Lactiplantibacillus plantarum OLL2712 Induces Autophagy via MYD88 and Strengthens Tight Junction Integrity to Promote the Barrier Function in Intestinal Epithelial Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Bacterial Cells
2.2. Cell Culture
2.3. Detection of Autophagy Activity Using DALGreen
2.4. Developing a Stable GFP-LC3-Expressing Caco-2 Strain
2.5. Detection of Autophagy Activity Using the Stable GFP-LC3-Expressing Caco-2 Strain
2.6. RNA Isolation and Quantitative Analysis by Real-Time Polymerase Chain Reaction (PCR)
2.7. Western Blotting
2.8. Simple Western Analysis
2.9. FD-4 Permeability Test
2.10. MUC2 Enzyme-Linked Immunosorbent Assay (ELISA)
2.11. Small Interfering RNA (siRNA) Knockdown Experiments
2.12. Statistical Analysis
3. Results
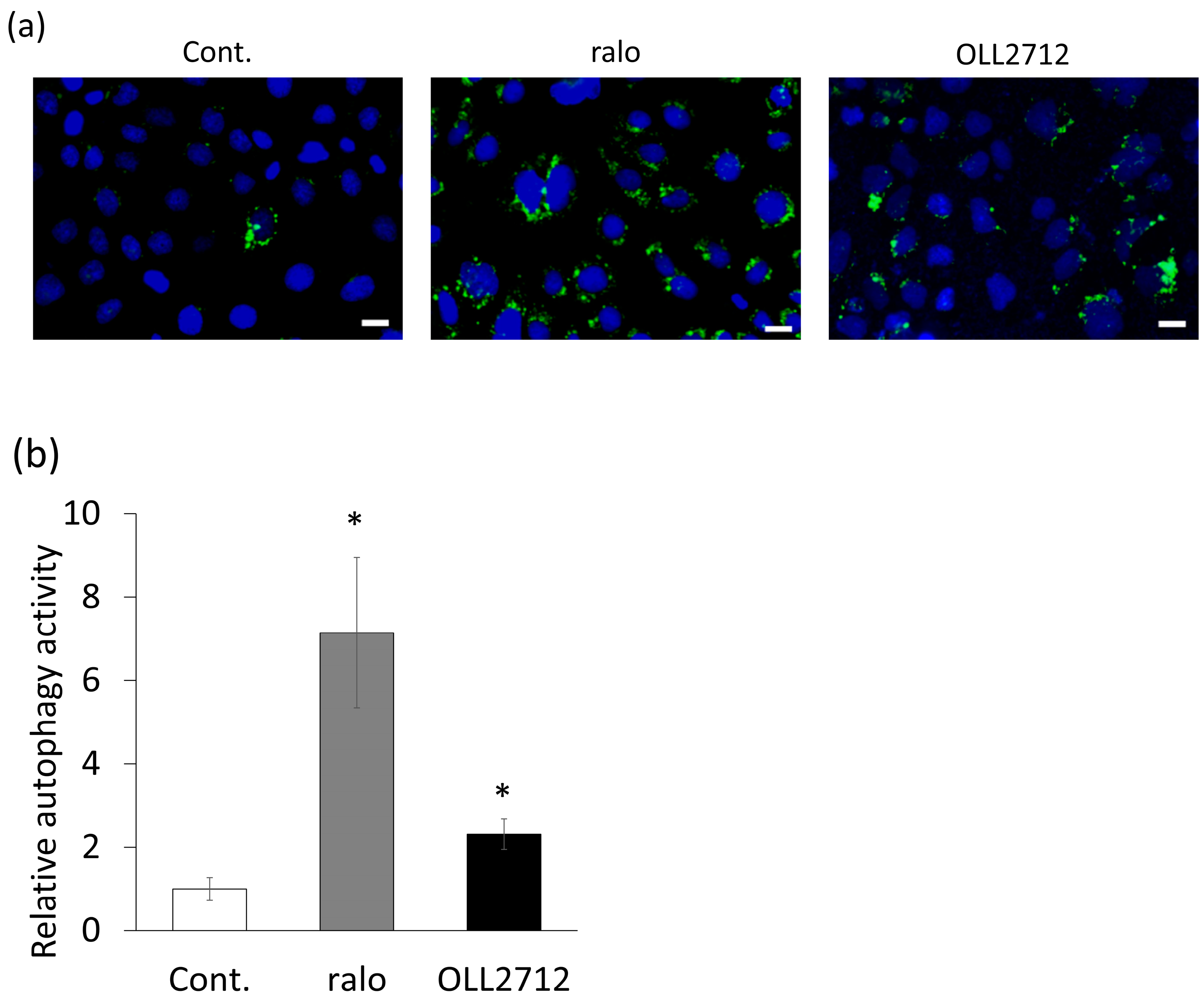
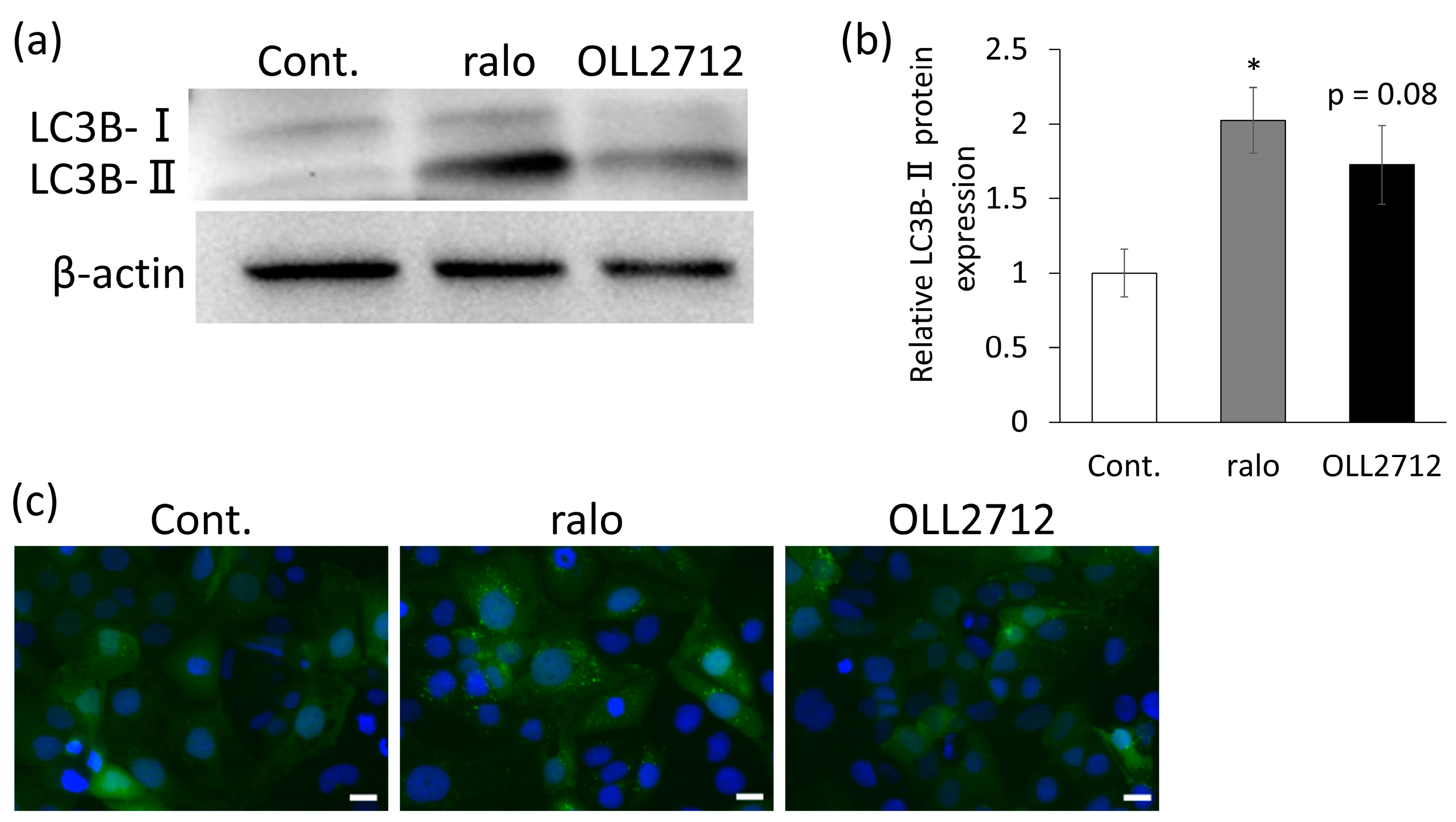
