Purification, Identification, and Inhibitory Mechanisms of a Novel ACE Inhibitory Peptide from Torreya grandis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Methods
2.2.1. Preparation of T. grandis Meal Protein Hydrolysate (TGMPH)
2.2.2. Purification of ACE Inhibitory Peptides
2.2.3. Measurement of ACE Inhibitory Activity
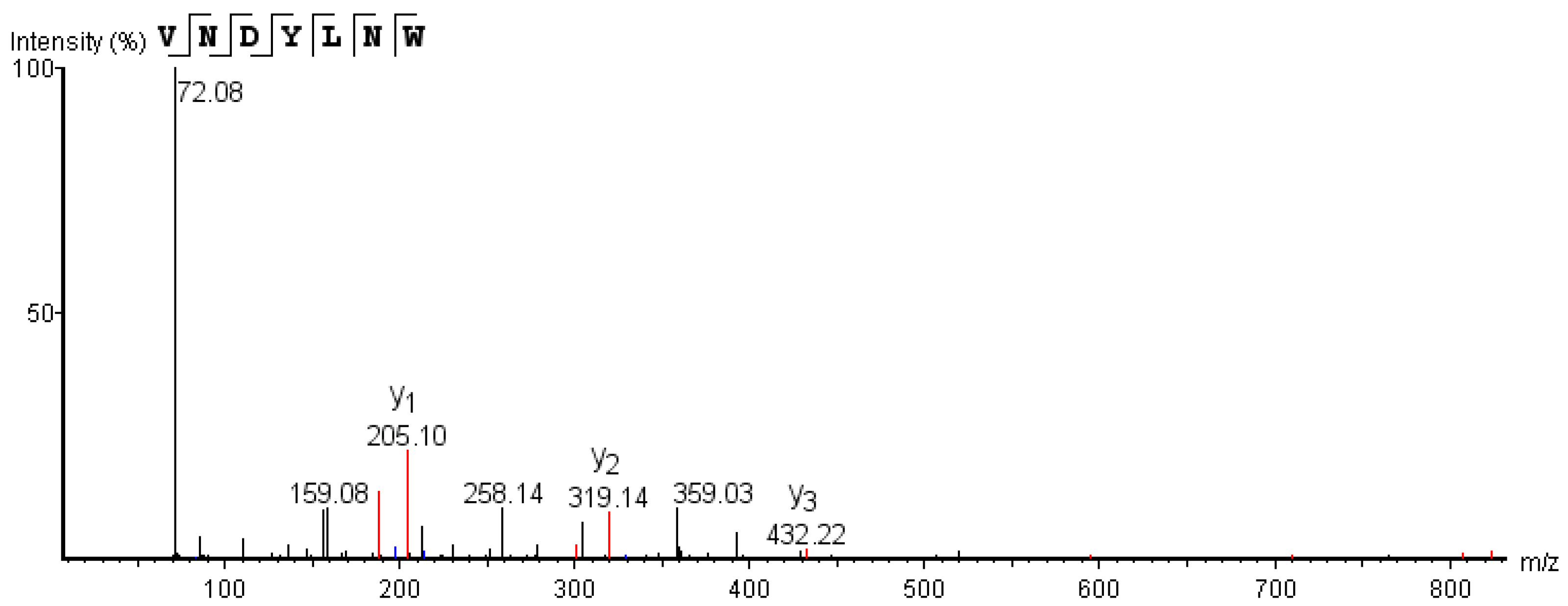
2.2.4. Identification of ACE Inhibitory Peptides
2.2.5. In Silico Prediction of ACE Inhibitory Peptides
2.2.6. Molecular Docking
2.2.7. Peptide Synthesis
2.2.8. Inhibitory Kinetics Study
2.2.9. Stability of VW-7 against Simulated Gastrointestinal Digestion
2.2.10. Cell Culture
2.2.11. Cell Viability Determination
2.2.12. Measurement of NO Levels
2.2.13. Statistical Analysis
3. Results and Discussion
3.1. Isolation of ACE Inhibitory Peptides from TGMPH
3.2. Identification and Screening of the Active ACE Inhibitory Peptides
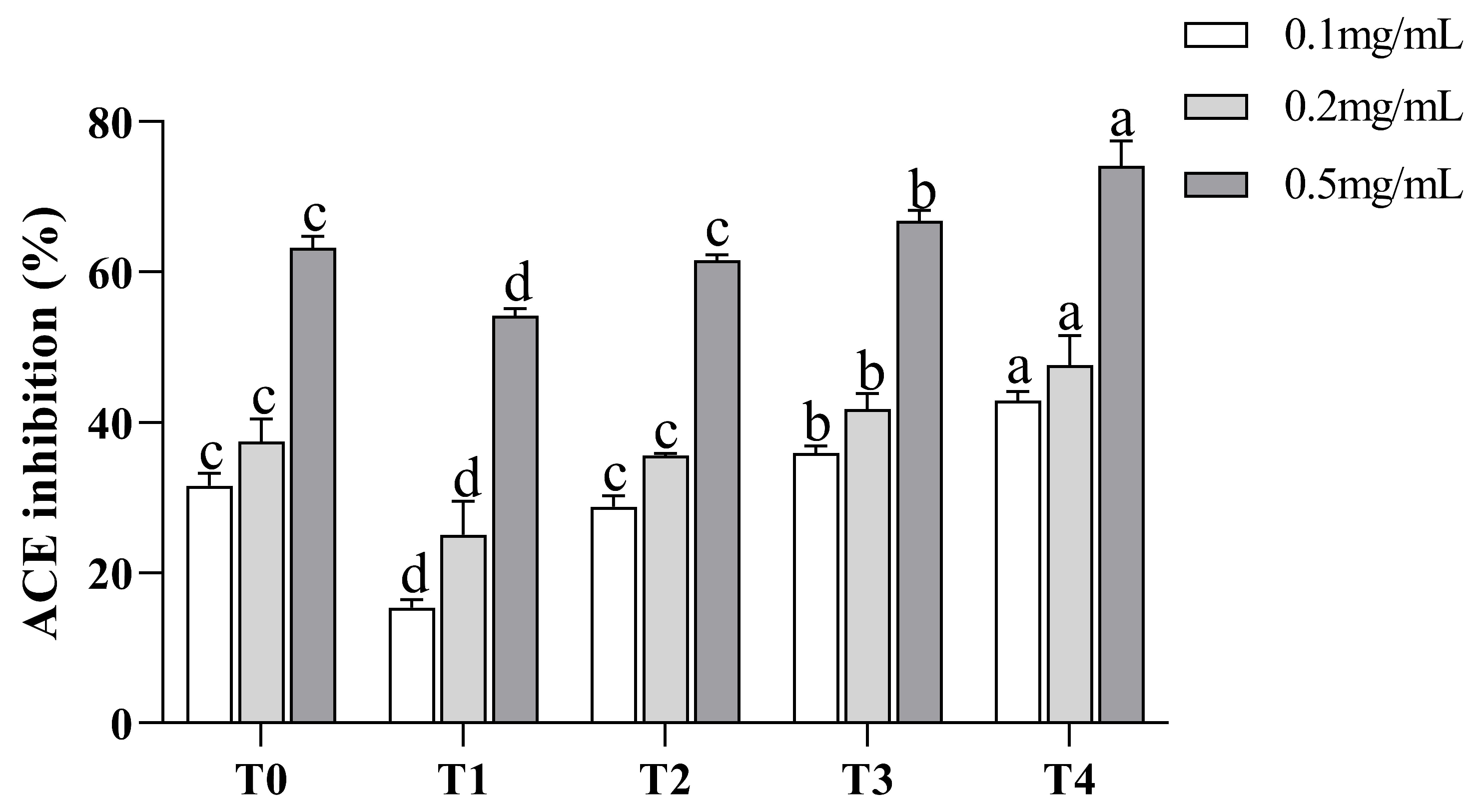
3.3. ACE Inhibitory Activity of Synthesized Peptide
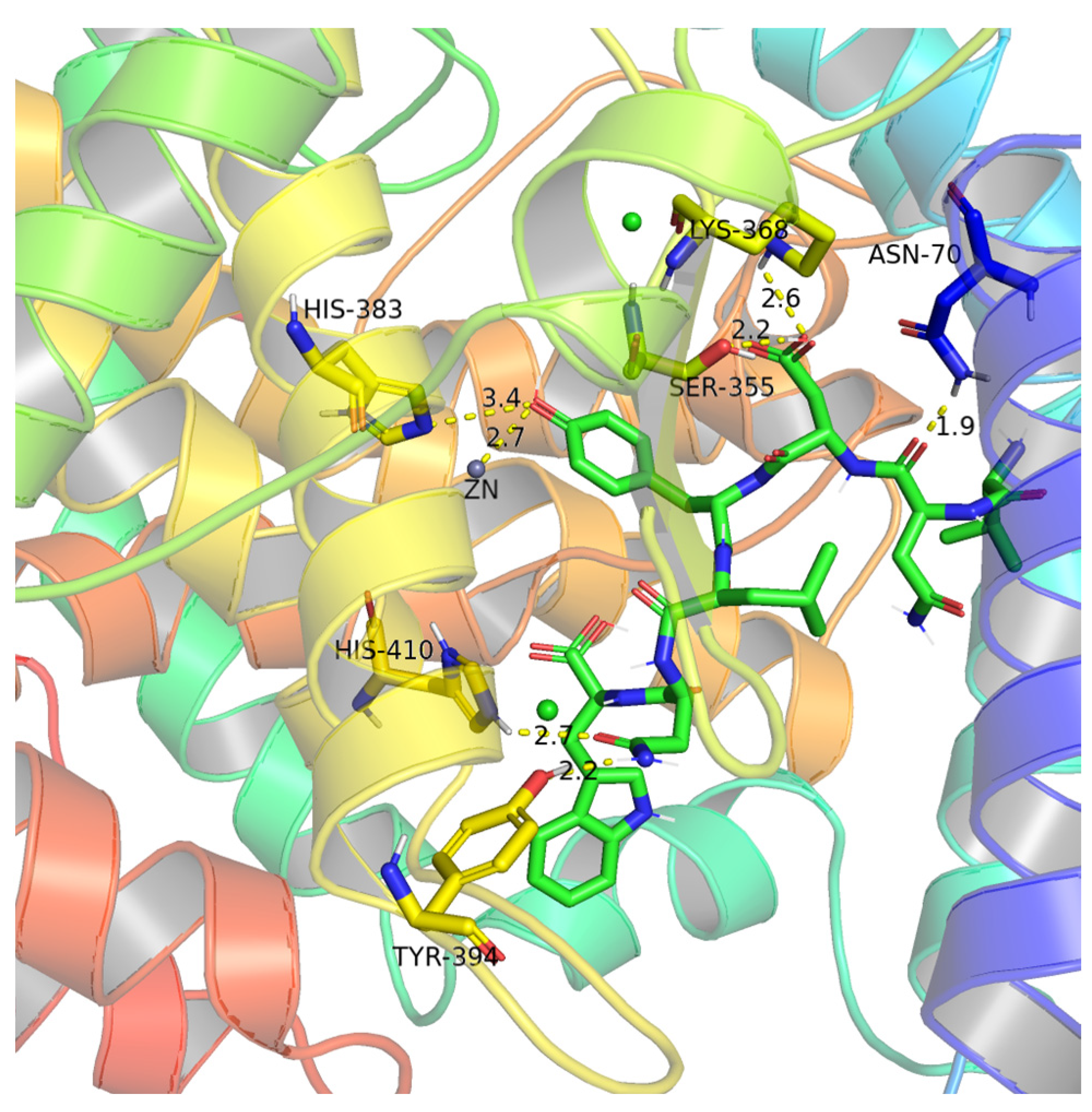
3.4. Binding Sites and Force Analysis of ACE and Peptides
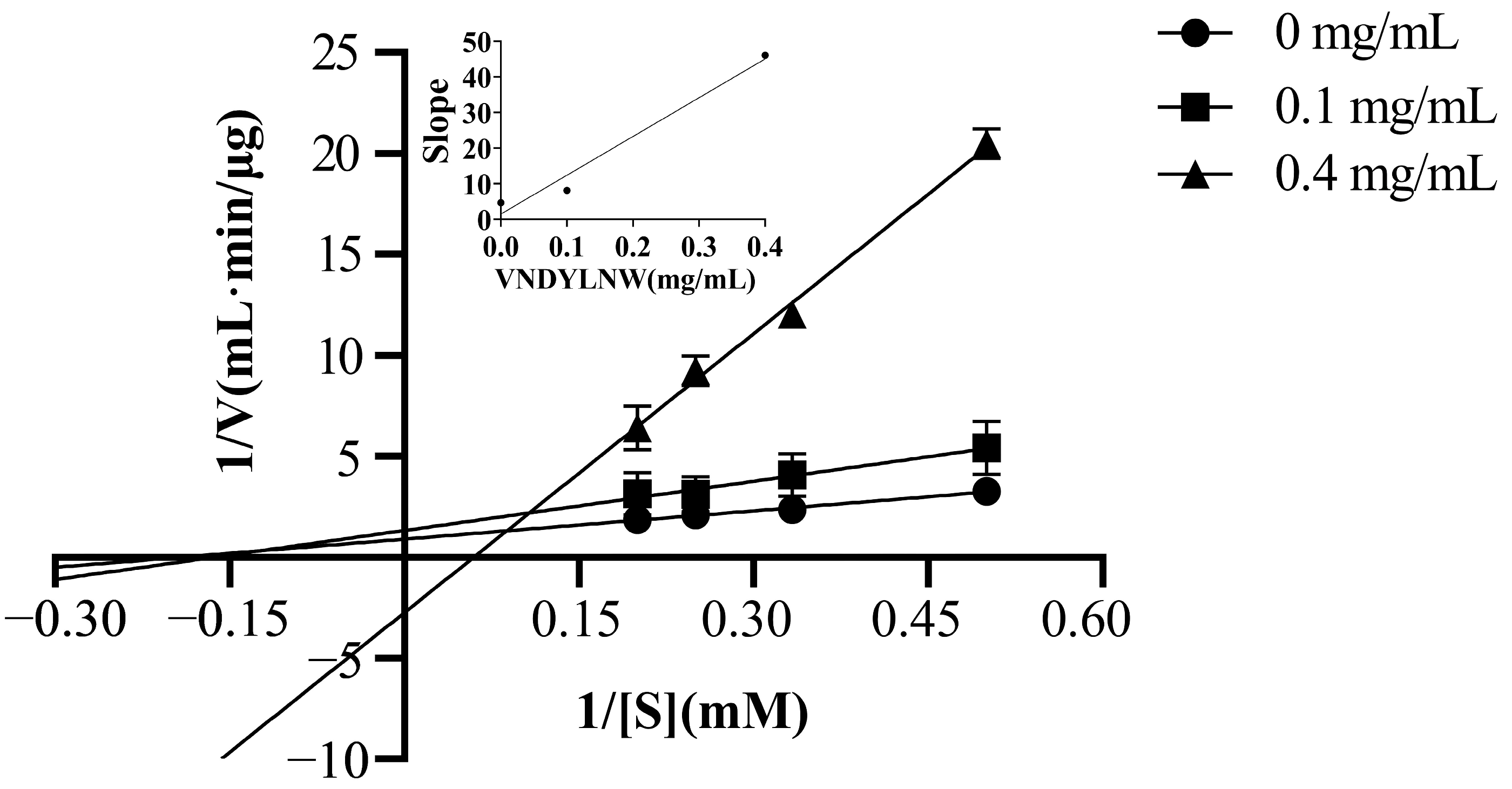
3.5. ACE Inhibition Mechanism of the Synthetic Peptides
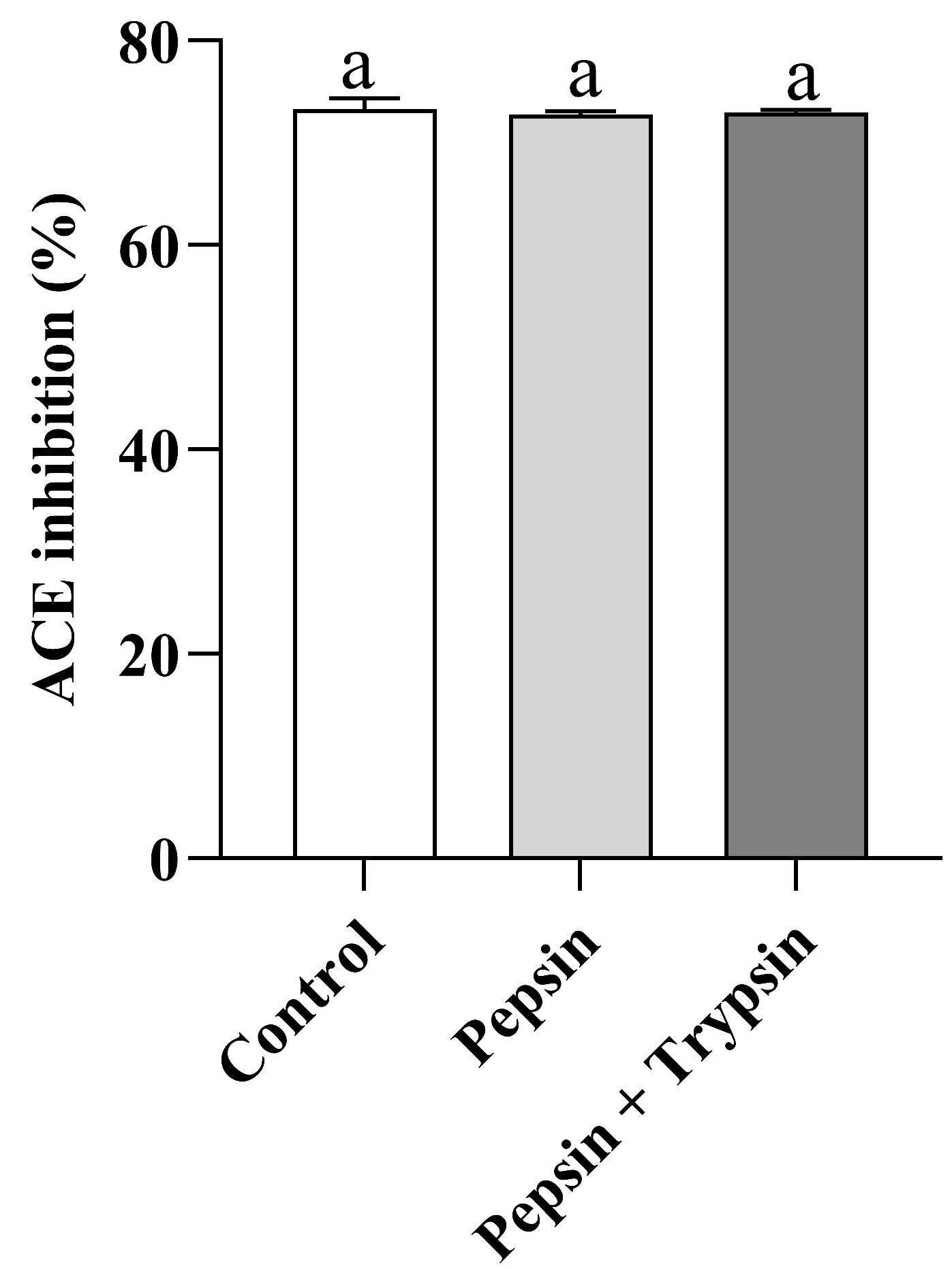
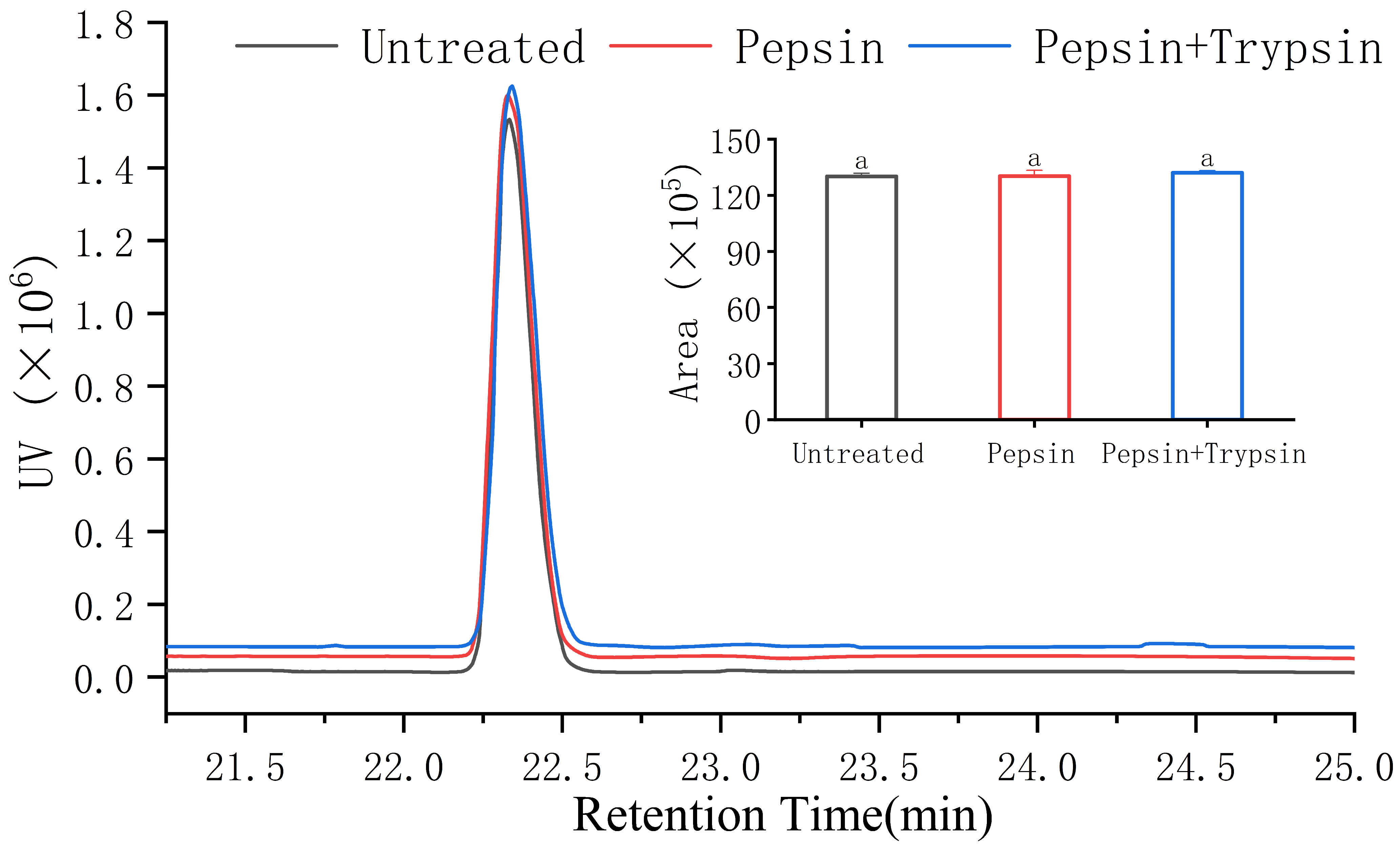
3.6. Gastrointestinal Digestion Stability of VW-7
3.7. Cell Viability Assay
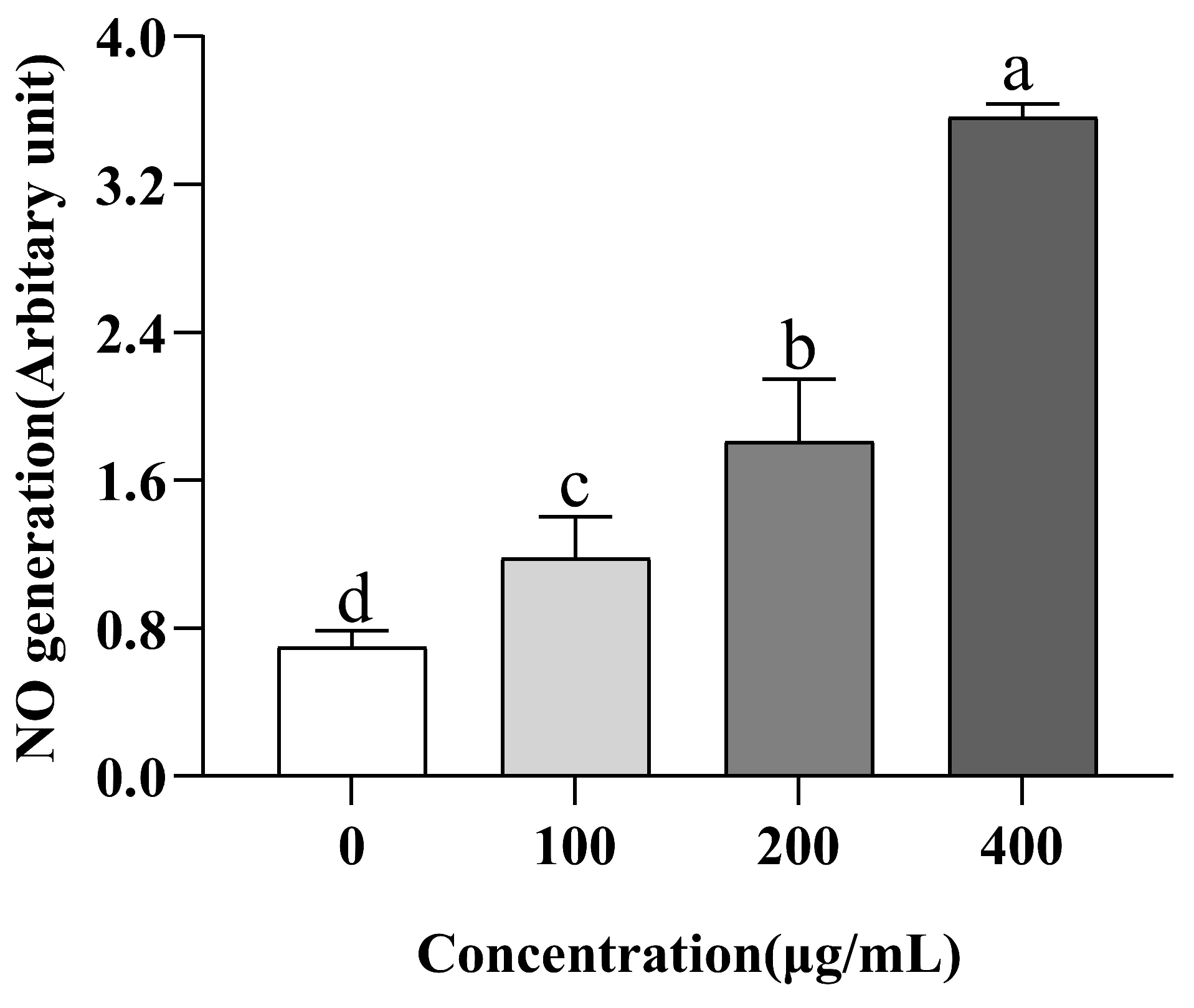
3.8. Evaluation of NO Production
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feng, Y.; Hu, Y.; Wu, J.; Chen, J.; Yrjälä, K.; Yu, W. Change in Microbial Communities, Soil Enzyme and Metabolic Activity in a Torreya grandis Plantation in Response to Root Rot Disease. For. Ecol. Manag. 2019, 432, 932–941. [Google Scholar] [CrossRef]
- Dong, D.; Wang, H.; Xu, F.; Xu, C.; Shao, X.; Li, H. Supercritical Carbon Dioxide Extraction, Fatty Acid Composition, Oxidative Stability, and Antioxidant Effect of Torreya grandis Seed Oil. JAOCS J. Am. Oil Chem. Soc. 2014, 91, 817–825. [Google Scholar] [CrossRef]
- He, Z.; Zhu, H.; Li, W.; Zeng, M.; Wu, S.; Chen, S.; Qin, F.; Chen, J. Chemical Components of Cold Pressed Kernel Oils from Different Torreya grandis Cultivars. Food Chem. 2016, 209, 196–202. [Google Scholar] [CrossRef]
