Hydrolyzed Bound Phenolics from Rice Bran Alleviate Hyperlipidemia and Improve Gut Microbiota Dysbiosis in High-Fat-Diet Fed Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Main Chemical and Reagents
2.2. Preparation of HBP from Rice Bran
2.3. Animal Experiment
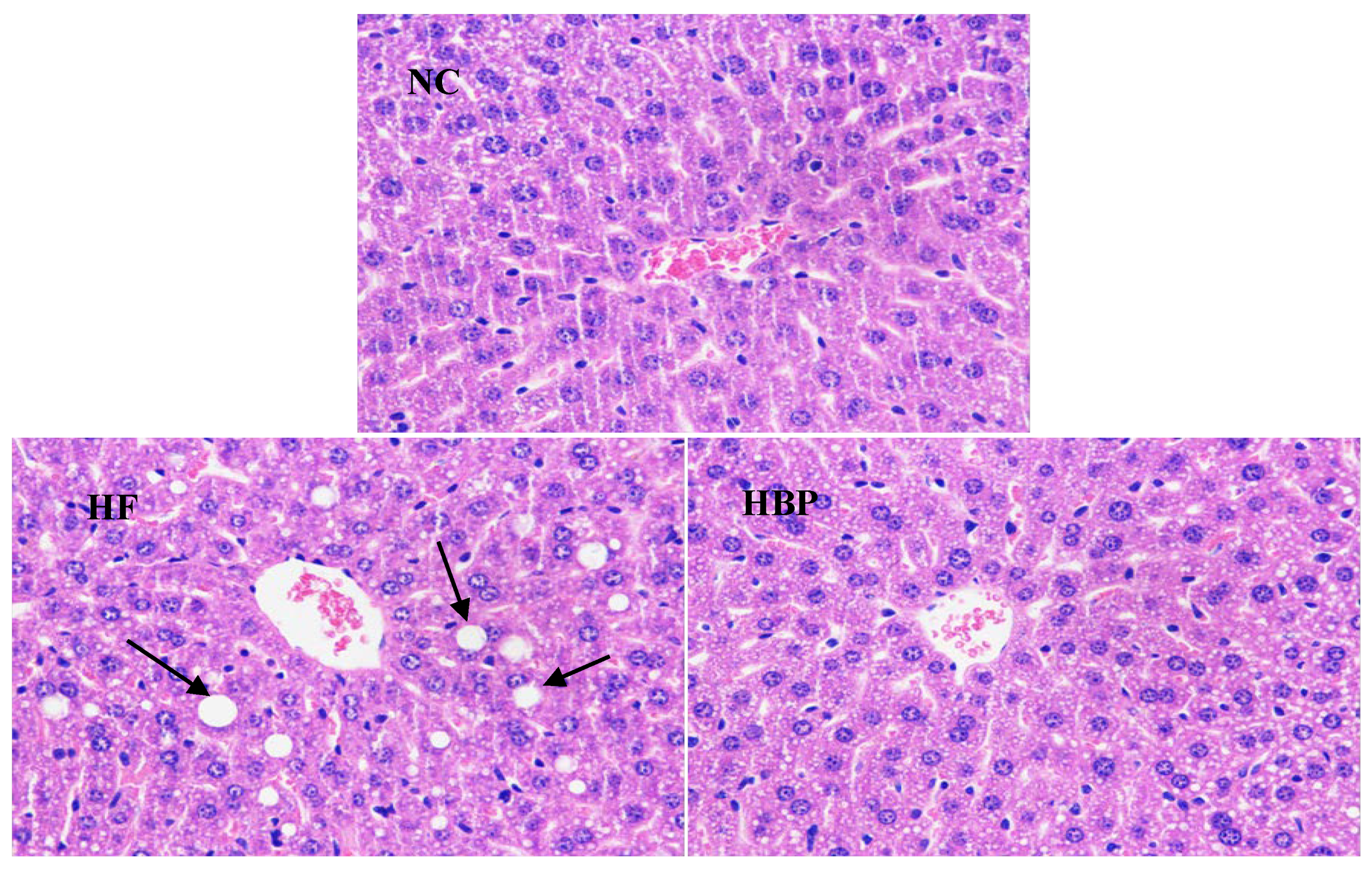
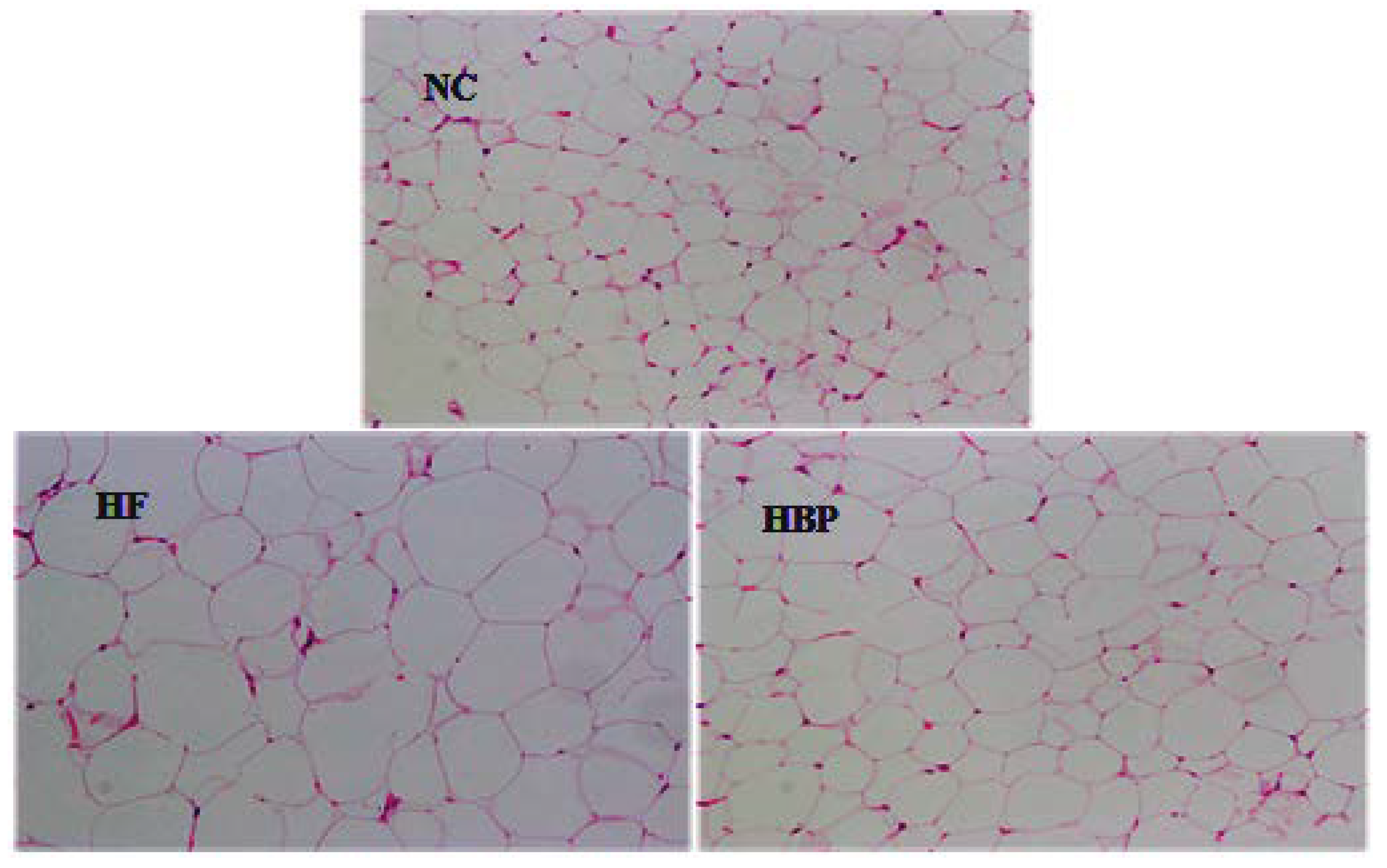
2.4. Histological Observation of the Liver and Epididymal Fat
2.5. Serumand Feces Biochemical Assays
2.6. Western Blot Analysis
2.7. Real Time PCR Analysis
2.8. Microbiota Analysis by 16S rRNA Gene Sequencing
2.9. Statistical Analysis
3. Results
3.1. Effects of HBP on the Serum and Feces Lipid Profiles of High-Fat-Diet Fed Mice
3.2. Effects of HBP on the Morphological Feasure of Liver and Visceral Adipose
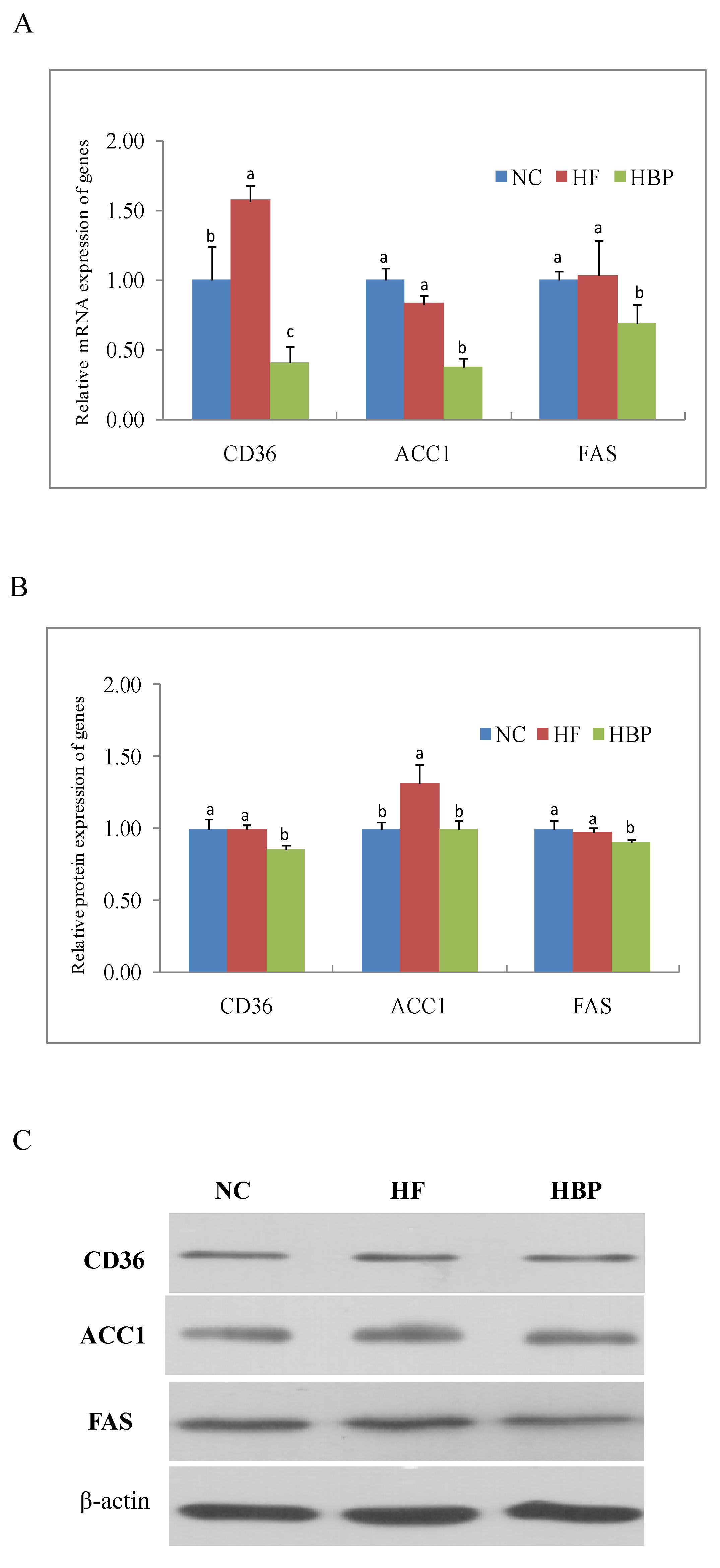
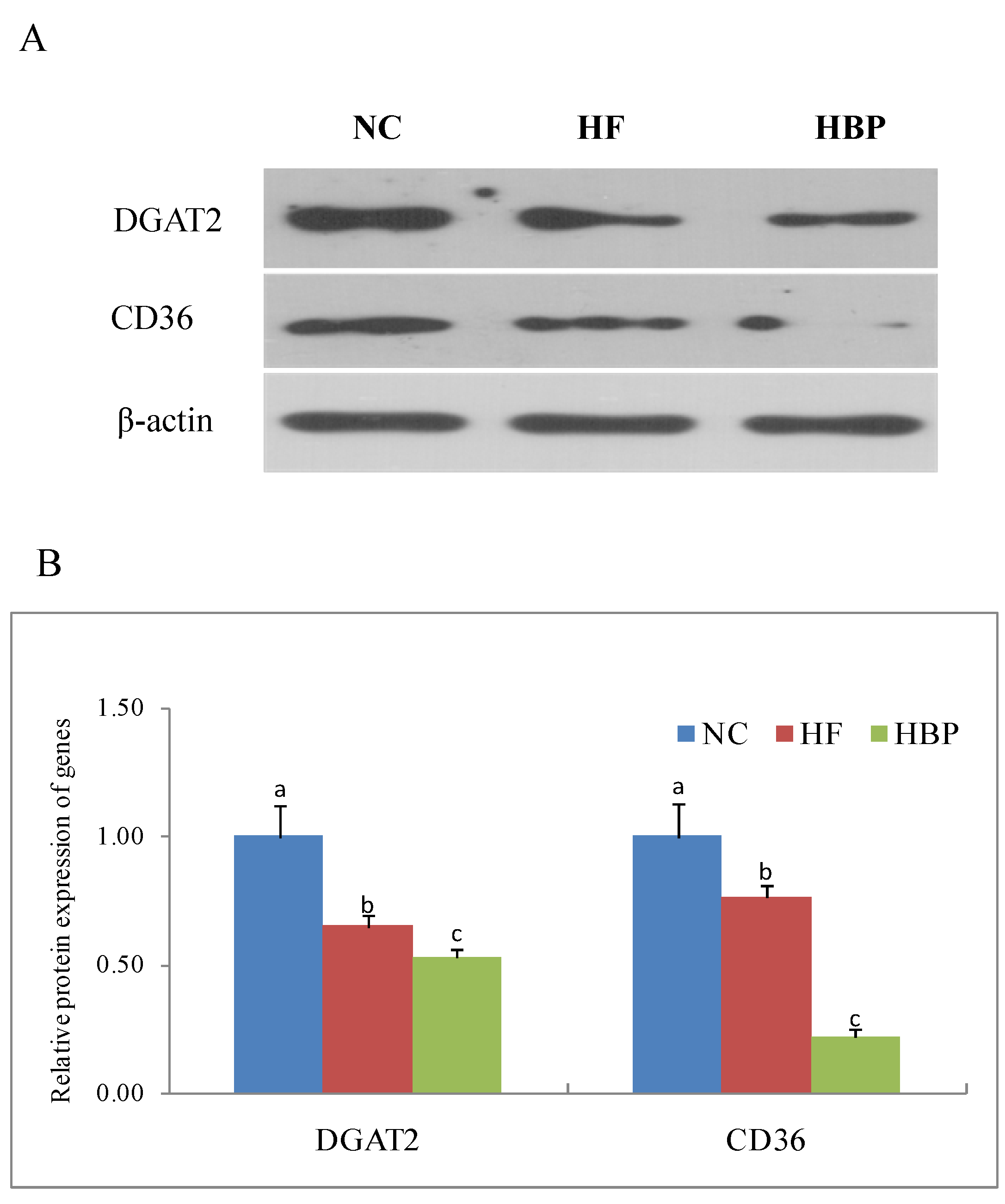
3.3. Effects of HBP on the Expression of Genes Related to Lipid Metabolism in the Liver
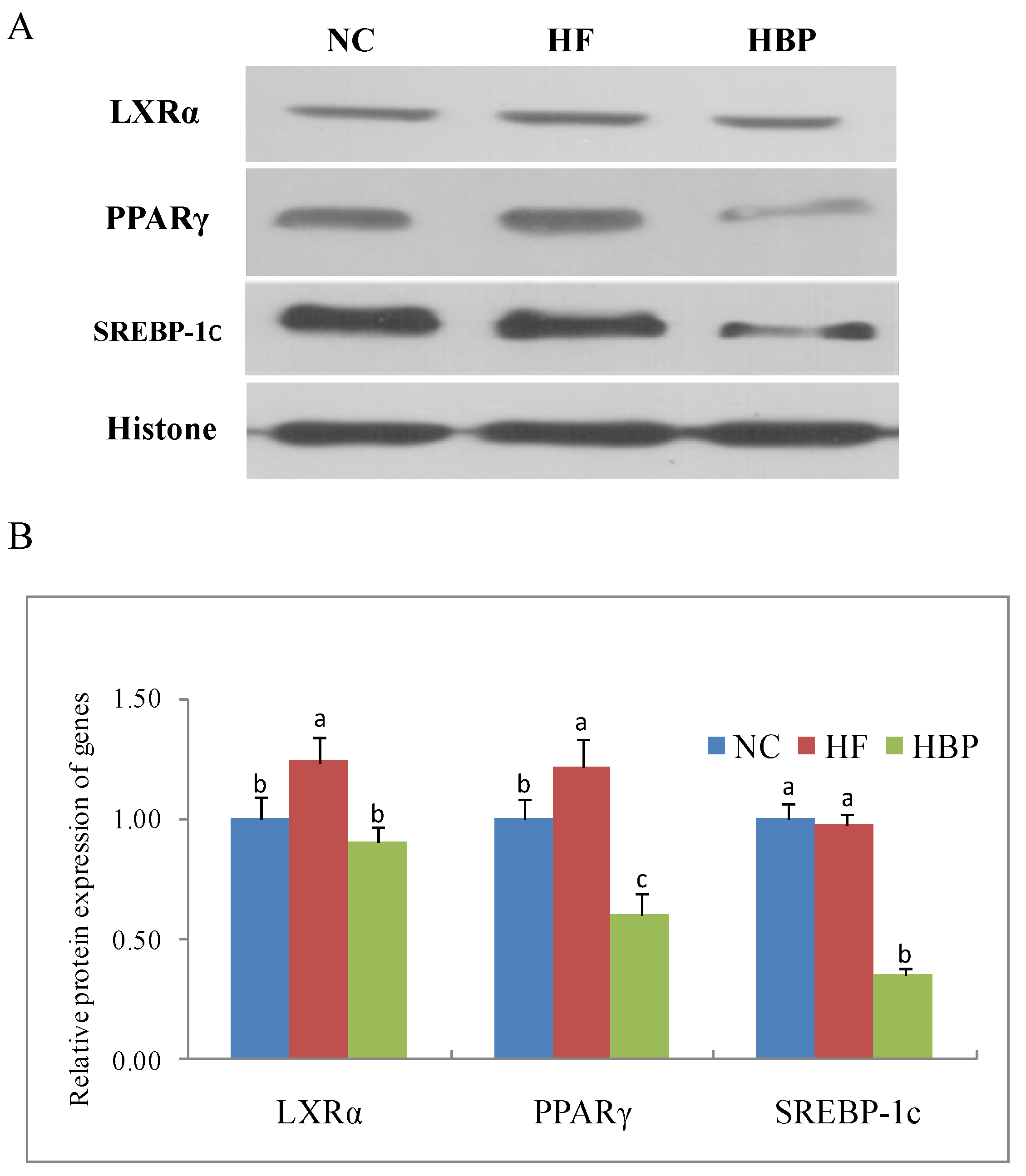
3.4. Effects of HBP on the Activation of Nuclear Receptors Related to Lipid Metabolism in the Liver
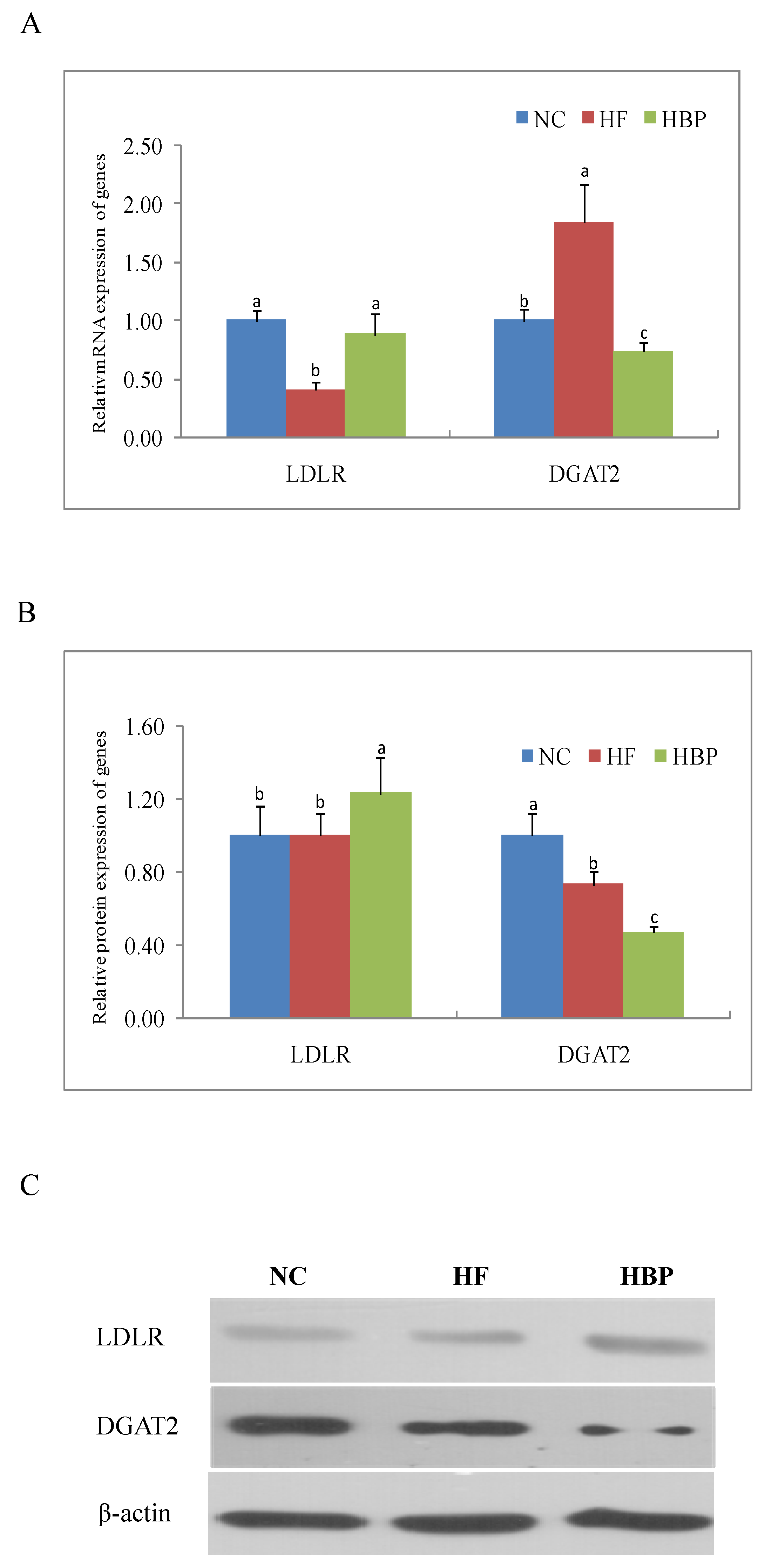
3.5. Effects of HBP on the Genes Related to Lipid Metabolism in the Gut
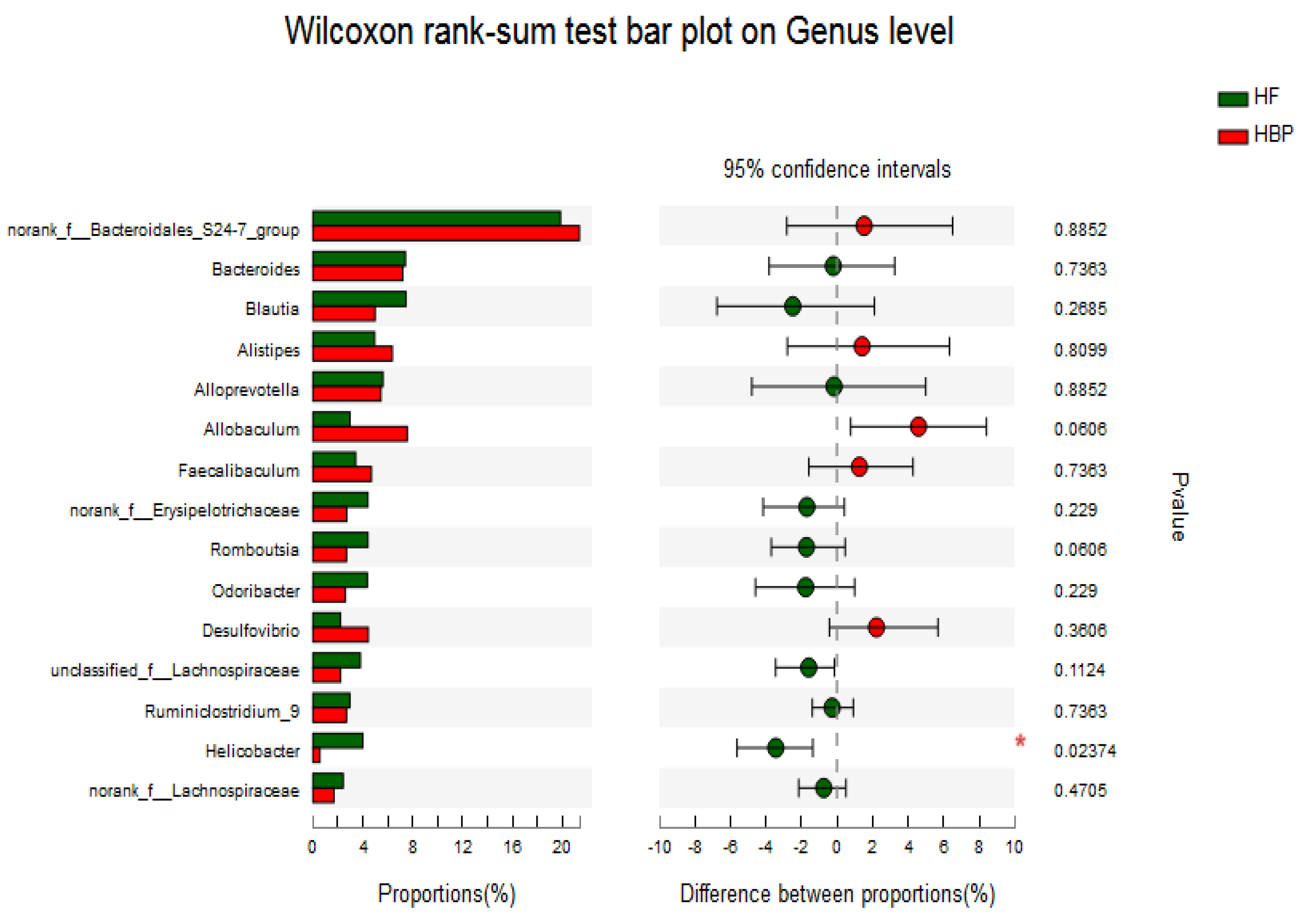
3.6. Assessment of Fecal Microbial Community Structure by 16S rRNA Sequencing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Bohn, T. Dietary factors affecting polyphenol bioavailability. Nutr. Rev. 2014, 72, 429–452. [Google Scholar] [CrossRef] [PubMed]
- Adom, K.K.; Liu, R.H. Antioxidant activity of grains. J. Agric. Food Chem. 2002, 50, 6182–6187. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Estrada, B.A.; Gutiérrez-UribeJ, A.; Serna-Saldívar, S.O. Bound phenolics in foods, a review. Food. Chem. 2014, 152, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.W.; Zhang, M.W.; Dong, L.H.; Jia, X.C.; Liu, L.; Ma, Y.X.; Huang, F.; Zhang, R.F. Phytochemical profile, bioactivity and prebiotic potential of bound phenolics released from rice bran dietary fiber during in vitro gastrointestinal digestion and colonic fermentation. J. Agr. Food Chem. 2019, 67, 12796–12805. [Google Scholar] [CrossRef]