3.1. OLL2712 Increases Autophagy in Intestinal Epithelial Cells
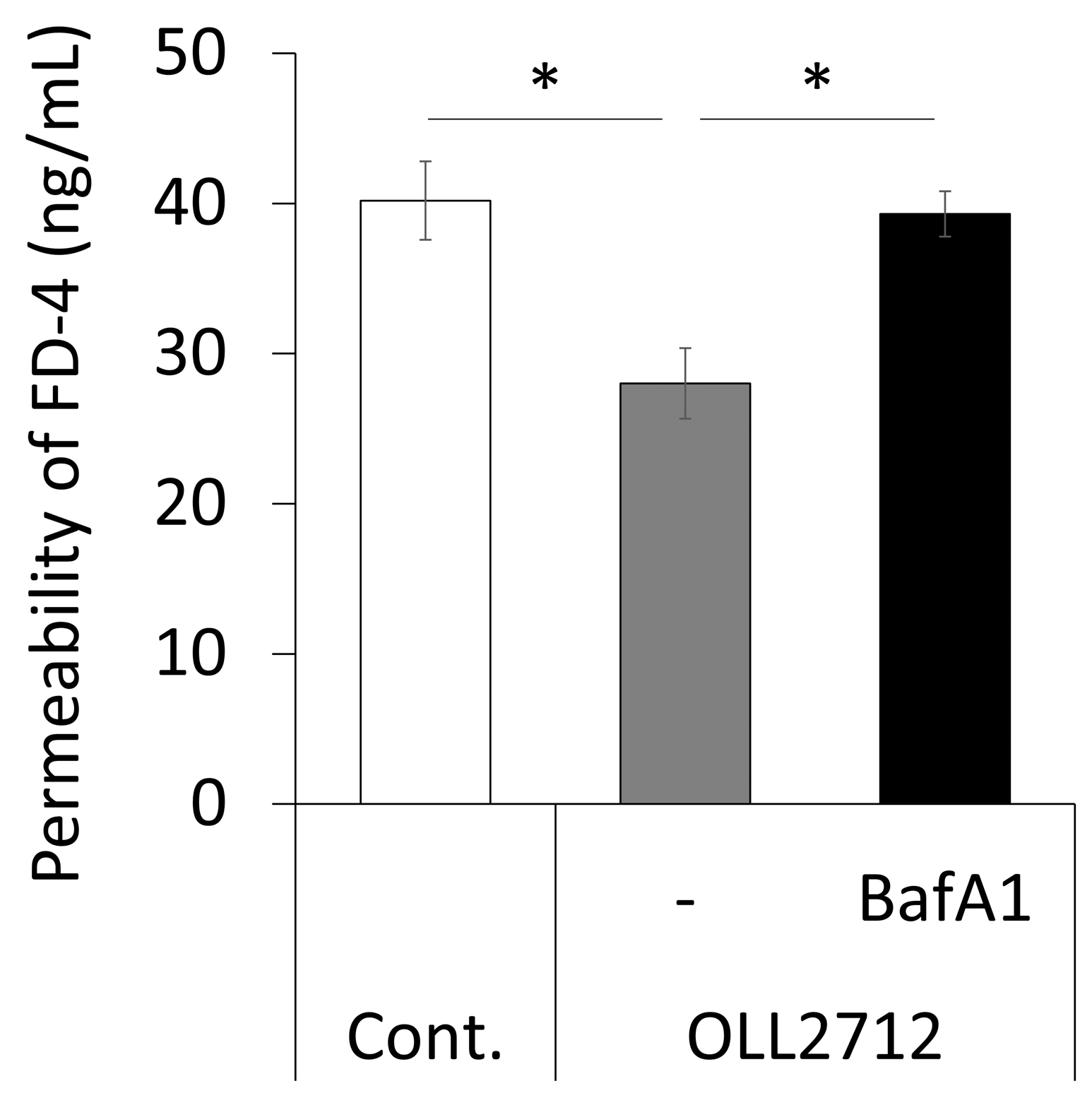
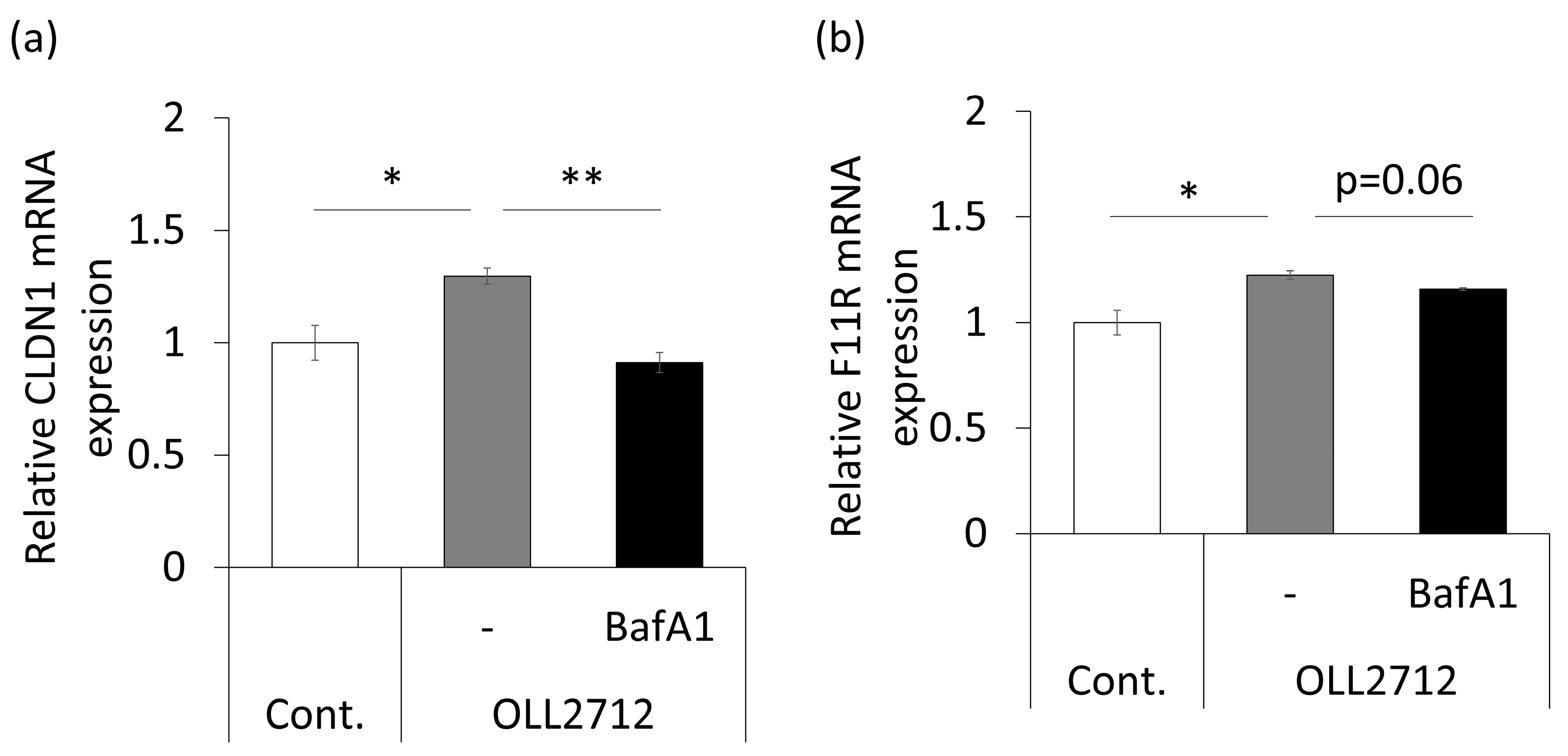
3.2. Autophagy Induced by OLL2712 in Intestinal Epithelial Cells Strengthens the Mucosal Barrier