- Wang, Z.; Bai, L.; Zhang, Y.; Zhao, K.; Wu, J.; Fu, W. Spatial Variation, Sources Identification and Risk Assessment of Soil Heavy Metals in a Typical Torreya grandis cv. Merrillii Plantation Region of Southeastern China. Sci. Total. Environ. 2022, 849, 157832. [Google Scholar] [CrossRef]
- Shi, L.K.; Zheng, L.; Mao, J.H.; Zhao, C.W.; Huang, J.H.; Liu, R.J.; Chang, M.; Jin, Q.Z.; Wang, X.G. Effects of the Variety and Oil Extraction Method on the Quality, Fatty Acid Composition and Antioxidant Capacity of Torreya grandis Kernel Oils. LWT 2018, 91, 398–405. [Google Scholar] [CrossRef]
- Yu, M.; Zeng, M.; Qin, F.; He, Z.; Chen, J. Physicochemical and Functional Properties of Protein Extracts from Torreya grandis Seeds. Food Chem. 2017, 227, 453–460. [Google Scholar] [CrossRef]
- Quan, W.; Zhang, C.; Wang, Z.; Zeng, M.; Qin, F.; He, Z.; Chen, J. Assessment Antioxidant Properties of Torreya grandis Protein Enzymatic Hydrolysates: Utilization of Industrial by-Products. Food Biosci. 2021, 43, 101325. [Google Scholar] [CrossRef]
- Luo, X.; Wu, S.; Xue, J.; Hu, H.; He, Z.; Liu, X.; Wu, F. The Bioactive Peptide Screening from Torreya grandis Meal Protein Hydrolysates. Food Biosci. 2021, 44, 101419. [Google Scholar] [CrossRef]
- Xingfei, L.; Shunshun, P.; Wenji, Z.; Lingli, S.; Qiuhua, L.; Ruohong, C.; Shili, S. Properties of ACE Inhibitory Peptide Prepared from Protein in Green Tea Residue and Evaluation of Its Anti-Hypertensive Activity. Process. Biochem. 2020, 92, 277–287. [Google Scholar] [CrossRef]
- Rui, X.; Boye, J.I.; Simpson, B.K.; Prasher, S.O. Angiotensin I-Converting Enzyme Inhibitory Properties of Phaseolus Vulgaris Bean Hydrolysates: Effects of Different Thermal and Enzymatic Digestion Treatments. Food Res. Int. 2012, 49, 739–746. [Google Scholar] [CrossRef]
- Oh, J.Y.; Kim, E.A.; Lee, H.; Kim, H.S.; Lee, J.S.; Jeon, Y.J. Antihypertensive Effect of Surimi Prepared from Olive Flounder (Paralichthys olivaceus) by Angiotensin-I Converting Enzyme (ACE) Inhibitory Activity and Characterization of ACE Inhibitory Peptides. Process. Biochem. 2019, 80, 164–170. [Google Scholar] [CrossRef]
- Ko, S.C.; Kang, M.C.; Kang, N.; Kim, H.S.; Lee, S.H.; Ahn, G.; Jung, W.K.; Jeon, Y.J. Effect of Angiotensin I-Converting Enzyme (ACE) Inhibition and Nitric Oxide (NO) Production of 6,6′-Bieckol, a Marine Algal Polyphenol and Its Anti-Hypertensive Effect in Spontaneously Hypertensive Rats. Process. Biochem. 2017, 58, 326–332. [Google Scholar] [CrossRef]
