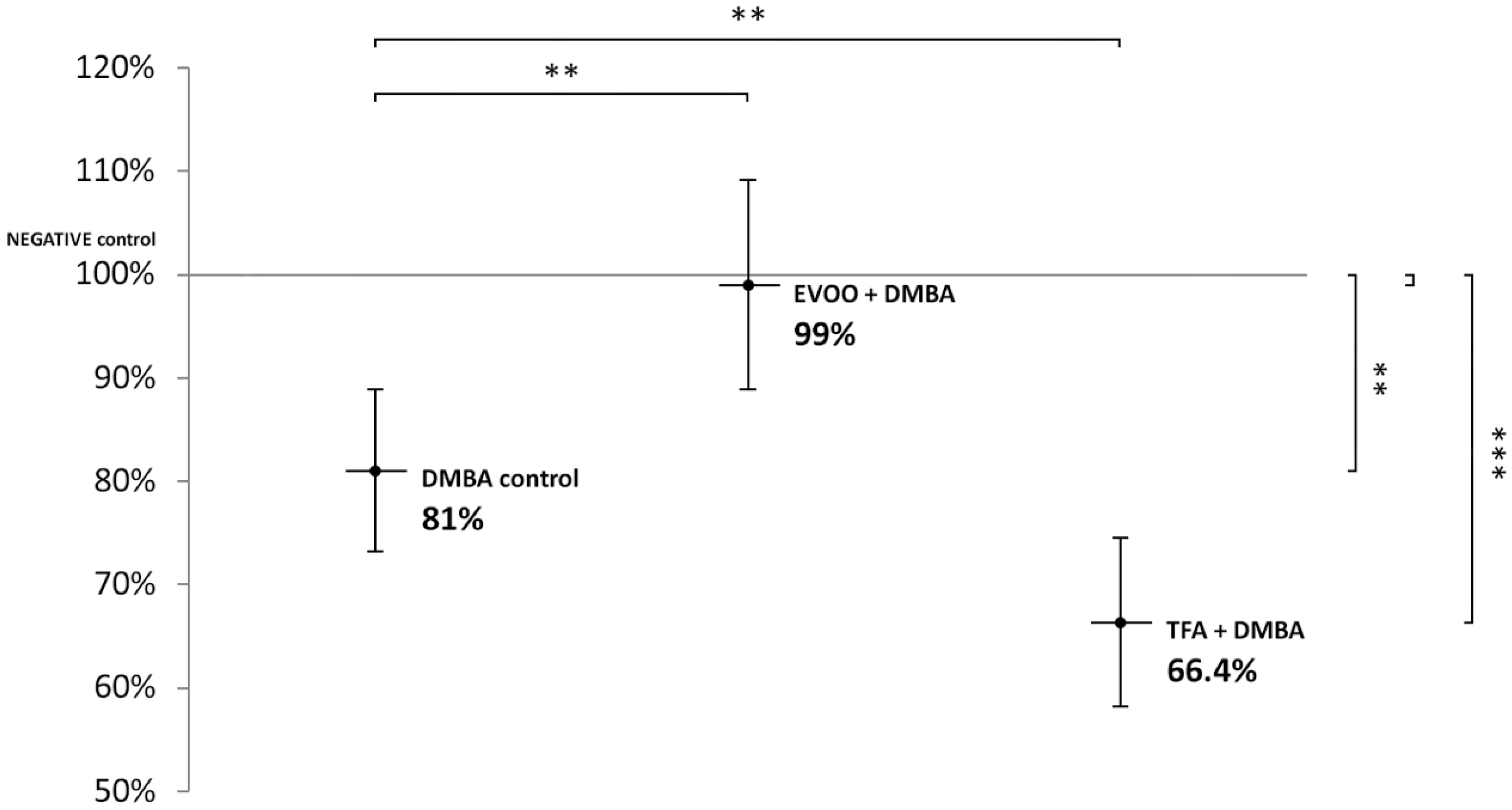
DMBA and DMBA + TFA-induced significant L1-RTP DNA hypomethylation in the liver, compared to the negative control. EVOO ameliorated the effect of DMBA (Figure 2).
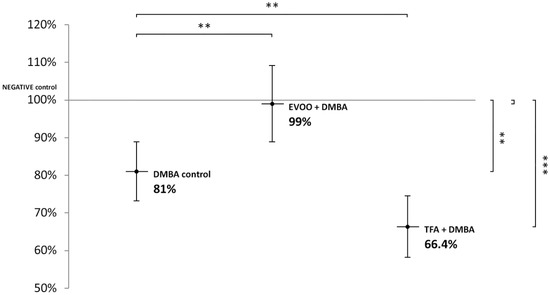
Figure 2.
L1-RTP DNA methylation pattern in the liver of CBA/Ca female mice (n = 6) exposed to the effect of DMBA, and the effect of EVOO or TFA coadministered with DMBA, expressed as the percentage of untreated control (** p < 0.01; *** p < 0.001).
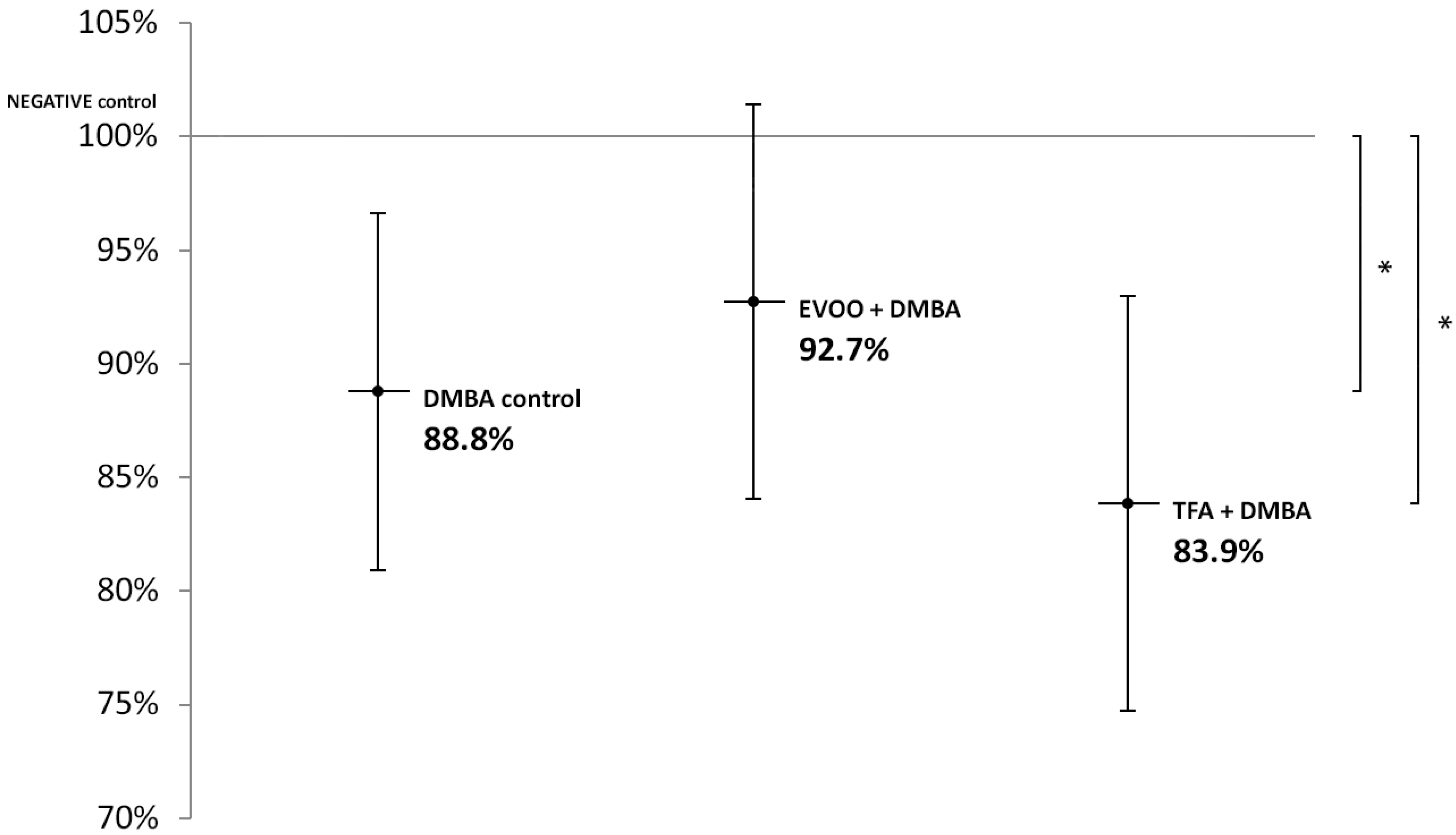
EVOO coadministered with DMBA could partly ameliorate the hypomethylating effect of DMBA. DMBA alone and DMBA + TFA caused significant L1-RTP DNA hypomethylation in the kidneys (Figure 3).
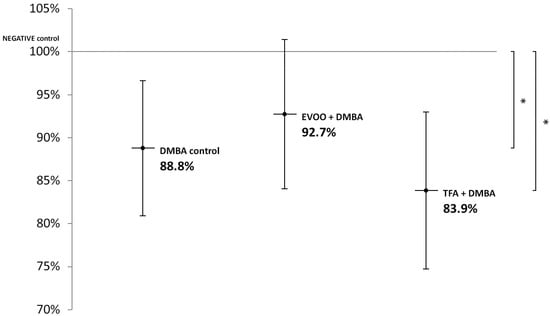
Figure 3.
L1-RTP DNA methylation pattern in the kidneys of CBA/Ca female mice (n = 6) exposed to the effects of DMBA and the effects of DMBA + EVOO or DMBA + TFA, expressed as the ratio of untreated control (* p < 0.05).
Thus, our observations showed that DMBA administered alone induced statistically significant L1-RTP DNA hypomethylation in all organs examined. DMBA could induce only a small, statistically not significant hypomethylation in all the three organs examined, if protective EVOO was added as well. As we expected, the combined effect induced by TFA and DMBA was significant and the highest degree of L1-RTP DNA hypomethylation in all three organs was observed. The numerical results of methylation level measurements are found in Appendix A’s Table A1, Table A2 and Table A3.
4. Discussion
Both DMBA and TFA generate ROS with partly overlapping molecular effects and signal transduction mechanisms [44,54,55].