- Shan, S.H.; Xie, Y.; Zhao, H.L.; Niu, J.P.; Zhang, S.; Zhang, X.L.; Li, Z.Y. Bound polyphenol extracted from jujube pulp triggers mitochondria-mediated apoptosis and cell cyclearrest of HepG2 cell in vitro and in vivo. J. Funct. Foods 2019, 53, 187–196. [Google Scholar] [CrossRef]
- Singh, R.S.G.; Negi, P.S.; Radha, C. Phenolic composition, antioxidant and antimicrobial activities of free and bound phenolic extracts of Moringaoleifera seed flour. J. Funct. Foods 2013, 5, 1883–1891. [Google Scholar] [CrossRef]
- Shi, J.Y.; Shan, S.H.; Li, H.Q.; Song, G.S.; Li, Z.Y. Anti-inflammatory effects of millet bran derived-bound polyphenols in LPS-induced HT-29 cell via ROS/miR-149/Akt/NF-κB signaling pathway. Oncotarget 2017, 8, 74582–74594. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.Y.; Shan, S.H.; Li, Z.W.; Li, H.Q.; Li, X.F.; Li, Z.Y. Bound polyphenol from foxtail millet bran induces apoptosis in HCT-116 cell through ROS generation. J. Funct. Foods 2015, 7, 958–968. [Google Scholar] [CrossRef]
- Shi, J.Y.; Shan, S.H.; Zhou, G.F.; Li, H.Q.; Song, G.S.; Li, Z.Y.; Yang, D.F. Bound polyphenol from foxtail millet bran exhibits an antiproliferative activity in HT-29 cells by reprogramming miR-149-mediated aerobicglycolysis. J. Funct. Foods 2019, 56, 246–254. [Google Scholar] [CrossRef]
- Lu, Y.; Shan, S.H.; Li, H.Q.; Shi, J.Y.; Zhang, X.L.; Li, Z.Y. Reversal effects of bound polyphenol from foxtail millet bran on multi-drug resistance in human HCT-8/fu colorectal cancer cell. J. Agric. Food Chem. 2018, 66, 5190–5199. [Google Scholar] [CrossRef] [PubMed]