- Tu, M.; Liu, H.; Cheng, S.; Xu, Z.; Wang, L.S.; Du, M. Identification and Analysis of Transepithelial Transport Properties of Casein Peptides with Anticoagulant and ACE Inhibitory Activities. Food Res. Int. 2020, 138, 109764. [Google Scholar] [CrossRef]
- Sutopo, C.C.Y.; Sutrisno, A.; Wang, L.F.; Hsu, J.L. Identification of a Potent Angiotensin-I Converting Enzyme Inhibitory Peptide from Black Cumin Seed Hydrolysate Using Orthogonal Bioassay-Guided Fractionations Coupled with in Silico Screening. Process. Biochem. 2020, 95, 204–213. [Google Scholar] [CrossRef]
- Ko, S.C.; Jung, W.K.; Kang, S.M.; Lee, S.H.; Kang, M.C.; Heo, S.J.; Kang, K.H.; Kim, Y.T.; Park, S.J.; Jeong, Y.; et al. Angiotensin I-Converting Enzyme (ACE) Inhibition and Nitric Oxide (NO)-Mediated Antihypertensive Effect of Octaphlorethol A Isolated from Ishige Sinicola: In Vitro Molecular Mechanism and in vivo SHR Model. J. Funct. Foods 2015, 18, 289–299. [Google Scholar] [CrossRef]
- Lin, K.; Ma, Z.; Ramachandran, M.; De Souza, C.; Han, X.; Zhang, L. ACE Inhibitory Peptide KYIPIQ Derived from Yak Milk Casein Induces Nitric Oxide Production in HUVECs and Diffuses via a Transcellular Mechanism in Caco-2 Monolayers. Process. Biochem. 2020, 99, 103–111. [Google Scholar] [CrossRef]
- Ye, S.; Chen, Q.; Li, D.; Zhou, H.; Chen, Y.; Meng, C.; Hong, J. Isolation and Identification of Novel Angiotensin I-Converting Enzyme (ACE) Inhibitory Peptides from Pony Seed and Evaluation of the Inhibitory Mechanisms. J. Funct. Foods 2022, 95, 105151. [Google Scholar] [CrossRef]
- García-Tejedor, A.; Gimeno-Alcañíz, J.V.; Tavárez, S.; Alonso, E.; Salom, J.B.; Manzanares, P. An Antihypertensive Lactoferrin Hydrolysate Inhibits Angiotensin I-Converting Enzyme, Modifies Expression of Hypertension-Related Genes and Enhances Nitric Oxide Production in Cultured Human Endothelial Cells. J. Funct. Foods 2015, 12, 45–54. [Google Scholar] [CrossRef]