4.1. Effect of ROS on the L1-RTP DNA Methylation and Aging
The damage caused by reactive oxygen species (ROS) generated during the decay of DMBA and TFA mainly contributes to global hypomethylation [44,54,55]. ROS depletes glutathione (GSH), S-adenosylmethionine (SAM) and S-adenosylhomocysteine (SAH) [31,56,57,58]. Decreases in GSH, SAM, and SAH levels cause global DNA hypomethylation [59,60], increase the risk of carcinogenesis [61,62], is associated with lipid peroxidation and cause age-related neurodegenerative diseases [63]. A decrease in SAH levels stimulates the DNMT1 enzyme [57] and contributes to the hypermethylation of CpG regions of tumor suppressor genes (for example P53) [31].
ROS also exerts harmful effects by activating secondary signaling pathways, for example, increases levels of interleukin 1β (IL1β), interleukin 6 (IL6), and tumor necrosis factor (TNF), and stimulates nuclear factor kappa B (NF-κB) [64,65], which indirectly increases the likelihood of malignant transformation [31,65,66,67], and is also directly proinflammatory [64,68,69]. TNF-α through IFN activation causes global DNA hypomethylation in aging cells [49,70]. Furthermore, when IL1β is present in high amounts, it stimulates additional inflammatory growth factors, namely TNF and matrix metalloproteinases (MMPs), etc. [71]. Both MMPs and TNF (in a redundant manner) promote malignant transformation of cells, as well as their progression [72], and activate NF-κB [71,73,74,75], thus forming a positive feedback loop. The mentioned interleukins and NF-κB mutually activate each other, and they also generate additional ROS [66,76], also forming a positive feedback loop.
Moreover, both DMBA and TFA activate the 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMG-CoAR) enzyme, which synthesizes cholesterol (for example in hepatocytes) that increases membrane rigidity [77,78]. For the sake of completeness, we need to mention that in the case of TFAs paradoxically, a decrease in cholesterol levels in Wistar rats has also been reported by Huang et al. [79]. With membrane rigidity and ROS formation, a positive association is presented within the phospholipid bilayer of the membrane [80] and ROS activity that elevates the risk of inflammation and malignant transformation [81,82]. For example, the increase of cholesterol levels in membranes favors the activation of the RAS oncogene family [78] both directly, through affecting the membrane rafts, and indirectly, via glycosylphosphatidylinositol (GPI) anchor proteins bound to membrane rafts [83,84].
The production of F2-isoprostane (F2-isoPs) increases up to 100-fold concentration in response to cholesterol and oxidative stress (predominantly lipid peroxidation) [81,85]. F2-isoPs distorts membrane fluidity and integrity [81]. Nevertheless, F2-isoPs increase the risk of carcinogenesis as well, for example by increasing proliferation [86]. Moreover, plasma free and total (free plus esterified) F2-isoPs increase with age (185% and 66%, respectively), but these increases are reduced by life-extending caloric restriction (50% and 23%, respectively) [87]. The levels of esterified F2-isoPs increase 68% with age in the liver, and 76% with age in the kidney, but caloric restriction modulated the age-related increase, reducing the esterified F2-isoPs levels 27% in the liver and 35% in the kidney [87].
4.2. Effect of DMBA on the L1-RTP DNA Methylation and Aging
DMBA caused significant L1-RTP DNA and oncogene (for example, RAS gene family) hypomethylation as well as hypermethylation of tumor suppressor genes (for example P53) compared to the control group via influencing DNMT enzymes [31,36,88]. Activated K-RAS hypermethylated the transcription factors of the tumor suppressor gene INK4-ARF, and thus silenced its expression [Struhl, 2014]. Its significance is that ARF/P53 signaling pathway is protective and has been shown to play an important role in slowing down aging [45], while P53 inhibits transposase enzyme [89] and hinders L1-RTP and presumably global DNA hypomethylation as well [31].
DMBA also activates the mitogen-activated protein kinase (MAPK) and Janus kinase (JAK) secondary signaling pathways [76], which activates the above-mentioned interleukins (and consequently NF-κB). These processes finally lead to global DNA hypomethylation [31] and accelerate aging, for example, by decreasing the expression of antitumorigenic microRNA-134 (miR-134) and P53 [67,75,90,91,92].
DMBA also significantly elevated mTORC1 gene expression and miR-9 level in the liver, spleen, and kidneys of CBA/CA female mice, compared to untreated controls [93]. DMBA activates the enzymes of glycolysis and lipogenesis [77]. Indeed, DMBA exposure in female Sprague-Dawley rats significantly elevated blood glucose levels compared to untreated controls [94]. The consequently released growth factors such as insulin and insulin-like growth factor (IGF) activate mTORC1 through phosphoinositide 3-kinase AKT-tuberous sclerosis-RHEB (PI3K-AKT-TSC-RHEB) signaling [95]. mTORC1 stimulates glycolysis and glucose uptake through modulating the transcription factor hypoxia-inducible factor (HIF1α) (Düvel 2010). HIF-1 increases glucose uptake and cell proliferation by increasing the expression of insulin-like growth factor 2 (IGF2) and C-MYC [96]. HIF-1 also induces inflammation by upregulating TNFα and cancer metastasis by upregulating fibronectin 1 [96]. However, increased activity of both mTOR and HIF-1 reduces life expectancy [96,97]. The expression level of miR-9 is increased by C-MYC, and miR-9 inhibits the progression of HCC as a tumor suppressor, but miR-9 also amplifies E-cadherin, which increases C-MYC expression, which increases miR-9 level, forming a positive feedback loop [93,98].
4.3. Effect of TFA on the L1-RTP DNA Methylation Pattern
TFA enters the cell membranes and increases their rigidity directly too leading to oxidative damage and inflammation [15]. Furthermore, TFAs decrease adiponectin and peroxisome proliferator-activated receptor gamma (PPAR-γ) activity [15,53]. If PPAR-γ is inactivated, it increases inflammatory response and hinders cholesterol transport, glucose, and fatty acid storage and promotes F2-isoPs formation [99]. Thus, the decrease of PPAR-γ activation results in a positive feedback loop with the mentioned harmful effects [53] [Smith, 2009], and it also hinders preadipocyte differentiation, thereby increasing the risk of developing malignant tumors and hinders tissue regeneration too [53,100].
Elaidic acid (trans-9-octadecenoic acid) (EA), induced global hypomethylation of THP-1 cells in vitro and activated proinflammatory (e.g., TNF-α, IL-6, C-reactive protein (CrP)) and adipogenic signaling pathways at concentrations of 50-200 μM [53,101,102]. Both trans-linoleic acid (trans, trans-9-12-octadecadienoic acid) (LA) and EA increase the levels of intercellular adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1) [64]. ICAM-1 and VCAM-1 also generate ROS, which activates NF-κB, which has a direct proinflammatory effect [64]. These oxidative and inflammatory damages are added to the effects of DMBA as mentioned earlier [55].
4.4. Protective Effect of OO
4.4.1. The Effect of Fat-Soluble Substances of OO on the LINE-1 DNA Methylation Pattern
The cell membrane fluidity enhancing effects of MUFA and PUFA promote DNA methylation via the above-mentioned secondary signaling pathways (for example by decreasing NF-κB) [67,103]. Theoretically, the saturated fatty acids, due to their membrane rigidity enhancing effects [104], could cause hypomethylation of L1-RTP DNA. In contrast, palmitic acid caused global hypermethylation [105] and reduced inflammation through the induction of the PPARγ gene [106] in human myocytes.
Oleic acid decreased the expression of TNF-α and IL1β and increased the anti-inflammatory IL10 in septic mice [107]. Furthermore, oleic acid can also stimulate PPAR [108], which activates antioxidant response, has anti-inflammatory and neuroprotective effects [107,109].
Oleic acid between 1 mM and 150 mM concentration allosterically activates the NAD-dependent deacetylase sirtuin-1 (SIRT1) [110], which is a regulator of mTOR [Ghosh, 2010]. SIRT1 inhibits the DNMT1 enzyme and through inhibition of DNMT3L protein, it blocks the gene expression of DNMT3A and DNMT3B enzymes too, [111]. For example, in MDA-MB-231 breast cancer cell line, SIRT1 reduced the inhibitory effect exerted by DNMT1 on tumor suppressor genes ERα and CDH1 [112]. (Interestingly, DNMT3 blocking effect was not accompanied by a decrease in the activity of the enzymes [111], but synergically with other chemopreventive agents, this could be still relevant).
PUFA, through its direct β-catenin inhibitory effect significantly reduced the expression of DMBA-induced C-MYC oncogene, compared to controls [44,52,113]. This is relevant, with respect to the DNA methylation pattern, is high since C-MYC induces oncogenic expression of the ten-eleven translocation methylcytosine dioxygenase 1 (TET1) gene, which codes for a DNA demethylating protein [114].
In addition, SIRT1 inhibits oxidative-stress-associated cellular aging [97], and C-MYC as well, through inhibition of β-catenin, which is important in the liver [67,115]. (In contrast, SIRT1 also inhibits P53, and hence SIRT1 may also act as an oncogene [115]). Moreover, oleic acid also prevented TNF-induced decline in insulin level by promoting the translocation of the transcription factor PPARγ into the nucleus, in a male KKAy type II diabetic mouse model [116].
4.4.2. Water-Soluble Substances of Olive Oil
Oleuropein and oleocanthal are water-soluble polyphenols of OO and are absorbed from the small intestine and reach the spleen and liver [117], where they exert a protective effect against ROS [118,119], mainly on the cell membrane [120].
Oleuropein can inhibit the activation of NF-κB [56,121] and increase the intracellular level of GSH, which is protective against the harmful effects of ROS [122,123,124]. Furthermore, oleuropein is also a PPARα agonist anti-inflammatory constituent [106,125].
Oleocanthal is a potent inhibitor of mTOR [126]. EVOO consumption significantly reduced the expression of mTORC1 gene both in the liver and the spleen of DMBA-treated CBA/Ca female mice [67]. Nanda et al. in Sprague-Dawley rats induced the DNMT1 enzyme by dimethylhydrazine and hypomethylated the promoters of NFκB, MMP-9, and VEGF, significantly increasing their gene expression compared to untreated controls, but these effects were counteracted by EVOO consumption [Nanda, 2019]. Indeed, the decrease in DNMT1 expression demethylates the promoter region of phosphatase and tensin homolog (PTEN), leading to the decrease of mTOR expression [127]. Although SAM, derived from methyl donor, stimulates mTOR through the SAMTOR protein [34], this effect is ultimately counteracted by EVOO [67].