- Roohinejad, S.; Omidizadeh, A.; Mirhosseini, H.; Rasti, B.; Saari, N.; Mustafa, S.; Yusof, R.M.; Hussin, A.S.M.; Hamid, A.; Manap, M.Y.A. Effect of hypocholesterolemic properties of brown rice varieties containing different gamma aminobutyric acid (GABA) levels on Sprague-Dawley male rats. J. Food Agric. Environ. 2009, 7, 197–203. [Google Scholar]
- Zhang, R.F.; Ma, Q.; Tong, X.; Liu, L.; Dong, L.H.; Huang, F.; Deng, Y.Y.; Jia, X.C.; Chi, J.W.; Zhang, M.W. Rice bran phenolic extract supplementation ameliorates impaired lipid metabolism in high-fat-diet fed mice through AMPK activation in liver. J. Funct. Foods 2020, 73, 104131. [Google Scholar] [CrossRef]
- Zhao, G.H.; Zhang, R.F.; Dong, L.H.; Huang, F.; Liu, L.; Deng, Y.Y.; Ma, Y.X.; Zhang, Y.; Wei, Z.C.; Xiao, J.; et al. A comparison of chemial composition, in vitro bioaccessibility and antioxidant activity of phenolics from rice bran and its dietary fibers. Molecules 2018, 23, 202. [Google Scholar] [CrossRef] [Green Version]
- Ti, H.H.; Li, Q.; Zhang, R.F.; Zhang, M.W.; Deng, Y.Y.; Wei, Z.C.; Chi, J.W.; Zhang, Y. Free and bound phenolic profiles and antioxidant activity of milled fractions of different indica rice varieties cultivated in southern China. Food Chem. 2014, 159, 166–174. [Google Scholar] [CrossRef]
- Son, M.J.; Rico, C.W.; Nam, S.H.; Mi, Y.K. Influence of oryzanol and ferulic acid on the lipid metabolism and antioxidative status in high fat-fed mice. J. Clin. Biochem. Nutr. 2010, 46, 150–156. [Google Scholar] [CrossRef] [Green Version]
- Gogoi, B.; Chatterjee, P.; Mukherjee, S.; Buragohain, A.K.; Bhattacharya, S.; Dasgupta, S. A polyphenol rescues lipid induced insulin resistance in skeletal muscle cells and adipocytes. Biochem. Biophys. Res. Commun. 2014, 452, 382–388. [Google Scholar] [CrossRef]