- Wang, C.X.; Song, C.C.; Liu, X.T.; Qiao, B.W.; Song, S.; Fu, Y.H. ACE Inhibitory Activities of Two Peptides Derived from Volutharpa Ampullacea Perryi Hydrolysate and Their Protective Effects on H2O2 Induced HUVECs Injury. Food Res. Int. 2022, 157, 111402. [Google Scholar] [CrossRef]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the Improved Discovery and Design of Functional Peptides: Common Features of Diverse Classes Permit Generalized Prediction of Bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.X.; Wang, Z.C.; Chen, J.X.; Li, H.R.; Wang, Y.B.; Ren, D.F.; Lu, J. Separation, Identification, and Molecular Docking of Tyrosinase Inhibitory Peptides from the Hydrolysates of Defatted Walnut (Juglans regia L.) Meal. Food Chem. 2021, 353, 129471. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; He, Y.; He, H.; Zhou, W.; Li, M.; Lu, A.; Che, T.; Shen, S. Purification Identification and Function Analysis of ACE Inhibitory Peptide from Ulva Prolifera Protein. Food Chem. 2023, 401, 134127. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Wang, S.; Wang, L.; Huang, D.; Chen, S. Identification and Characterization of an Angiotensin-I Converting Enzyme Inhibitory Peptide from Enzymatic Hydrolysate of Rape (Brassica napus L.) Bee Pollen. LWT 2021, 147, 111502. [Google Scholar] [CrossRef]
- Sompinit, K.; Lersiripong, S.; Reamtong, O.; Pattarayingsakul, W.; Patikarnmonthon, N.; Panbangred, W. In Vitro Study on Novel Bioactive Peptides with Antioxidant and Antihypertensive Properties from Edible Rhizomes. LWT 2020, 134, 110227. [Google Scholar] [CrossRef]
- Natesh, R.; Schwager, S.L.U.; Sturrock, E.D.; Acharya, K.R. Crystal Structure of the Human Angiotensin-Converting Enzyme-Lisinopril Complex. Nature 2003, 421, 551–554. [Google Scholar] [CrossRef]
- Lin, Z.; Lai, J.; He, P.; Pan, L.; Zhang, Y.; Zhang, M.; Wu, H. Screening, ACE-Inhibitory Mechanism and Structure-Activity Relationship of a Novel ACE-Inhibitory Peptide from Lepidium meyenii (Maca) Protein Hydrolysate. Food Biosci. 2023, 52, 102374. [Google Scholar] [CrossRef]
- Ma, T.; Fu, Q.; Mei, Q.; Tu, Z.; Zhang, L. Extraction Optimization and Screening of Angiotensin-Converting Enzyme Inhibitory Peptides from Channa Striatus through Bioaffinity Ultrafiltration Coupled with LC-Orbitrap-MS/MS and Molecular Docking. Food Chem. 2021, 354, 129589. [Google Scholar] [CrossRef]