4.5. L1-RTP DNA Methylation Patterns
ROS induce elevated blood glucose level, which is reflected in age-dependent biomarkers of renal damage, such as oxidant-sensitive heme oxygenase, advanced glycation end product (AGE), and F2-isoPs [128]. However, F2-isoPs, when added in vitro to renal mesangial cells (under high glucose levels, to which DMBA also contributes [94]; see above), increased the gene expression of TGF-β by activating protein kinase-C (PKC) [129]. TGF-β induced both expression and activity of DNA methyltransferases (DNMT) -1, -3A, and -3B in ovarian cancer cells [130], while in vitro phosphorylation of DNMT1 by PKCζ reduced its methyltransferase activity [131]. TGFβ, as a tumor suppressor, acts as a double-edged sword and activates anti-inflammatory signaling, but when its receptor loses function during malignant transformation, it indirectly acts as an immunosuppressant, promoting vascularization and metastasis, and thus enhances the malignancy of carcinomas [132] as mentioned earlier [20].
EVOO significantly decreased the DMBA-induced L1-RTP DNA hypomethylation both in the liver and spleen but not in the kidneys of experimental animals. This may be related to the fact that hypomethylation of L1-RTP DNA is not common even in RCC [133]. TFA tends to incorporate into the kidneys in smaller amounts than into the liver [79]. In Wistar rats, Huang et al. measured 1.2 mg/g TFA in the liver and only 0.6 mg/g TFA in the kidneys after their 16 weeks of consumption of a diet containing 4.5% TFA [79]. Indeed, lipid sensitivity of organs and hypomethylation of the L1-RTP DNA segment are associated in the case of TFA exposure [134].
4.5.1. L1-RTP DNA Methylation Pattern in the Liver and Spleen
The trans-3-hexadecenoic acid significantly increased the mTOR gene expression in the liver of DMBA treated mice group, even compared to the increase induced by DMBA exposure [67,77]. This can be explained by the fact that TFA inhibits the activity of CAT, SOD, and GSH peroxidase enzymes in lipid-sensitive liver and spleen [78,134]. Furthermore, TFA depletes antioxidant molecules (for example, GSH), which mainly protects against hepatotoxic processes [58]. Thereby TFA indirectly promotes the above-mentioned inflammation, tumor formation, and global DNA hypomethylation [31]. In addition, the elevated F2-isoPs levels under DMBA and TFA damage enhance the proliferation of stellate cells in the liver [86].
In nonalcoholic steatohepatitis (NASH) diseases, which include liver fibrosis and liver cancer, the composition of the cell membrane and the PPARα and the methylation pattern of DNA is also important [105,135]. Oleuropein as a PPARα agonist exerts hepatoprotective effects, such as reducing triglyceride levels [125]. Indeed, in hepatocellular carcinomas (HCC) the adenomatous polyposis coli (APC) and RASSF1 tumor suppressor genes were hypermethylated and the MEST gene was hypomethylated [136]. Both APC and RASSF1 slows cell proliferation—the former inhibits β-catenin, while the latter induces a cell cycle arrest mechanism by inhibiting cyclin D1, while MEST phosphorylates and thereby activates the transcription factor CREB, which enhances the expression of the C-FOS proto-oncogene [137]. Its importance is that in healthy aging, exons 1 and 4 of the C-FOS gene are hypermethylated, but both liver cirrhosis and liver carcinogenesis are accompanied by hypomethylation [138]. In an in vivo rat model, the DMBA and corn oil induced hypermethylation of RASSF1 promoter, but it was ameliorated by EVOO through decreasing DNMT1 enzyme’s activity [139]. Even in ApcMin/+ mice (that spontaneously develop intestinal polyps), the OO-enriched diet reduced polyp number and volume through a reduction of proliferation as well as proapoptotic effect by inhibiting fatty acid synthase and HMGCoA reductase gene expression [140]. Intriguingly, the secoiridoid polyphenol content of EVOO activated through C-FOS pathway the AP-1 (activator protein-1) transcription factors, which in this context were not associated with tumorigenesis but rather with growth inhibition and/or differentiation of breast cancer cells [141]. The predominant antiaging effect of EVOO secoiridoids was exerted through inhibiting mTOR and not by decreasing C-FOS activity [141].
PPARγ also regulates inflammatory factors in the liver, but the promoter of PPARγ is hypermethylated both in liver inflammation and liver fibrosis, and thus its expression is reduced [142]—although the oleuropein content of EVOO can counteract it [106]. Moreover, in rats fed with a high-fat diet, EVOO prevented hyperglycemia, insulinemia, apoptosis of pancreatic β-cells, and improved insulin resistance [143].
4.5.2. L1-RTP DNA Methylation Pattern in the Kidneys
The result of the kidneys, namely that the DMBA (or DMBA+TFA) induced L1-RTP DNA hypomethylation was weaker than in the other examined organs, could be explained by decreased DMBA damage through the generally silenced TSPYL5 gene [144] and by the generally increased expression of the antioxidant and anti-inflammatory lactoferrin (LTF) gene in the kidneys [144,145].
Both the expression of the TSPYL5 gene and the amount of TSPYL5 protein decrease with age [144] because both the DNMT1 (also indirectly activated by DMBA [31]) and the DNMT3B enzymes can cause hypermethylation of the promoter region of the TSPYL5 gene [146]. The TSPYL5 inhibits the activity of ubiquitin-specific protease 7 (USP7), which is the deubiquitylase enzyme for the P53 [147]. In summary, TSPYL5 reduces the activity of USP7 toward P53, resulting in increased P53 degradation through ubiquitylation [147]. Thus, ultimately, the decrease of TSPYL5, which inhibits the P53 and P21 tumor suppressors, may be the cause of the reduction of risk of mutation in the kidneys, compared to other organs. Indeed, P53 expression is slightly increased due to DMBA treatment in CBA/Ca mice in comparison to corn oil control [52] and P53 promotes both global and L1-RTP DNA hypermethylation by inhibiting LINE-1 transposons [89,148,149]. For the sake of completeness, it should be mentioned that TSPYL5 gene hypermethylation also occurs in HCC cells [150] as a protective mechanism.
LTF is generally highly expressed in the human kidneys, increasing further with age and is in vivo protective against DMBA generated ROS damage [144,145,151]. However, LTF from the viewpoint of senescence, as a double-edged sword, not only suppresses ROS-induced senescence of human mesenchymal stem cells (hMSCs) but also activates NF-κB through the Toll-like receptor 4 pathway [56,152].
The difference between the result in the kidneys and in other studied organs are explained by the fact that OO does not induce oxidative stress in the kidneys, but does in the other examined organs [153]. Thus, in the kidneys, the expression of the stress-dependent P53 gene was only slightly increased due to DMBA treatment [Budan, 2009; Kouka, 2020], while P53 could have theoretically stimulate global and L1-RTP DNA methylation, as mentioned earlier [89,148,149].
The effect of EVOO on methylation pattern may also contribute to the decrease in TSPYL5 expression and to the increase in LTF gene expression, which explains the reduced possibility of somatic mutation proportional to aging observed in the KIRC and the KIRP cell lines [Horvath, 2013], which is supported by the methylation pattern of the renal L1-RTP DNA in this study (Figure 3).
5. Conclusions
Both DMBA treatment and DMBA added combined with TFA caused significant L1-RTP DNA hypomethylation in the liver, spleen, and kidneys of CBA/Ca mice. According to the literature, DMBA forms DNA adducts and thereby inhibits tumor suppressor genes (for example, P53), activates oncogenes (for example, RAS, C-MYC, BCL-2, NOTCH), and alters microRNA (for example miR-9, miR-124, miR-132; miR-134) patterns leading to global hypomethylation [29,31,52,67,93,154].
Both DMBA and TFA manifest a dominant oxidative stress source by generating ROS and exerts proinflammatory effect, with mostly overlapping molecular biological features, namely depleting antioxidants (for example, GSH, SAM, SAH) promoting inflammatory signaling pathways (for example, IL-1β, IL-6, TNF, NF-κB, mTOR), and causing ultimately L1-RTP DNA hypomethylation [31,67].
Especially important is that according to the literature, TFA decreases PPAR-γ activity [Ali Abd El-Aal, 2019; Smith, 2009], which could otherwise ameliorate the harmful effect of DMBA [155], but if one is exposed to both agents, the synergically deleterious effect of DMBA and TFA exacerbates L1-RTP DNA hypomethylation, as reflected in the results of the present study. Moreover, TFA administration combined with DMBA further increased the significant L1-RTP DNA hypomethylation due to increased oxidative stress as well as increased adipogenic secondary signal transducers induction. Since aging and L1-RTP DNA methylation are similar in human and mouse species [156], the results are also of human relevance [157].
EVOO exerts antioxidant and anti-inflammatory effects directly on cell membranes, and through the regulation of secondary signal transporters [11,12,67], DMBA decreased significantly; additionally, combined DMBA + TFA-induced L1-RTP DNA hypomethylation was observed in the liver and spleen but not significantly in the kidneys of CBA/Ca mice. EVOO induces the PPARγ gene [106], and thereby, theoretically, it could decrease the mentioned synergic damage of DMBA combined with TFA.