- Wang, Y.X.; Chen, X.L.; Huang, Z.Q.; Chen, D.W.; Yu, B.; Yu, J.; Chen, H.; He, J.; Luo, Y.H.; Zheng, P. Dietary ferulic acid supplementation improves antioxidant capacity and lipid metabolism in weaned piglets. Nutrients 2020, 12, 3811. [Google Scholar] [CrossRef]
- Zhang, X.W.; Dong, L.H.; Jia, X.C.; Liu, L.; Chi, J.W.; Huang, F.; Ma, Q.; Zhang, M.W.; Zhang, R.F. Bound phenolics ensure the anti-hyperglycemic effect of rice bran dietary fiber in db/db mice via activating the insulin signaling pathway in skeletal muscle and altering gut microbiota. J. Agric. Food Chem. 2020, 68, 4387–4398. [Google Scholar] [CrossRef]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V. Cellular fatty acid metabolism and cancer. Cell. Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef] [Green Version]
- Ge, J.F.; Walewski, J.L.; Anglade, D.; Berk, P.D. Regulation of hepatocellular fatty acid uptake in mouse models of fatty liver disease with and without functional leptin signaling: Roles of NF-κB and SREBP-1c and the effects of spexin. Semin. Liver Dis. 2016, 36, 360–372. [Google Scholar] [CrossRef]
- Koonen, D.P.Y.; Jacobs, R.L.; Febbraio, M.; Young, M.E.; Soltys, C.L.M.; Ong, H.; Vance, D.E.; Dyck, J.R. Increased hepatic CD36 expression contributes to dyslipidemia associated with diet-induced obesity. Diabetes 2007, 56, 2863–2871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, M.; Houten, S.M.; Wang, L.; Moschetta, A.; Auwerx, J. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J. Clin. Investig. 2004, 113, 1408–1418. [Google Scholar] [CrossRef] [Green Version]
- Kalaany, N.Y.; Gauthier, K.C.; Zavacki, A.M.; Mammen, P.P.; Kitazume, T.; Peterson, J.A.; Horton, J.D.; Garry, D.J.; Bianco, A.; Mangelsdorf, D.J. LXRs regulate the balance between fat storage and oxidation. Cell Metab. 2005, 1, 231–244. [Google Scholar] [CrossRef] [Green Version]