- Xie, D.; Du, L.; Lin, H.; Su, E.; Shen, Y.; Xie, J.; Wei, D. In Vitro-in Silico Screening Strategy and Mechanism of Angiotensin I-Converting Enzyme Inhibitory Peptides from α-Lactalbumin. LWT 2022, 156, 112984. [Google Scholar] [CrossRef]
- Hao, X.; Xia, Y.; Wang, Y.; Zhang, X.; Liu, L. The Addition of Probiotic Promotes the Release of ACE-I Peptide of Cheddar Cheese: Peptide Profile and Molecular Docking. Int. Dairy J. 2023, 137, 105507. [Google Scholar] [CrossRef]
- Gao, Y.F.; Liu, M.Q.; Li, Z.H.; Zhang, H.L.; Hao, J.Q.; Liu, B.H.; Li, X.Y.; Yin, Y.Q.; Wang, X.H.; Zhou, Q.; et al. Purification and Identification of Xanthine Oxidase Inhibitory Peptides from Enzymatic Hydrolysate of α-Lactalbumin and Bovine Colostrum Casein. Food Res. Int. 2023, 169, 112882. [Google Scholar] [CrossRef]
- Chen, J.; Yu, X.; Chen, Q.; Wu, Q.; He, Q. Screening and Mechanisms of Novel Angiotensin-I-Converting Enzyme Inhibitory Peptides from Rabbit Meat Proteins: A Combined in Silico and in Vitro Study. Food Chem. 2022, 370, 131070. [Google Scholar] [CrossRef]
- Chen, P.; Li, L.; Huo, X.; Qiao, L.; Zhang, Y.; Chen, Z.; Wang, L. New Angiotensin-Converting Enzyme Inhibitory Peptide from Coix Prolamin and Its Influence on the Gene Expression of Renin-Angiotensin System in Vein Endothelial Cells. J. Cereal Sci. 2020, 96, 103099. [Google Scholar] [CrossRef]
- Lee, S.Y.; Hur, S.J. Antihypertensive Peptides from Animal Products, Marine Organisms, and Plants. Food Chem. 2017, 228, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Ledesma, B.; Del Mar Contreras, M.; Recio, I. Antihypertensive Peptides: Production, Bioavailability and Incorporation into Foods. Adv. Colloid Interface Sci. 2011, 165, 23–35. [Google Scholar] [CrossRef]
- Zhang, Y.; He, S.; Rui, X.; Simpson, B.K. Interactions of C. Frondosa-Derived Inhibitory Peptides against Angiotensin I-Converting Enzyme (ACE), α-Amylase and Lipase. Food Chem. 2022, 367, 130695. [Google Scholar] [CrossRef]
- Yu, Z.; Guo, H.; Shiuan, D.; Xia, C.; Zhao, W.; Ding, L.; Zheng, F.; Liu, J. Interaction Mechanism of Egg White- Derived ACE Inhibitory Peptide TNGIIR with ACE and Its Effect on the Expression of ACE and AT1 Receptor. Food Sci. Hum. Wellness 2020, 9, 52–57. [Google Scholar] [CrossRef]