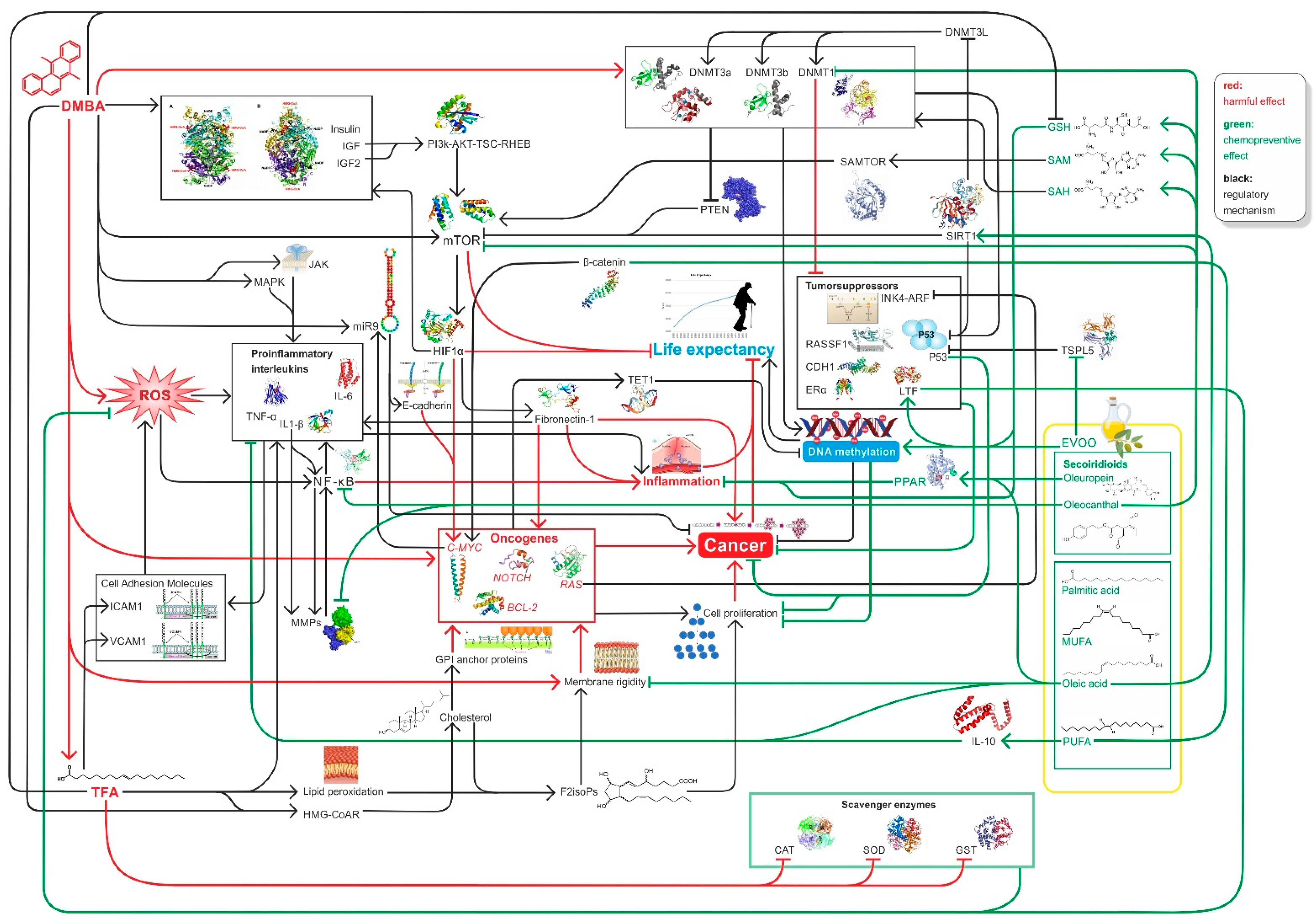
In summary, high EVOO intake with diet decreases the likelihood of cancer and increases life expectancy because EVOO can counteract DMBA and TFA-induced damage by improving global DNA methylation pattern, while decreasing hyperglycemia, mTOR activity, and inducing SIRT1 function among other [11,12,97,103,106,158] (Figure 4).
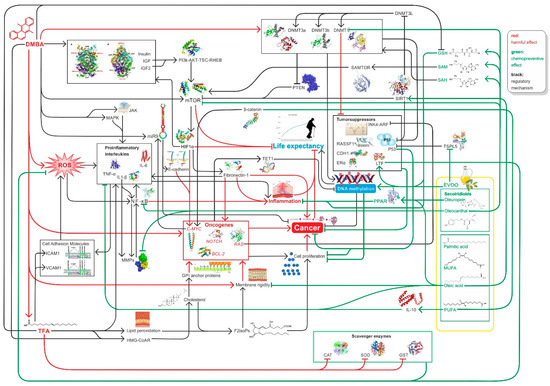
Figure 4.
Summary of relevant factors influencing inflammation, carcinogenesis DNA methylation, and ageing in connection with our study.
Author Contributions
Conceptualization, R.M. and T.V.; Data curation, A.T., R.D. and T.V.; Formal analysis, A.T., A.D. and T.V.; Funding acquisition, I.K.; Investigation, L.S., R.M., A.T., R.D. and T.V.; Methodology, L.S., R.M., A.D., R.D., Z.R., J.L.S., D.M. and F.B.; Project administration, L.S., R.M., D.M., A.S., F.B. and I.K.; Resources, L.S., Z.R., J.L.S., K.A., A.S. and F.B.; Software, A.T., R.D. and Z.R.; Supervision, Z.R., I.H., A.S., F.B., and I.K.; Validation, A.D., K.A., D.M., A.S. and F.B.; Visualization, J.L.S., K.A., D.M., I.H. and F.B.; Writing—original draft, D.M. and F.B.; Writing—review & editing, I.H., D.M., F.B. and I.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the European Union’s Horizon 2020 program, with grant numbers NEURAM Nr. 712821 and HCEMM Nr. 739593, as well as the Ministry of Innovation and Technology of Hungary Thematic Excellence Program (TKP-BIOImaging at Semmelweis University). The APC was funded by the European Union’s grant HCEMM Nr. 739593.
Institutional Review Board Statement
The animal study protocol was approved by University of Pécs, Animal Welfare Committee; and granted by the Hungarian Notified Body, ie. the National Food Chain Security Authority on 2.1.2018, with ethical approval number: BA02/2000-79/2017.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available upon reasonable request from the corresponding authors.
Acknowledgments
This work was supported by the European Union’s Horizon 2020 OPEN FET RIA (NEURAM, No, 712821), the Higher Education Institutional Excellence Programme of the Ministry for Innovation and Technology in Hungary, within the framework of the “Innovation for the sustainable life and environment” thematic programme of the University of Pecs. The research leading to these results has received funding from the European Union’s Horizon 2020 Teaming grant scheme with contract Nr. 739593 (HCEMM), as well as by the Hungarian Ministry of Innovation and Technology’s Thematic Excellence Program (TKP-BIOImaging at Semmelweis University). The authors express their special thanks to Péter Lajosházi for valuable technical assistance preparing Figure 4.
Conflicts of Interest
The Authors declare no conflict of interest.
Appendix A

Table A1.
LINE-1 methylation pattern in the spleen of CBA/Ca female mice (n = 6) exposed to the effect of DMBA and to the effect of EVOO or TFA co-administered with DMBA, expressed as the percentage of untreated control.
Table A1.
LINE-1 methylation pattern in the spleen of CBA/Ca female mice (n = 6) exposed to the effect of DMBA and to the effect of EVOO or TFA co-administered with DMBA, expressed as the percentage of untreated control.
| DMBA Control | DMBA + EVOO | DMBA + TFA | |
|---|---|---|---|
| mean LINE-1 methylation | 86.1% | 94.5% | 75.5% |
| distribution | 6.3% | 6.2% | 6.9% |
| p-value | 0.0180 | 0.2852 | 0.0007 |

Table A2.
LINE-1 methylation pattern in the liver of CBA/Ca female mice (n = 6) exposed to the effect of DMBA and to the effect of EVOO or TFA co-administered with DMBA, expressed as the percentage of untreated control.
Table A2.
LINE-1 methylation pattern in the liver of CBA/Ca female mice (n = 6) exposed to the effect of DMBA and to the effect of EVOO or TFA co-administered with DMBA, expressed as the percentage of untreated control.
| DMBA Control | DMBA + EVOO | DMBA + TFA | |
|---|---|---|---|
| mean LINE-1 methylation | 81.0% | 99.0% | 66.4% |
| distribution | 7.9% | 10.1% | 8.2% |
| p-value | 0.0042 | 0.8635 | 0.0001 |

Table A3.
LINE-1 methylation pattern in the kidneys of CBA/Ca female mice (n = 6) exposed to the effects of DMBA and to the effects of DMBA + EVOO or DMBA + TFA, expressed as the ratio of untreated control.
Table A3.
LINE-1 methylation pattern in the kidneys of CBA/Ca female mice (n = 6) exposed to the effects of DMBA and to the effects of DMBA + EVOO or DMBA + TFA, expressed as the ratio of untreated control.
| DMBA Control | DMBA + Olive Oil | DMBA + TFA | |
|---|---|---|---|
| mean LINE-1 methylation | 88.8% | 92.7% | 83.9% |
| distribution | 7.9% | 8.7% | 9.1% |
| p-value | 0.0444 | 0.1861 | 0.0117 |
References
- Ehrlich, M. DNA methylation in cancer: Too much, but also too little. Oncogene 2002, 21, 5400–5413. [Google Scholar] [CrossRef] [PubMed]
- Li, C. Global surveillance of trans-fatty acids. Prev. Chronic Dis. 2019, 16, 190121. [Google Scholar] [CrossRef] [PubMed]
- Craig-Schmidt, M.C. World-wide consumption of trans fatty acids. Atheroscler. Suppl. 2006, 7, 1–4. [Google Scholar] [CrossRef]
- Mondal, P.; Natesh, J.; Penta, D.; Meeran, S.M. Progress and promises of epigenetic drugs and epigenetic diets in cancer prevention and therapy: A clinical update. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, B.B.; Bhardwaj, A.; Aggarwal, R.S.; Seeram, N.P.; Shishodia, S.; Takada, Y. Role of resveratrol in prevention and therapy of cancer: Preclinical and clinical studies. Anticancer Res. 2004, 24, 2783–2840. [Google Scholar]
- Budán, F.; Szabó, I.; Varjas, T.; Nowrasteh, G.; Dávid, T.; Gergely, P.; Varga, Z.; Molnár, K.; Kádár, B.; Orsós, Z. Mixtures of Uncaria and Tabebuia extracts are potentially chemopreventive in CBA/Ca mice: A long-term experiment. Phytother. Res. 2011, 25, 493–500. [Google Scholar] [CrossRef]
- Cui, Y.; Morgenstern, H.; Greenland, S.; Tashkin, D.P.; Mao, J.T.; Cai, L.; Cozen, W.; Mack, T.M.; Lu, Q.Y.; Zhang, Z.F. Dietary flavonoid intake and lung cancer—A population-based case-control study. Cancer 2008, 112, 2241–2248. [Google Scholar] [CrossRef]
- Narayanan, B.A. Chemopreventive agents alters global gene expression pattern: Predicting their mode of action and targets. Curr. Cancer Drug Targets 2006, 6, 711–727. [Google Scholar] [CrossRef]
- Varjas, T.; Nowrasteh, G.; Budán, F.; Nadasi, E.; Horváth, G.; Makai, S.; Gracza, T.; Cseh, J.; Ember, I. Chemopreventive effect of Panax ginseng. Phytother. Res. Int. J. Devoted Pharmacol. Toxicol. Eval. Nat. Product Deriv. 2009, 23, 1399–1403. [Google Scholar]
- Varjas, T.; Nowrasteh, G.; Budán, F.; Horváth, G.; Cseh, J.; Gyöngyi, Z.; Makai, S.; Ember, I. The effect of fenugreek on the gene expression of arachidonic acid metabolizing enzymes. Phytother. Res. 2011, 25, 221–227. [Google Scholar] [CrossRef]
- Fernández del Río, L.; Gutiérrez-Casado, E.; Varela-López, A.; Villalba, J.M. Olive oil and the hallmarks of aging. Molecules 2016, 21, 163. [Google Scholar] [CrossRef] [PubMed]
- Giovannelli, L. Beneficial effects of olive oil phenols on the aging process: Experimental evidence and possible mechanisms of action. Nutr. Aging 2012, 1, 207–223. [Google Scholar] [CrossRef]
- Andreadou, I.; Iliodromitis, E.K.; Mikros, E.; Constantinou, M.; Agalias, A.; Magiatis, P.; Skaltsounis, A.L.; Kamber, E.; Tsantili-Kakoulidou, A.; Kremastinos, D.T. The olive constituent oleuropein exhibits anti-ischemic, antioxidative, and hypolipidemic effects in anesthetized rabbits. J. Nutr. 2006, 136, 2213–2219. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.J.; Goodman, S.J.; Kobor, M.S. DNA methylation and healthy human aging. Aging Cell 2015, 14, 924–932. [Google Scholar] [CrossRef]
- Abd El-Aal, Y.A.; Abdel-Fattah, D.M.; Ahmed, K.E.-D. Some biochemical studies on trans fatty acid-containing diet. Diabetes Metab. Syndr. Clin. Res. Rev. 2019, 13, 1753–1757. [Google Scholar] [CrossRef]
- Zhuang, P.; Zhang, Y.; He, W.; Chen, X.; Chen, J.; He, L.; Mao, L.; Wu, F.; Jiao, J. Dietary fats in relation to total and cause-specific mortality in a prospective cohort of 521 120 individuals with 16 years of follow-up. Circ. Res. 2019, 124, 757–768. [Google Scholar] [CrossRef]
- Anjom-Shoae, J.; Sadeghi, O.; Larijani, B.; Esmaillzadeh, A. Dietary intake and serum levels of trans fatty acids and risk of breast cancer: A systematic review and dose-response meta-analysis of prospective studies. Clin. Nutr. 2020, 39, 755–764. [Google Scholar] [CrossRef]