- Shao, W.; Espenshade, P. Expanding roles for SREBP in metabolism. Cell Metab. 2012, 16, 414–419. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.D.; Chen, W.D.; Moore, D.D.; Huang, W.D. FXR: A metabolic regulator and cell protector. Cell Res. 2008, 18, 1087–1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mwangi, S.M.; Peng, S.; Nezami, B.G.; Thorn, N.; Farris III, A.B.; Jain, S.; Laroui, H.; Merlin, D.; Anania, F.; Srinivasan, S. Glial cell line-derived neurotrophic factor protects against high-fat diet-induced hepatic steatosis by suppressing hepatic PPARγ expression. Am. J. Physiol.-Gastrointest. Liver Physiol. 2016, 310, G103–G116. [Google Scholar]
- Zhou, J.; Febbraio, M.; Wada, T.; Zhai, Y.G.; Kuruba, R.; He, J.H.; Lee, J.H.; Khadem, S.; Ren, S.R.; Li, S.; et al. Hepatic fatty acid transporter CD36 is a common target of LXR, PXR, and PPARγ in promoting steatosis. Gastroenterology 2008, 134, 556–567. [Google Scholar] [CrossRef]
- Yen, C.L.E.; Stone, S.J.; Koliwad, S.; Harris, C.; Jr, R.V.F. Thematic Review Series: Glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J. Lipid Res. 2008, 49, 2283–2301. [Google Scholar] [CrossRef] [Green Version]
- Abumrad, N.A.; Davidson, N.O. Role of the gut in lipidhomeostasis. Physiol. Rev. 2012, 92, 1061–1085. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Yang, Y.K.; Braunstein, E.; Georgeson, K.E.; Harmon, C.M. Gut expression and regulation of FAT/CD36: Possible role in fatty acid transport in rat enterocytes. Am. J. Physiol. Endocrinol. Metab. 2001, 281, E916–E923. [Google Scholar] [CrossRef] [Green Version]
- Pepino, M.Y.; Kuda, O.; Samovski, D.; Abumrad, N.A. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu. Rev. Nutr. 2014, 34, 281–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Axling, U.; Olsson, C.; Xu, J.; Fernandez, C.; Larsson, S.; Ström, K.; Ahrné, S.; Holm, C.; Molin, G.; Berger, K. Green tea powder and Lactobacillus plantarum affect gut microbiota, lipid metabolism and inflammation in high-fat fed C57BL/6J mice. Nutr. Metab. 