- Urbizo-Reyes, U.; Liceaga, A.M.; Reddivari, L.; Kim, K.H.; Anderson, J.M. Enzyme Kinetics, Molecular Docking, and in Silico Characterization of Canary Seed (Phalaris canariensis L.) Peptides with ACE and Pancreatic Lipase Inhibitory Activity. J. Funct. Foods 2022, 88, 104892. [Google Scholar] [CrossRef]
- Renjuan, L.; Xiuli, Z.; Liping, S.; Yongliang, Z. Identification, in Silico Screening, and Molecular Docking of Novel ACE Inhibitory Peptides Isolated from the Edible Symbiot Boletus Griseus-Hypomyces Chrysospermus. LWT 2022, 169, 114008. [Google Scholar] [CrossRef]
- Li, M.; Fan, W.; Xu, Y. Identification of Angiotensin Converting Enzyme (ACE) Inhibitory and Antioxidant Peptides Derived from Pixian Broad Bean Paste. LWT 2021, 151, 112221. [Google Scholar] [CrossRef]
- Tu, M.; Wang, C.; Chen, C.; Zhang, R.; Liu, H.; Lu, W.; Jiang, L.; Du, M. Identification of a Novel ACE-Inhibitory Peptide from Casein and Evaluation of the Inhibitory Mechanisms. Food Chem. 2018, 256, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Feng, C.; Hong, H.; Zhang, Y.; Luo, Z.; Wang, Q.; Luo, Y.; Tan, Y. Novel ACE Inhibitory Peptides Derived from Whey Protein Hydrolysates: Identification and Molecular Docking Analysis. Food Biosci. 2022, 48, 101737. [Google Scholar] [CrossRef]
- Forghani, B.; Zarei, M.; Ebrahimpour, A.; Philip, R.; Bakar, J.; Abdul Hamid, A.; Saari, N. Purification and Characterization of Angiotensin Converting Enzyme-Inhibitory Peptides Derived from Stichopus Horrens: Stability Study against the ACE and Inhibition Kinetics. J. Funct. Foods 2016, 20, 276–290. [Google Scholar] [CrossRef]
- Zhang, P.; Roytrakul, S.; Sutheerawattananonda, M. Production and Purification of Glucosamine and Angiotensin-I Converting Enzyme (ACE) Inhibitory Peptides from Mushroom Hydrolysates. J. Funct. Foods 2017, 36, 72–83. [Google Scholar] [CrossRef]
- Wongngam, W.; Hamzeh, A.; Tian, F.; Roytrakul, S.; Yongsawatdigul, J. Purification and Molecular Docking of Angiotensin Converting Enzyme-Inhibitory Peptides Derived from Corn Gluten Meal Hydrolysate and from in Silico Gastrointestinal Digestion. Process. Biochem. 2023, 129, 113–120. [Google Scholar] [CrossRef]
- Villadóniga, C.; Cantera, A.M.B. New ACE-Inhibitory Peptides Derived from α-Lactalbumin Produced by Hydrolysis with Bromelia Antiacantha Peptidases. Biocatal. Agric. Biotechnol. 2019, 20, 101258. [Google Scholar] [CrossRef]