- Mozaffarian, D.; Katan, M.B.; Ascherio, A.; Stampfer, M.J.; Willett, W.C. Trans fatty acids and cardiovascular disease. N. Eng. J. Med. 2006, 354, 1601–1613. [Google Scholar] [CrossRef] [PubMed]
- Alfin-Slater, R.B.; Wells, A.F.; Aftergood, L.; Deuel, H.J., Jr. Nutritive value and safety of hydrogenated vegetable fats as evaluated by long-term feeding experiments with rats. J. Nutr. 1957, 63, 241–261. [Google Scholar] [CrossRef]
- Katabathina, V.S.; Vikram, R.; Nagar, A.M.; Tamboli, P.; Menias, C.O.; Prasad, S.R. Mesenchymal neoplasms of the kidney in adults: Imaging spectrum with radiologic-pathologic correlation. Radiographics 2010, 30, 1525–1540. [Google Scholar] [CrossRef]
- Beltrán, G.; Del Rio, C.; Sánchez, S.; Martínez, L. Influence of harvest date and crop yield on the fatty acid composition of virgin olive oils from cv. Picual. J. Agric. Food Chem. 2004, 52, 3434–3440. [Google Scholar] [CrossRef] [PubMed]
- Lipworth, L.; Martínez, M.a.E.; Angell, J.; Hsieh, C.-C.; Trichopoulos, D. Olive oil and human cancer: An assessment of the evidence. Prev. Med. 1997, 26, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Owen, R.; Giacosa, A.; Hull, W.; Haubner, R.; Spiegelhalder, B.; Bartsch, H. The antioxidant/anticancer potential of phenolic compounds isolated from olive oil. Eur. J. Cancer 2000, 36, 1235–1247. [Google Scholar] [CrossRef]
- Pelucchi, C.; Bosetti, C.; Negri, E.; Lipworth, L.; La Vecchia, C. Olive oil and cancer risk: An update of epidemiological findings through 2010. Curr. Pharm. Des. 2011, 17, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Fortes, C.; Forastiere, F.; Farchi, S.; Mallone, S.; Trequattrinni, T.; Anatra, F.; Schmid, G.; Perucci, C.A. The protective effect of the Mediterranean diet on lung cancer. Nutr. Cancer 2003, 46, 30–37. [Google Scholar] [CrossRef]
- Bosetti, C.; La Vecchia, C.; Talamini, R.; Negri, E.; Levi, F.; Dal Maso, L.; Franceschi, S. Food groups and laryngeal cancer risk: A case-control study from Italy and Switzerland. Int. J. Cancer 2002, 100, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, M.T.; Buntinx, F.; Kellen, E.; Van Dongen, M.C.; Dagnelie, P.C.; Muls, E.; Zeegers, M.P. Consumption of animal products, olive oil and dietary fat and results from the Belgian case–control study on bladder cancer risk. Eur. J. Cancer 2011, 47, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Anisimov, V.N. Carcinogenesis and aging 20 years after: Escaping horizon. Mech. Ageing Dev. 2009, 130, 105–121. [Google Scholar] [CrossRef]
- Ember, I.; Gyöngyi, Z.; Kiss, I.; Ghodratollah, N.; Arany, I. The possible relationship between onco/suppressor gene expression and carcinogen exposure in vivo: Evaluation of a potential biomarker in preventive and predictive medicine. Anticancer Res. 2002, 22, 2109–2116. [Google Scholar]
- Perjési, P.; Gyöngyi, Z.; Bayer, Z. Effect of E-2-(4’-methoxybenzylidene)-1-benzosuberone on the 7, 12-dimethylbenz [alpha] anthracene-induced onco/suppressor gene action in vivo II: A 48-hour experiment. Anticancer Res. 2000, 20, 1839–1848. [Google Scholar] [PubMed]
- Szabo, L.; Molnar, R.; Tomesz, A.; Deutsch, A.; Darago, R.; Nowrasteh, G.; Varjas, T.; Nemeth, B.; Budan, F.; Kiss, I. The effects of flavonoids, green tea polyphenols and coffee on DMBA induced LINE-1 DNA hypomethylation. PLoS ONE 2021, 16, e0250157. [Google Scholar] [CrossRef] [PubMed]
- Weidner, C.I.; Lin, Q.; Koch, C.M.; Eisele, L.; Beier, F.; Ziegler, P.; Bauerschlag, D.O.; Jöckel, K.-H.; Erbel, R.; Mühleisen, T.W. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014, 15, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Baba, Y.; Yagi, T.; Sawayama, H.; Hiyoshi, Y.; Ishimoto, T.; Iwatsuki, M.; Miyamoto, Y.; Yoshida, N.; Baba, H. Long interspersed element-1 methylation level as a prognostic biomarker in gastrointestinal cancers. Digestion 2018, 97, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Orozco, J.M.; Saxton, R.A.; Condon, K.J.; Liu, G.Y.; Krawczyk, P.A.; Scaria, S.M.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 2017, 358, 813–818. [Google Scholar] [CrossRef]
- Fraga, M.F.; Agrelo, R.; Esteller, M. Cross-talk between aging and cancer: The epigenetic language. Annu. N. Y. Acad. Sci. 2007, 1100, 60–74. [Google Scholar] [CrossRef] [PubMed]
- Laird, P.W. Oncogenic mechanisms mediated by DNA methylation. Mol. Med. Today 1997, 3, 223–229. [Google Scholar] [CrossRef]
- Li, M.; Liu, W.; Yuan, T.; Bai, R.; Liu, G.-H.; Zhang, W.; Qu, J. DNA methylome: Unveiling your biological age. Protein Cell 2013, 4, 723–725. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lim, U.; Song, M. DNA methylation as a biomarker of aging in epidemiologic studies. Cancer Epigenetics Precis. Med. 2018, 1856, 219–231. [Google Scholar]
- Snir, S.; vonHoldt, B.M.; Pellegrini, M. A statistical framework to identify deviation from time linearity in epigenetic aging. PLoS Comput. Biol. 2016, 12, e1005183. [Google Scholar] [CrossRef]
- Zampieri, M.; Ciccarone, F.; Calabrese, R.; Franceschi, C.; Bürkle, A.; Caiafa, P. Reconfiguration of DNA methylation in aging. Mech. Ageing Dev. 2015, 151, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Ardeljan, D.; Taylor, M.S.; Ting, D.T.; Burns, K.H. The human long interspersed element-1 retrotransposon: An emerging biomarker of neoplasia. Clin. Chem. 2017, 63, 816–822. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, W.; Erichsen, L.; Ott, P.; Schulz, W.A.; Fischer, J.C.; Arauzo-Bravo, M.J.; Bendhack, M.L.; Hassan, M.; Santourlidis, S. Aging-associated distinctive DNA methylation changes of LINE-1 retrotransposons in pure cell-free DNA from human blood. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tangkijvanich, P.; Hourpai, N.; Rattanatanyong, P.; Wisedopas, N.; Mahachai, V.; Mutirangura, A. Serum LINE-1 hypomethylation as a potential prognostic marker for hepatocellular carcinoma. Clinica Chimica Acta 2007, 379, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Budán, F.; Varjas, T.; Nowrasteh, G.; Varga, Z.; Boncz, I.; Cseh, J.; Prantner, I.; Antal, T.; Pazsit, E.; GŐBEL, G. Early Modification of c-myc, Ha-ras and p53 Expressions by N-Methyl-N-nitrosourea. In Vivo 2008, 22, 793–797. [Google Scholar]
- Matheu, A.; Maraver, A.; Klatt, P.; Flores, I.; Garcia-Cao, I.; Borras, C.; Flores, J.M.; Viña, J.; Blasco, M.A.; Serrano, M. Delayed ageing through damage protection by the Arf/p53 pathway. Nature 2007, 448, 375–379. [Google Scholar] [CrossRef]
- Ono, T.; Takahashi, N.; Okada, S. Age-associated changes in DNA methylation and mRNA level of the c-myc gene in spleen and liver of mice. Mutat. Res./DNAging 1989, 219, 39–50. [Google Scholar] [CrossRef]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.-B.; Gao, Y. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Hunt, N.J.; Kang, S.W.S.; Lockwood, G.P.; Le Couteur, D.G.; Cogger, V.C. Hallmarks of aging in the liver. Comput. Struct. Biotechnol. J. 2019, 17, 1151–1161. [Google Scholar] [CrossRef]
- Marques-Rocha, J.L.; Milagro, F.I.; Mansego, M.L.; Mourão, D.M.; Martínez, J.A.; Bressan, J. LINE-1 methylation is positively associated with healthier lifestyle but inversely related to body fat mass in healthy young individuals. Epigenetics 2016, 11, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Mackenbach, J.P.; Valverde, J.R.; Bopp, M.; Brønnum-Hansen, H.; Deboosere, P.; Kalediene, R.; Kovács, K.; Leinsalu, M.; Martikainen, P.; Menvielle, G. Determinants of inequalities in life expectancy: An international comparative study of eight risk factors. Lancet Public Health 2019, 4, e529–e537. [Google Scholar] [CrossRef]
- Ghazani, S.; Marangoni, A. Nutrition and food grains in Encyclopedia of Food Grains; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Budán, F.; Varjas, T.; Nowrasteh, G.; Prantner, I.; Varga, Z.; Ember, Á.; Cseh, J.; Gombos, K.; Pázsit, E.; Gőbel, G. Early Modification of c-myc, Ha-ras and p53 Expressions by Chemical Carcinogens (DMBA, MNU). In Vivo 2009, 23, 591–598. [Google Scholar]
- Smith, B.K.; Robinson, L.E.; Nam, R.; Ma, D.W. Trans-fatty acids and cancer: A mini-review. Br. J. Nutr. 2009, 102, 1254–1266. [Google Scholar] [CrossRef]