2012, 9, 105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.L.; Chen, S.G.; Zhao, Y.Y.; Luo, C.X.; Li, J.; Gao, Y.Q. Total phenolic contents of 33 fruits and their antioxidant capacities before and after in vitro digestion. Ind. Crop. Prod. 2014, 57, 150–157. [Google Scholar] [CrossRef]
- Schwiertz, A.; Taras, D.; Schäfer, K.; Beijer, S.; Bos, N.A.; Donus, C.; Hardt, P.D. Microbiota and SCFA in lean and overweight healthy subjects. Obesity 2010, 18, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.L.; Dang, X.Y.; Dong, J.L.; Hu, X.Z. Effects of oat β-glucan and barley β-glucan on fecal characteristics, intestinal microflora, and intestinal bacterial metabolites in rats. J. Agr. Food Chem. 2012, 60, 11301–11308. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.Y.; Xiao, X.H.; Zhang, Q.; Zheng, J.; Li, M.; Yu, M.; Wang, X.J.; Deng, M.Q.; Zhai, X.; Li, R.R. Improved glucose and lipid metabolism in the early life of female offspring by maternal dietary genistein is associated with alterations in the gut microbiota. Front. Endocrinol. 2018, 9, 516. [Google Scholar] [CrossRef]
- Martiínez, I.; Wallace, G.; Zhang, C.; Legge, R.; Benson, A.K.; Carr, T.P.; Moriyama, E.N.; Walter, J. Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl. Environ. Microbiol. 2009, 75, 4175–4184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.H.; Zhang, M.H.; Wang, S.Y.; Han, R.J.; Cao, Y.F.; Hua, W.Y.; Mao, Y.J.; Zhang, X.J.; Pang, X.Y.; Wei, C.C.; et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010, 4, 232–241. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhao, Y.F.; Zhang, M.H.; Pang, X.Y.; Xu, J.; Kang, C.Y.; Li, M.; Zhang, C.H.; Zhang, Z.G.; Zhang, Y.F.; et al. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS ONE 2012, 7, e42529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koren, O.; Spor, A.; Felin, J.; Fåk, F.; Stombaugh, J.; Tremaroli, V.; Behre, C.J.; Knight, R.; Fagerberg, B.; Ley, R.E.; et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. USA 2011, 108, 4592–4598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kverka, M.; Zakostelska, Z.; Klimesova, K.; Sokol, D.; Hudcovic, T.; Hrncir, T.; Rossmann, P.; Mrazek, J.; Kopecny, J.; Verdu, E.F.; et al. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clin. Exp. Immunol. 2011, 163, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.Y.; Zhang, F.; Cao, R.H.; Ni, X.J.; Xin, Z.Q.; Deng, J.P.; Wu, G.Y.; Ren, W.K.; Yin, Y.L.; Deng, B.C. Cecropin A alleviates inflammation through modulating the gut microbiota of C57BL/6 mice with DSS-induced IBD. Front. Microbiol. 2019, 10, 1595. [Google Scholar] [CrossRef] [PubMed]
- Gerritsen, J.; Timmerman, H.M.; Fuentes, S.; van Minnen, L.P.; Panneman, H.; Konstantinov, S.R.; Rombouts, F.M.; Gooszen, H.G.; Akkermans, L.M.A.; Smidt, H.; et al. Correlation between protection against sepsis by probiotic therapy and stimulation of a novel bacterial phylotype. Appl. Environ. Microbiol. 2011, 77, 7749–7756. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Target Genes | Primer Sequences (5′–3′) | Product Length | |
|---|---|---|---|
| GAPDH | Forward | AGGAGCGAGACCCCACTAACA | 247 bp |
| Reverse | AGGGGGGCTAAGCAGTTGGT | ||
| CD36 | Forward | CCCAGATGACGTGGCAAAGA | 159 bp |
| Reverse | GAAGGCTCAAAGATGGCTCCA | ||
| ACC1 | Forward | GCTAAACCAGCACTCCCGATTC | 279 bp |
| Reverse | GCTGGAGAAGCCACAGTGAAATC | ||
| FAS | Forward | AGGAGGTGGTGATAGCCGGTA | 298 bp |
| Reverse | CGGAGTGAGGCTGGGTTGATA | ||
| LDLR | Forward | AGACTCATGCAGCAGGAACGA | 271 bp |
| Reverse | GAAGTCATCCTGGGAGCACGT | ||
| DGAT2 | Forward | TAAAGGATCTGCCCTGTCACG | 163 bp |
| Reverse | CAGGAAGGATAGGACCCATTGTA | ||
| NC | HF | HBP | |
|---|---|---|---|