- Wong, F.C.; Xiao, J.; Ong, M.G.L.; Pang, M.J.; Wong, S.J.; Teh, L.K.; Chai, T.T. Identification and Characterization of Antioxidant Peptides from Hydrolysate of Blue-Spotted Stingray and Their Stability against Thermal, PH and Simulated Gastrointestinal Digestion Treatments. Food Chem. 2019, 271, 614–622. [Google Scholar] [CrossRef]
- Wang, K.; Luo, Q.; Hong, H.; Liu, H.; Luo, Y. Novel Antioxidant and ACE Inhibitory Peptide Identified from Arthrospira Platensis Protein and Stability against Thermal/PH Treatments and Simulated Gastrointestinal Digestion. Food Res. Int. 2021, 139, 109908. [Google Scholar] [CrossRef]
- Ahmed, T.; Sun, X.; Udenigwe, C.C. Role of Structural Properties of Bioactive Peptides in Their Stability during Simulated Gastrointestinal Digestion: A Systematic Review. Trends Food Sci. Technol. 2022, 120, 265–273. [Google Scholar] [CrossRef]
- Zhang, Y.; Pan, D.; Yang, Z.; Gao, X.; Dang, Y. Angiotensin I-Converting Enzyme (ACE) Inhibitory and Dipeptidyl Peptidase-4 (DPP-IV) Inhibitory Activity of Umami Peptides from Ruditapes Philippinarum. LWT 2021, 144, 111265. [Google Scholar] [CrossRef]
- Zhao, Y.Q.; Zhang, L.; Tao, J.; Chi, C.F.; Wang, B. Eight Antihypertensive Peptides from the Protein Hydrolysate of Antarctic Krill (Euphausia superba): Isolation, Identification, and Activity Evaluation on Human Umbilical Vein Endothelial Cells (HUVECs). Food Res. Int. 2019, 121, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Xi, Z.; Li, J.; Gao, T.; Liao, R.; Wang, S.; Li, X.; Tang, Y.; Wang, Z.; Hou, S.; et al. Vasodilator and Hypotensive Effects of the Spider Peptide Lycosin-I in vitro and in vivo. Peptides 2018, 99, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.Y.; Je, J.G.; Lee, H.G.; Kim, E.A.; Kang, S.I.; Lee, J.S.; Jeon, Y.J. Anti-Hypertensive Activity of Novel Peptides Identified from Olive Flounder (Paralichthys olivaceus) Surimi. Foods 2020, 9, 647. [Google Scholar] [CrossRef] [PubMed]



| Time (Min) | A (%) | B (%) |
|---|---|---|
| 0 | 95 | 5 |
| 25 | 35 | 65 |
| 30 | 5 | 95 |
| 35 | 5 | 95 |
| 40 | 95 | 5 |
| Peptide Sequence | Binding Energy (kcal/mol) | PeptideRanker Score | Toxicity Prediction |
|---|---|---|---|
| FFFN | −9.6 | 0.99 | Non-Toxin |