- Furlan, D.; Trapani, D.; Berrino, E.; Debernardi, C.; Panero, M.; Libera, L.; Sahnane, N.; Riva, C.; Tibiletti, M.G.; Sessa, F. Oxidative DNA damage induces hypomethylation in a compromised base excision repair colorectal tumourigenesis. Br. J. Cancer 2017, 116, 793–801. [Google Scholar] [CrossRef]
- Storz, P. Reactive oxygen species in tumor progression. Front. Biosci. 2005, 10, 1881–1896. [Google Scholar] [CrossRef]
- Park, L.K.; Friso, S.; Choi, S.-W. Nutritional influences on epigenetics and age-related disease. Proc. Nutr. Soc. 2012, 71, 75–83. [Google Scholar] [CrossRef]
- Ponnaluri, V.C.; Estève, P.-O.; Ruse, C.I.; Pradhan, S. S-adenosylhomocysteine hydrolase participates in DNA methylation inheritance. J. Mol. Biol. 2018, 430, 2051–2065. [Google Scholar] [CrossRef]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. and Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef]
- Karouzakis, E.; Gay, R.E.; Gay, S.; Neidhart, M. Increased recycling of polyamines is associated with global DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 2012, 64, 1809–1817. [Google Scholar] [CrossRef]
- Lertratanangkoon, K.; Wu, C.J.; Savaraj, N.; Thomas, M.L. Alterations of DNA methylation by glutathione depletion. Cancer Lett. 1997, 120, 149–156. [Google Scholar] [CrossRef]
- Tibbetts, A.S.; Appling, D.R. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu. Review Nutr. 2010, 30, 57–81. [Google Scholar] [CrossRef]
- Beetch, M.; Stefanska, B. DNA Methylation in Anti-cancer Effects of Dietary Catechols and Stilbenoids: An Overview of Underlying Mechanisms. In Handbook of Nutrition, Diet, and Epigenetics; Patel, V., Preedy, V., Eds.; Springer: Cham, Switzerland, 2019; pp. 1819–1844. [Google Scholar]
- Zhu, Y.; Carvey, P.M.; Ling, Z. Age-related changes in glutathione and glutathione-related enzymes in rat brain. Brain Res. 2006, 1090, 35–44. [Google Scholar] [CrossRef]
- Bryk, D.; Zapolska-Downar, D.; Malecki, M.; Hajdukiewicz, K.; Sitkiewicz, D. Trans fatty acids induce a proinflammatory response in endothelial cells through ROS-dependent nuclear factor-κB activation. J. Physiol. Pharmacol. 2011, 62, 229. [Google Scholar]
- RM, P.E.P.; Stein, I.; Bramovitch, R.; Amit, S.; Kasem, S.; Gutkovich-Pyest, E.; Urieli-Shoval, S.; Galun, E.; Ben-Neriah, Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature 2004, 431, 461–466. [Google Scholar]
- De Souza, V.R.; Cabrera, W.K.; Galvan, A.; Ribeiro, O.G.; De Franco, M.; Vorraro, F.; Starobinas, N.; Massa, S.; Dragani, T.A.; Ibanez, O.M. Aryl hydrocarbon receptor polymorphism modulates DMBA-induced inflammation and carcinogenesis in phenotypically selected mice. Int. J. Cancer 2009, 124, 1478–1482. [Google Scholar] [CrossRef]
- Molnar, R.; Szabo, L.; Tomesz, A.; Deutsch, A.; Darago, R.; Ghodratollah, N.; Varjas, T.; Nemeth, B.; Budan, F.; Kiss, I. In vivo effects of olive oil and trans-fatty acids on miR-134, miR-132, miR-124-1, miR-9-3 and mTORC1 gene expression in a DMBA-treated mouse model. PLoS ONE 2021, 16, e0246022. [Google Scholar] [CrossRef]
- Hendrayani, S.-F.; Al-Harbi, B.; Al-Ansari, M.M.; Silva, G.; Aboussekhra, A. The inflammatory/cancer-related IL-6/STAT3/NF-κB positive feedback loop includes AUF1 and maintains the active state of breast myofibroblasts. Oncotarget 2016, 7, 41974. [Google Scholar] [CrossRef]
- Kyriakis, J.M.; Avruch, J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 2001, 81, 807–869. [Google Scholar] [CrossRef]
- De Cecco, M.; Ito, T.; Petrashen, A.P.; Elias, A.E.; Skvir, N.J.; Criscione, S.W.; Caligiana, A.; Brocculi, G.; Adney, E.M.; Boeke, J.D. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 2019, 566, 73–78. [Google Scholar] [CrossRef]
- Apte, R.N.; Dotan, S.; Elkabets, M.; White, M.R.; Reich, E.; Carmi, Y.; Song, X.; Dvozkin, T.; Krelin, Y.; Voronov, E. The involvement of IL-1 in tumorigenesis, tumor invasiveness, metastasis and tumor-host interactions. Cancer Metastasis Rev. 2006, 25, 387–408. [Google Scholar] [CrossRef]
- Stetler-Stevenson, W.G.; Yu, A.E. Proteases in invasion: Matrix metalloproteinases. Semin. Cancer Biol. 2001, 11, 143–152. [Google Scholar] [CrossRef]
- Yeh, C.-B.; Hsieh, M.-J.; Hsieh, Y.-H.; Chien, M.-H.; Chiou, H.-L.; Yang, S.-F. Antimetastatic effects of norcantharidin on hepatocellular carcinoma by transcriptional inhibition of MMP-9 through modulation of NF-kB activity. PLoS ONE 2012, 7, e31055. [Google Scholar] [CrossRef]
- Wajant, H. The Role of TNF in Cancer. Death Recept. Cogn. Ligands Cancer 2009, 49, 1–15. [Google Scholar]
- Webster, G.A.; Perkins, N.D. Transcriptional cross talk between NF-κB and p53. Mol. Cell. Biol. 1999, 19, 3485–3495. [Google Scholar] [CrossRef]
- Brasier, A.R. The nuclear factor-κB–interleukin-6 signalling pathway mediating vascular inflammation. Cardiovasc. Res. 2010, 86, 211–218. [Google Scholar] [CrossRef]
- Haque, M.W.; Bose, P.; Siddique, M.U.M.; Sunita, P.; Lapenna, A.; Pattanayak, S.P. Taxifolin binds with LXR (α & β) to attenuate DMBA-induced mammary carcinogenesis through mTOR/Maf-1/PTEN pathway. Biomed. Pharmacother. 2018, 105, 27–36. [Google Scholar]
- Oteng, A.-B.; Kersten, S. Mechanisms of action of trans fatty acids. Adv. Nutr. 2020, 11, 697–708. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, B.; Pace, R.D.; Yoon, S. Trans fat intake lowers total cholesterol and high-density lipoprotein cholesterol levels without changing insulin sensitivity index in Wistar rats. Nutr. Res. 2009, 29, 206–212. [Google Scholar] [CrossRef]
- Pervaiz, S.; Taneja, R.; Ghaffari, S. Oxidative stress regulation of stem and progenitor cells. Antioxidants Redox Signal. 2009, 11, 2777–2789. [Google Scholar] [CrossRef]
- Morrow, J.D.; Awad, J.A.; Boss, H.J.; Blair, I.A.; Roberts, L.J. Non-cyclooxygenase-derived prostanoids (F2-isoprostanes) are formed in situ on phospholipids. Proc. Natl. Acad. Sci. USA 1992, 89, 10721–10725. [Google Scholar] [CrossRef]
- Clandinin, M.; Cheema, S.; Field, C.; Garg, M.; Venkatraman, J.; Clandinin, T. Dietary fat: Exogenous determination of membrane structure and cell function. FASEB J. 1991, 5, 2761–2769. [Google Scholar] [CrossRef]
- Ma, D.W. Lipid mediators in membrane rafts are important determinants of human health and disease. Appl. Physiol. Nutr. Metab. 2007, 32, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Prior, I.A.; Hancock, J.F. Compartmentalization of Ras proteins. J. Cell Sci. 2001, 114, 1603–1608. [Google Scholar] [CrossRef] [PubMed]
- Czerska, M.; Zieliński, M.; Gromadzińska, J. Isoprostanes–A novel major group of oxidative stress markers. Int. J. Occup. Med. Environ. Health 2016, 29, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Comporti, M.; Signorini, C.; Arezzini, B.; Vecchio, D.; Monaco, B.; Gardi, C. F2-isoprostanes are not just markers of oxidative stress. Free Radic. Biol. Med. 2008, 44, 247–256. [Google Scholar] [CrossRef]
- Ward, W.F.; Qi, W.; Remmen, H.V.; Zackert, W.E.; Roberts, L.J.; Richardson, A. Effects of age and caloric restriction on lipid peroxidation: Measurement of oxidative stress by F2-isoprostane levels. J. Gerontol. Ser. A Biol. Sci. Med. Sc. 2005, 60, 847–851. [Google Scholar] [CrossRef]
- Watson, R.E.; Curtin, G.M.; Doolittle, D.J.; Goodman, J.I. Progressive alterations in global and GC-rich DNA methylation during tumorigenesis. Toxicol. Sci. 2003, 75, 289–299. [Google Scholar] [CrossRef][Green Version]
- Tiwari, B.; Jones, A.E.; Abrams, J.M. Transposons, p53 and genome security. Trends Genet. 2018, 34, 846–855. [Google Scholar] [CrossRef]
- Rufini, A.; Tucci, P.; Celardo, I.; Melino, G. Senescence and aging: The critical roles of p53. Oncogene 2013, 32, 5129–5143. [Google Scholar] [CrossRef]
- Shuang, T.; Wang, M.; Zhou, Y.; Shi, C.; Wang, D. NF-κB1, c-Rel, and ELK1 inhibit miR-134 expression leading to TAB1 upregulation in paclitaxel-resistant human ovarian cancer. Oncotarget 2017, 8, 24853. [Google Scholar] [CrossRef]
- Wu, D.; Prives, C. Relevance of the p53–MDM2 axis to aging. Cell Death Differ. 2018, 25, 169–179. [Google Scholar] [CrossRef]
- Tomesz, A.; Szabo, L.; Molnar, R.; Deutsch, A.; Darago, R.; Mathe, D.; Budan, F.; Ghodratollah, N.; Varjas, T.; Nemeth, B. Effect of 7, 12-Dimethylbenz (α) anthracene on the Expression of miR-330, miR-29a, miR-9-1, miR-9-3 and the mTORC1 Gene in CBA/Ca Mice. In Vivo 2020, 34, 2337–2343. [Google Scholar] [CrossRef] [PubMed]