| Serum | |||
| TG (mmol/L) | 1.68 ± 0.12 b | 2.31 ± 0.19 a | 1.73 ± 0.19 b |
| TCH (mmol/L) | 3.72 ± 0.14 c | 5.13 ± 0.33 a | 4.12 ± 0.18 b |
| LDL-C (mmol/L) | 2.01 ± 0.20 b | 2.60 ± 0.22 a | 1.55 ± 0.14 c |
| HDL-C (mmol/L) | 1.45 ± 0.14 | 1.44 ± 0.16 | 1.38 ± 0.07 |
| FFA (mmol/L) | 1.08 ± 0.08 b | 1.54 ± 0.12 a | 0.93 ± 0.07 c |
| Feces | |||
| TG (mg/g dry feces) | 4.34 ± 0.24 b | 5.15 ± 0.12 a | 4.70 ± 0.35 ab |
| TCH (mg/g dry feces) | 2.31 ± 0.26 | 2.36 ± 0.23 | 2.29 ± 0.18 |
| TBA (μmol/g dry feces) | 26.45 ± 0.85 b | 28.36 ± 1.76 b | 39.33 ± 1.82 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, G.; Zhang, R.; Huang, F.; Dong, L.; Liu, L.; Jia, X.; Chi, J.; Ma, Y.; Deng, M.; Chen, Y.; et al. Hydrolyzed Bound Phenolics from Rice Bran Alleviate Hyperlipidemia and Improve Gut Microbiota Dysbiosis in High-Fat-Diet Fed Mice. Nutrients 2022, 14, 1277. https://doi.org/10.3390/nu14061277
Zhao G, Zhang R, Huang F, Dong L, Liu L, Jia X, Chi J, Ma Y, Deng M, Chen Y, et al. Hydrolyzed Bound Phenolics from Rice Bran Alleviate Hyperlipidemia and Improve Gut Microbiota Dysbiosis in High-Fat-Diet Fed Mice. Nutrients. 2022; 14(6):1277. https://doi.org/10.3390/nu14061277
Chicago/Turabian StyleZhao, Guanghe, Ruifen Zhang, Fei Huang, Lihong Dong, Lei Liu, Xuchao Jia, Jianwei Chi, Yongxuan Ma, Mei Deng, Yanxia Chen, and et al. 2022. "Hydrolyzed Bound Phenolics from Rice Bran Alleviate Hyperlipidemia and Improve Gut Microbiota Dysbiosis in High-Fat-Diet Fed Mice" Nutrients 14, no. 6: 1277. https://doi.org/10.3390/nu14061277
APA StyleZhao, G., Zhang, R., Huang, F., Dong, L., Liu, L., Jia, X., Chi, J., Ma, Y., Deng, M., Chen, Y., Ma, Q., & Zhang, M. (2022). Hydrolyzed Bound Phenolics from Rice Bran Alleviate Hyperlipidemia and Improve Gut Microbiota Dysbiosis in High-Fat-Diet Fed Mice. Nutrients, 14(6), 1277. https://doi.org/10.3390/nu14061277