| YTKLEPR | −8.4 | 0.12 | Non-Toxin |
| DYLYH | −9 | 0.42 | Non-Toxin |
| FDKLLSPR | −9 | 0.58 | Non-Toxin |
| KALGAP | −9.1 | 0.32 | Non-Toxin |
| FFDK | −8.6 | 0.89 | Non-Toxin |
| YSSAL | −8.7 | 0.32 | Non-Toxin |
| YSTAL | −9 | 0.31 | Non-Toxin |
| FFDLK | −8.8 | 0.87 | Non-Toxin |
| VVPF | −8.7 | 0.58 | Non-Toxin |
| LLSPR | −7.7 | 0.45 | Non-Toxin |
| VYLQ | −8.5 | 0.12 | Non-Toxin |
| QLFVK | −8.3 | 0.32 | Non-Toxin |
| WMLV | −8.5 | 0.84 | Non-Toxin |
| FDKL | −8.4 | 0.67 | Non-Toxin |
| LLSSPR | −7.9 | 0.41 | Non-Toxin |
| LLAA | −7 | 0.25 | Non-Toxin |
| FFNVN | −9.2 | 0.69 | Non-Toxin |
| LRLL | −7.3 | 0.57 | Non-Toxin |
| VNDYLYH | −9.7 | 0.34 | Non-Toxin |
| VGYL | −8 | 0.42 | Non-Toxin |
| SVPNWESH | −9.4 | 0.34 | Non-Toxin |
| TYLY | −9 | 0.29 | Non-Toxin |
| YTKLERP | −8.6 | 0.11 | Non-Toxin |
| GHPLVQ | −8.5 | 0.25 | Non-Toxin |
| AVVHY | −8.8 | 0.1 | Non-Toxin |
| GYGL | −8.5 | 0.79 | Non-Toxin |
| MLPH | −7.5 | 0.72 | Non-Toxin |
| YTKLEP | −9 | 0.1 | Non-Toxin |
| FDLK | −7.7 | 0.58 | Non-Toxin |
| NRNPPSLL | −8.8 | 0.57 | Non-Toxin |
| YLRPN | −8.7 | 0.48 | Non-Toxin |
| AYRDF | −10.5 | 0.79 | Non-Toxin |
| SFMLPH | −8.3 | 0.88 | Non-Toxin |
| LHNPNPY | −9.6 | 0.45 | Non-Toxin |
| LSLF | −7.2 | 0.77 | Non-Toxin |
| VNDYLNW | −10 | 0.6 | Non-Toxin |
| Parameter | 0 mg/mL | 0.1 mg/mL | 0.4 mg/mL |
|---|---|---|---|
| Vmax (μg/mL·min) | 1.12 | 0.76 | 0.36 |
| Km (mM) | 5.21 | 6.14 | 16.79 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, F.; Luo, X.; Zhang, Y.; Wang, P.; Chang, Y.; He, Z.; Liu, X. Purification, Identification, and Inhibitory Mechanisms of a Novel ACE Inhibitory Peptide from Torreya grandis. Nutrients 2023, 15, 2374. https://doi.org/10.3390/nu15102374
Wu F, Luo X, Zhang Y, Wang P, Chang Y, He Z, Liu X. Purification, Identification, and Inhibitory Mechanisms of a Novel ACE Inhibitory Peptide from Torreya grandis. Nutrients. 2023; 15(10):2374. https://doi.org/10.3390/nu15102374
Chicago/Turabian StyleWu, Fenghua, Xiaohui Luo, Yongzhu Zhang, Peng Wang, Yinzi Chang, Zhiping He, and Xingquan Liu. 2023. "Purification, Identification, and Inhibitory Mechanisms of a Novel ACE Inhibitory Peptide from Torreya grandis" Nutrients 15, no. 10: 2374. https://doi.org/10.3390/nu15102374