- Padlianah; Arif, M.; Yustisia, I. Blood chemistry profiles of DMBA-induced mammary tumor in female sprague dawley rats. In Proceedings of the International Conference on Bioinformatics and Nano-Medicine from Natural Resources for Biomedical Research: 3rd Annual Scientific Meeting for Biomedical Sciences, Malang, Indonesia, 21–23 November 2018; p. 020043. [Google Scholar]
- Mao, Z.; Zhang, W. Role of mTOR in glucose and lipid metabolism. Int. J. Mol. Sci. 2018, 19, 2043. [Google Scholar] [CrossRef] [PubMed]
- Yeo, E.-J. Hypoxia and aging. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, B.P.; Sinclair, D.A. Small molecule SIRT1 activators for the treatment of aging and age-related diseases. Trends Pharmacol. Sci. 2014, 35, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Khew-Goodall, Y.; Goodall, G.J. Myc-modulated miR-9 makes more metastases. Nat. Cell Biol. 2010, 12, 209–211. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, M.S.; Bilodeau, J.-F.; Larose, J.; Greffard, K.; Julien, P.; Barbier, O.; Rudkowska, I. Modulation of the biomarkers of inflammation and oxidative stress by ruminant trans fatty acids and dairy proteins in vascular endothelial cells (HUVEC). Prostaglandins Leukot. Essent. Fat. Acids 2017, 126, 64–71. [Google Scholar] [CrossRef]
- Zhang, K.; Yang, X.; Zhao, Q.; Li, Z.; Fu, F.; Zhang, H.; Zheng, M.; Zhang, S. Molecular mechanism of stem cell differentiation into adipocytes and adipocyte differentiation of malignant tumor. Stem Cells Int. 2020, 2020, 8892300. [Google Scholar] [CrossRef]
- Flores-Sierra, J.; Arredondo-Guerrero, M.; Cervantes-Paz, B.; Rodríguez-Ríos, D.; Alvarado-Caudillo, Y.; Nielsen, F.C.; Wrobel, K.; Wrobel, K.; Zaina, S.; Lund, G. The trans fatty acid elaidate affects the global DNA methylation profile of cultured cells and in vivo. Lipids Health Dis. 2016, 15, 1–7. [Google Scholar] [CrossRef]
- Menaa, F.; Menaa, A.; Menaa, B.; Tréton, J. Trans-fatty acids, dangerous bonds for health? A background review paper of their use, consumption, health implications and regulation in France. Eur. J. Nutr. 2013, 52, 1289–1302. [Google Scholar] [CrossRef]
- Papsdorf, K.; Brunet, A. Linking lipid metabolism to chromatin regulation in aging. Trends Cell Biol. 2019, 29, 97–116. [Google Scholar] [CrossRef]
- Hąc-Wydro, K.; Wydro, P. The influence of fatty acids on model cholesterol/phospholipid membranes. Chem. Phys. Lipids 2007, 150, 66–81. [Google Scholar] [CrossRef] [PubMed]
- Silva-Martínez, G.A.; Rodríguez-Ríos, D.; Alvarado-Caudillo, Y.; Vaquero, A.; Esteller, M.; Carmona, F.J.; Moran, S.; Nielsen, F.C.; Wickström-Lindholm, M.; Wrobel, K. Arachidonic and oleic acid exert distinct effects on the DNA methylome. Epigenetics 2016, 11, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Youssef, J.; Badr, M. Role of peroxisome proliferator-activated receptors in inflammation control. J. Biomed. Biotechnol. 2004, 2004, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Medeiros-de-Moraes, I.M.; Gonçalves-de-Albuquerque, C.F.; Kurz, A.R.; Oliveira, F.M.d.J.; Abreu, V.H.P.d.; Torres, R.C.; Carvalho, V.F.; Estato, V.; Bozza, P.T.; Sperandio, M. Omega-9 oleic acid, the main compound of olive oil, mitigates inflammation during experimental sepsis. Oxidative Med. Cell. Longev. 2018, 2018, 6053492. [Google Scholar] [CrossRef]
- Bento-Abreu, A.; Tabernero, A.; Medina, J.M. Peroxisome proliferator-activated receptor-alpha is required for the neurotrophic effect of oleic acid in neurons. J. Neurochem. 2007, 103, 871–881. [Google Scholar] [CrossRef]
- Fidaleo, M.; Fanelli, F.; Paola Ceru, M.; Moreno, S. Neuroprotective properties of peroxisome proliferator-activated receptor alpha (PPARα) and its lipid ligands. Curr. Med. Chem. 2014, 21, 2803–2821. [Google Scholar] [CrossRef]
- Najt, C.P.; Khan, S.A.; Heden, T.D.; Witthuhn, B.A.; Perez, M.; Heier, J.L.; Mead, L.E.; Franklin, M.P.; Karanja, K.K.; Graham, M.J. Lipid droplet-derived monounsaturated fatty acids traffic via PLIN5 to allosterically activate SIRT1. Mol. Cell 2020, 77, 810–824. e8. [Google Scholar] [CrossRef]
- Heo, J.; Lim, J.; Lee, S.; Jeong, J.; Kang, H.; Kim, Y.; Kang, J.W.; Yu, H.Y.; Jeong, E.M.; Kim, K. Sirt1 regulates DNA methylation and differentiation potential of embryonic stem cells by antagonizing Dnmt3l. Cell Rep. 2017, 18, 1930–1945. [Google Scholar] [CrossRef]
- Peng, L.; Yuan, Z.; Ling, H.; Fukasawa, K.; Robertson, K.; Olashaw, N.; Koomen, J.; Chen, J.; Lane, W.S.; Seto, E. SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities. Mol. Cell. Biol. 2011, 31, 4720–4734. [Google Scholar] [CrossRef]
- Notarnicola, M.; Tutino, V.; De Nunzio, V.; Dituri, F.; Caruso, M.G.; Giannelli, G. Dietary ω-3 polyunsaturated fatty acids inhibit tumor growth in transgenic ApcMin/+ mice, correlating with CB1 receptor up-regulation. Int. J. Mol. Sci. 2017, 18, 485. [Google Scholar] [CrossRef]
- Poole, C.J.; Lodh, A.; Choi, J.-H.; Van Riggelen, J. MYC deregulates TET1 and TET2 expression to control global DNA (hydroxy) methylation and gene expression to maintain a neoplastic phenotype in T-ALL. Epigenetics Chromatin 2019, 12, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.; Gu, W. SIRT1: Regulator of p53 deacetylation. Genes Cancer 2013, 4, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Vassiliou, E.K.; Gonzalez, A.; Garcia, C.; Tadros, J.H.; Chakraborty, G.; Toney, J.H. Oleic acid and peanut oil high in oleic acid reverse the inhibitory effect of insulin production of the inflammatory cytokine TNF-α both in vitro and in vivo systems. Lipids Health Dis. 2009, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bermúdez, B.; Pacheco, Y.M.; López, S.; Abia, R.; Muriana, F.J. Digestion and absorption of olive oil. Grasas y aceites 2004, 55, 1–10. [Google Scholar]
- Nakbi, A.; Tayeb, W.; Dabbou, S.; Issaoui, M.; Grissa, A.K.; Attia, N.; Hammami, M. Dietary olive oil effect on antioxidant status and fatty acid profile in the erythrocyte of 2, 4-D-exposed rats. Lipids Health Dis. 2010, 9, 1–10. [Google Scholar] [CrossRef]
- Seiquer, I.; Rueda, A.; Olalla, M.; Cabrera-Vique, C. Assessing the bioavailability of polyphenols and antioxidant properties of extra virgin argan oil by simulated digestion and Caco-2 cell assays. Comparative study with extra virgin olive oil. Food Chem. 2015, 188, 496–503. [Google Scholar] [CrossRef]
- Ferreri, C.; Masi, A.; Sansone, A.; Giacometti, G.; Larocca, A.V.; Menounou, G.; Scanferlato, R.; Tortorella, S.; Rota, D.; Conti, M. Fatty acids in membranes as homeostatic, metabolic and nutritional biomarkers: Recent advancements in analytics and diagnostics. Diagnostics 2017, 7, 1. [Google Scholar] [CrossRef]
- Hornedo-Ortega, R.; Cerezo, A.B.; De Pablos, R.M.; Krisa, S.; Richard, T.; García-Parrilla, M.C.; Troncoso, A.M. Phenolic compounds characteristic of the mediterranean diet in mitigating microglia-mediated neuroinflammation. Front. Cell. Neurosci. 2018, 373. [Google Scholar] [CrossRef]
- Giusti, L.; Angeloni, C.; Barbalace, M.C.; Lacerenza, S.; Ciregia, F.; Ronci, M.; Urbani, A.; Manera, C.; Digiacomo, M.; Macchia, M. A proteomic approach to uncover neuroprotective mechanisms of oleocanthal against oxidative stress. Int. J. Mol. Sci. 2018, 19, 2329. [Google Scholar] [CrossRef]
- Kouka, P.; Tsakiri, G.; Tzortzi, D.; Dimopoulou, S.; Sarikaki, G.; Stathopoulos, P.; Veskoukis, A.S.; Halabalaki, M.; Skaltsounis, A.-L.; Kouretas, D. The polyphenolic composition of extracts derived from different greek extra virgin olive oils is correlated with their antioxidant potency. Oxidative Med. Cell. Longev. 2019, 2019, 1870965. [Google Scholar] [CrossRef]
- López-Miranda, J.; Pérez-Jiménez, F.; Ros, E.; De Caterina, R.; Badimón, L.; Covas, M.I.; Escrich, E.; Ordovás, J.M.; Soriguer, F.; Abia, R. Olive oil and health: Summary of the II international conference on olive oil and health consensus report, Jaén and Córdoba (Spain) 2008. Nutr. Metab. Cardiovasc. Dis. 2010, 20, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Malliou, F.; Andreadou, I.; Gonzalez, F.J.; Lazou, A.; Xepapadaki, E.; Vallianou, I.; Lambrinidis, G.; Mikros, E.; Marselos, M.; Skaltsounis, A.-L. The olive constituent oleuropein, as a PPARα agonist, markedly reduces serum triglycerides. J. Nutr. Biochem. 2018, 59, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Khanfar, M.A.; Bardaweel, S.K.; Akl, M.R.; El Sayed, K.A. Olive oil-derived oleocanthal as potent inhibitor of mammalian target of rapamycin: Biological evaluation and molecular modeling studies. Phytother. Res. 2015, 29, 1776–1782. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Chen, H.; Ren, S.; Xia, M.; Zhu, J.; Liu, Y.; Zhang, L.; Tang, L.; Sun, L.; Liu, H. Aberrant DNA methylation of mTOR pathway genes promotes inflammatory activation of immune cells in diabetic kidney disease. Kidney Int. 2019, 96, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Giacco, F.; Brownlee, M. Oxidative stress and diabetic complications. Circ. Res. 2010, 107, 1058–1070. [Google Scholar] [CrossRef]
- Badr, K.F.; Abi-Antoun, T.E. Isoprostanes and the kidney. Antioxid. Redox Signal. 2005, 7, 236–243. [Google Scholar] [CrossRef]
- Cardenas, H.; Vieth, E.; Lee, J.; Segar, M.; Liu, Y.; Nephew, K.P.; Matei, D. TGF-β induces global changes in DNA methylation during the epithelial-to-mesenchymal transition in ovarian cancer cells. Epigenetics 2014, 9, 1461–1472. [Google Scholar] [CrossRef]
- Lavoie, G.; Estève, P.-O.; Laulan, N.B.; Pradhan, S.; St-Pierre, Y. PKC isoforms interact with and phosphorylate DNMT1. BMC Biol. 2011, 9, 1–15. [Google Scholar] [CrossRef]
- Massagué, J.; Blain, S.W.; Lo, R.S. TGFβ signaling in growth control, cancer, and heritable disorders. Cell 2000, 103, 295–309. [Google Scholar] [CrossRef]
- Florl, A.R.; Löwer, R.; Schmitz-Dräger, B.; Schulz, W. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br. J. Cancer 1999, 80, 1312–1321. [Google Scholar] [CrossRef]
- Kelley, D.S.; Bartolini, G.L.; Newman, J.W.; Vemuri, M.; Mackey, B.E. Fatty acid composition of liver, adipose tissue, spleen, and heart of mice fed diets containing t10, c12-, and c9, t11-conjugated linoleic acid. Prostaglandins Leukot. Essent. Fatty Acids 2006, 74, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Hwang, J.-T.; Shin, E.J.; Hur, H.J.; Kang, K.; Choi, H.-K.; Chung, M.-Y.; Chung, S.; Sung, M.J.; Park, J.-H. Genome-wide analysis of DNA methylation identifies novel differentially methylated regions associated with lipid accumulation improved by ethanol extracts of Allium tubersosum and Capsella bursa-pastoris in a cell model. PLoS ONE 2019, 14, e0217877. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.-P.; Ancey, P.-B.; Esposti, D.D.; Cros, M.-P.; Sklias, A.; Scoazec, J.-Y.; Durantel, D.; Hernandez-Vargas, H.; Herceg, Z. Aberrant DNA methylation of imprinted loci in hepatocellular carcinoma and after in vitro exposure to common risk factors. Clin. Epigenetics 2015, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sheng, M.; McFadden, G.; Greenberg, M.E. Membrane depolarization and calcium induce c-fos transcription via phosphorylation of transcription factor CREB. Neuron 1990, 4, 571–582. [Google Scholar] [CrossRef]
- Choi, E.; Uyeno, S.; Nishida, N.; Okumoto, T.; Fujimura, S.; Aoki, Y.; Nata, M.; Sagisaka, K.; Fukuda, Y.; Nakao, K. Alterations of c-fos gene methylation in the processes of aging and tumorigenesis in human liver. Mutat. Res./Fundam. Mol. Mech. Mutagenesis 1996, 354, 123–128. [Google Scholar] [CrossRef]
- Rodríguez-Miguel, C.; Moral, R.; Escrich, R.; Vela, E.; Solanas, M.; Escrich, E. The role of dietary extra virgin olive oil and corn oil on the alteration of epigenetic patterns in the rat DMBA-induced breast cancer model. PLoS ONE 2015, 10, e0138980. [Google Scholar] [CrossRef]
- Barone, M.; Notarnicola, M.; Caruso, M.G.; Scavo, M.P.; Viggiani, M.T.; Tutino, V.; Polimeno, L.; Pesetti, B.; Di Leo, A.; Francavilla, A. Olive oil and omega-3 polyunsaturated fatty acids suppress intestinal polyp growth by modulating the apoptotic process in ApcMin/+ mice. Carcinogenesis 2014, 35, 1613–1619. [Google Scholar] [CrossRef]
- Menendez, J.A.; Joven, J.; Aragonès, G.; Barrajón-Catalán, E.; Beltrán-Debón, R.; Borrás-Linares, I.; Camps, J.; Corominas-Faja, B.; Cufí, S.; Fernández-Arroyo, S. Xenohormetic and anti-aging activity of secoiridoid polyphenols present in extra virgin olive oil: A new family of gerosuppressant agents. Cell Cycle 2013, 12, 555–578. [Google Scholar] [CrossRef]
- Zhao, Q.; Fan, Y.C.; Zhao, J.; Gao, S.; Zhao, Z.H.; Wang, K. DNA methylation patterns of peroxisome proliferator-activated receptor gamma gene associated with liver fibrosis and inflammation in chronic hepatitis B. J. Viral Hepat. 2013, 20, 430–437. [Google Scholar] [CrossRef]
- Jurado-Ruiz, E.; Álvarez-Amor, L.; Varela, L.M.; Berná, G.; Parra-Camacho, M.S.; Oliveras-Lopez, M.J.; Martínez-Force, E.; Rojas, A.; Hmadcha, A.; Soria, B. Extra virgin olive oil diet intervention improves insulin resistance and islet performance in diet-induced diabetes in mice. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Rowland, J.; Akbarov, A.; Eales, J.; Xu, X.; Dormer, J.P.; Guo, H.; Denniff, M.; Jiang, X.; Ranjzad, P.; Nazgiewicz, A. Uncovering genetic mechanisms of kidney aging through transcriptomics, genomics, and epigenomics. Kidney Int. 2019, 95, 624–635. [Google Scholar] [CrossRef] [PubMed]
- Teng, C.T. Lactoferrin gene expression and regulation: An overview. Biochem. and Cell Biol. 2002, 80, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.R.; Bryan, J.N.; Esebua, M.; Amos-Landgraf, J.; May, T.J. Testis specific Y-like 5: Gene expression, methylation and implications for drug sensitivity in prostate carcinoma. BMC Cancer 2017, 17, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Epping, M.T.; Meijer, L.A.; Krijgsman, O.; Bos, J.L.; Pandolfi, P.P.; Bernards, R. TSPYL5 suppresses p53 levels and function by physical interaction with USP7. Nat. Cell Biol. 2011, 13, 102–108. [Google Scholar] [CrossRef]
- Nasr, A.F.; Nutini, M.; Palombo, B.; Guerra, E.; Alberti, S. Mutations of TP53 induce loss of DNA methylation and amplification of the TROP1 gene. Oncogene 2003, 22, 1668–1677. [Google Scholar] [CrossRef]
- Tiwari, B.; Jones, A.E.; Caillet, C.J.; Das, S.; Royer, S.K.; Abrams, J.M. p53 directly represses human LINE1 transposons. Genes Dev. 2020, 34, 1439–1451. [Google Scholar] [CrossRef]
- Qiu, X.; Hu, B.; Huang, Y.; Deng, Y.; Wang, X.; Zheng, F. Hypermethylation of ACP1, BMP4, and TSPYL5 in hepatocellular carcinoma and their potential clinical significance. Dig. Dis. Sci. 2016, 61, 149–157. [Google Scholar] [CrossRef]
- Letchoumy, P.V.; Chandra Mohan, K.; Stegeman, J.; Gelboin, H.; Hara, Y.; Nagini, S. In vitro antioxidative potential of lactoferrin and black tea polyphenols and protective effects in vivo on carcinogen activation, DNA damage, proliferation, invasion, and angiogenesis during experimental oral carcinogenesis. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2008, 17, 193–203. [Google Scholar] [CrossRef]
- Ando, K.; Hasegawa, K.; Shindo, K.I.; Furusawa, T.; Fujino, T.; Kikugawa, K.; Nakano, H.; Takeuchi, O.; Akira, S.; Akiyama, T. Human lactoferrin activates NF-κB through the Toll-like receptor 4 pathway while it interferes with the lipopolysaccharide-stimulated TLR4 signaling. FEBS J. 2010, 277, 2051–2066. [Google Scholar] [CrossRef]
- Kouka, P.; Tekos, F.; Papoutsaki, Z.; Stathopoulos, P.; Halabalaki, M.; Tsantarliotou, M.; Zervos, I.; Nepka, C.; Liesivuori, J.; Rakitskii, V.N. Olive oil with high polyphenolic content induces both beneficial and harmful alterations on rat redox status depending on the tissue. Toxicol. Rep. 2020, 7, 421–432. [Google Scholar] [CrossRef]
- Sobinoff, A.P.; Mahony, M.; Nixon, B.; Roman, S.D.; McLaughlin, E.A. Understanding the Villain: DMBA-induced preantral ovotoxicity involves selective follicular destruction and primordial follicle activation through PI3K/Akt and mTOR signaling. Toxicol. Sci. 2011, 123, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Tikoo, K.; Kumar, P.; Gupta, J. Rosiglitazone synergizes anticancer activity of cisplatin and reduces its nephrotoxicity in 7, 12-dimethyl benz {a} anthracene (DMBA) induced breast cancer rats. BMC Cancer 2009, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.; Van Meter, M.; Ablaeva, J.; Ke, Z.; Gonzalez, R.S.; Taguchi, T.; De Cecco, M.; Leonova, K.I.; Kogan, V.; Helfand, S.L. LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab. 2019, 29, 871–885.e5. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Lee, Y.D.; Wagers, A.J. Stem cell aging: Mechanisms, regulators and therapeutic opportunities. Nat. Med. 2014, 20, 870–880. [Google Scholar] [CrossRef]
- Fontana, L.; Partridge, L.; Longo, V.D. Extending healthy life span—from yeast to humans. Science 2010, 328, 321–326. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).