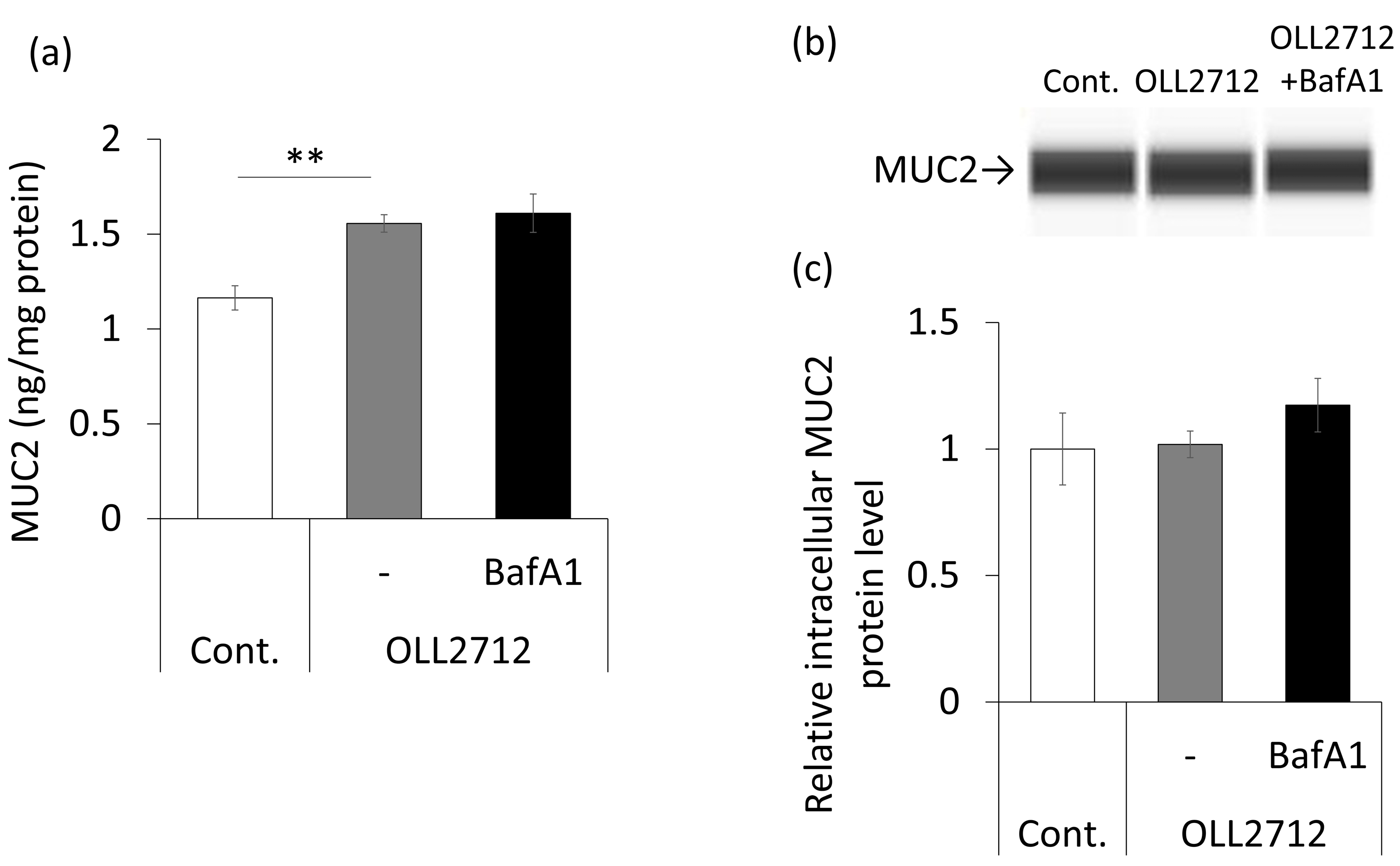
3.3. OLL2712 Promotes Mucin Secretion in an Autophagy-Independent Pathway
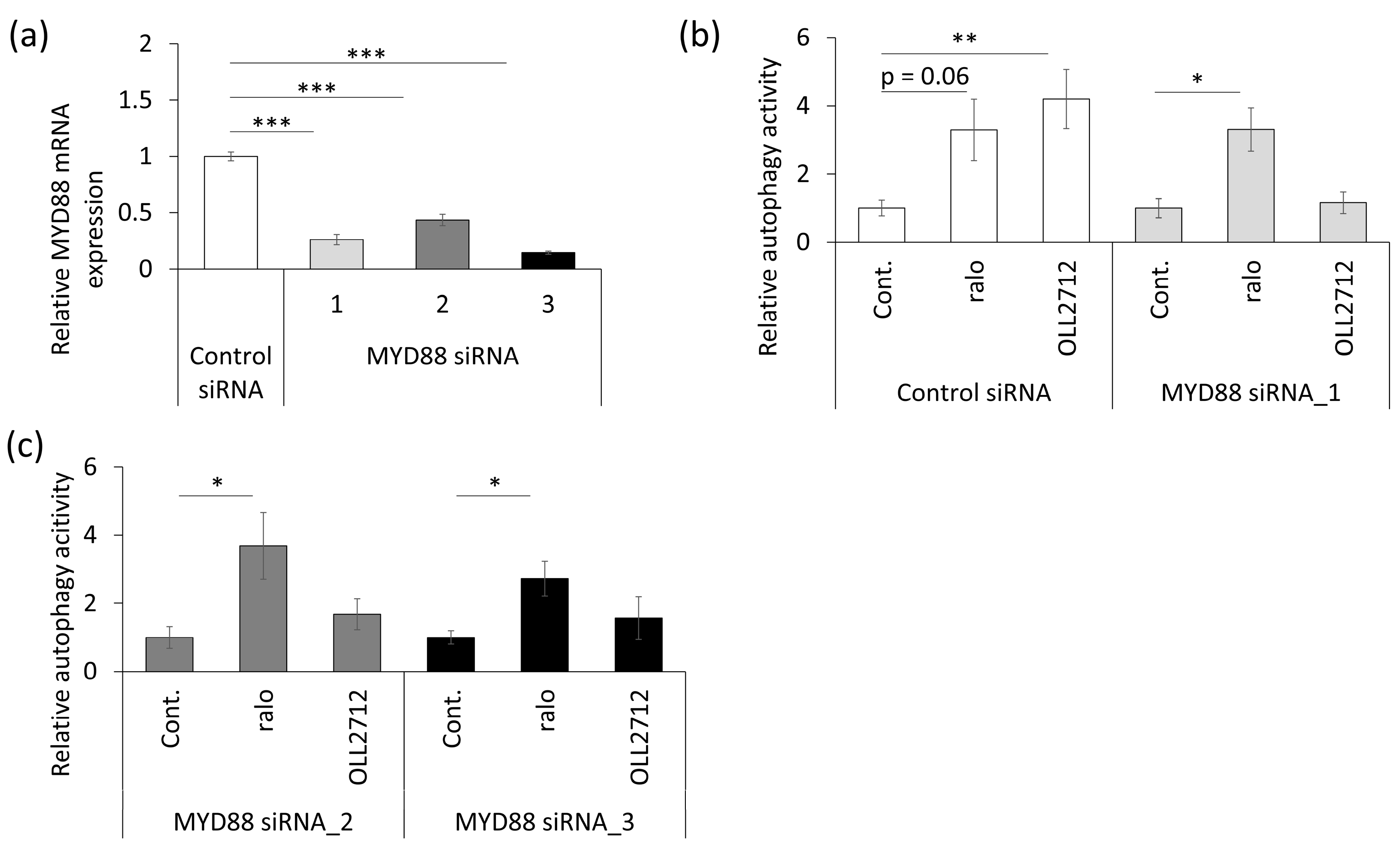
3.4. OLL2712 Induces Autophagy via MYD88
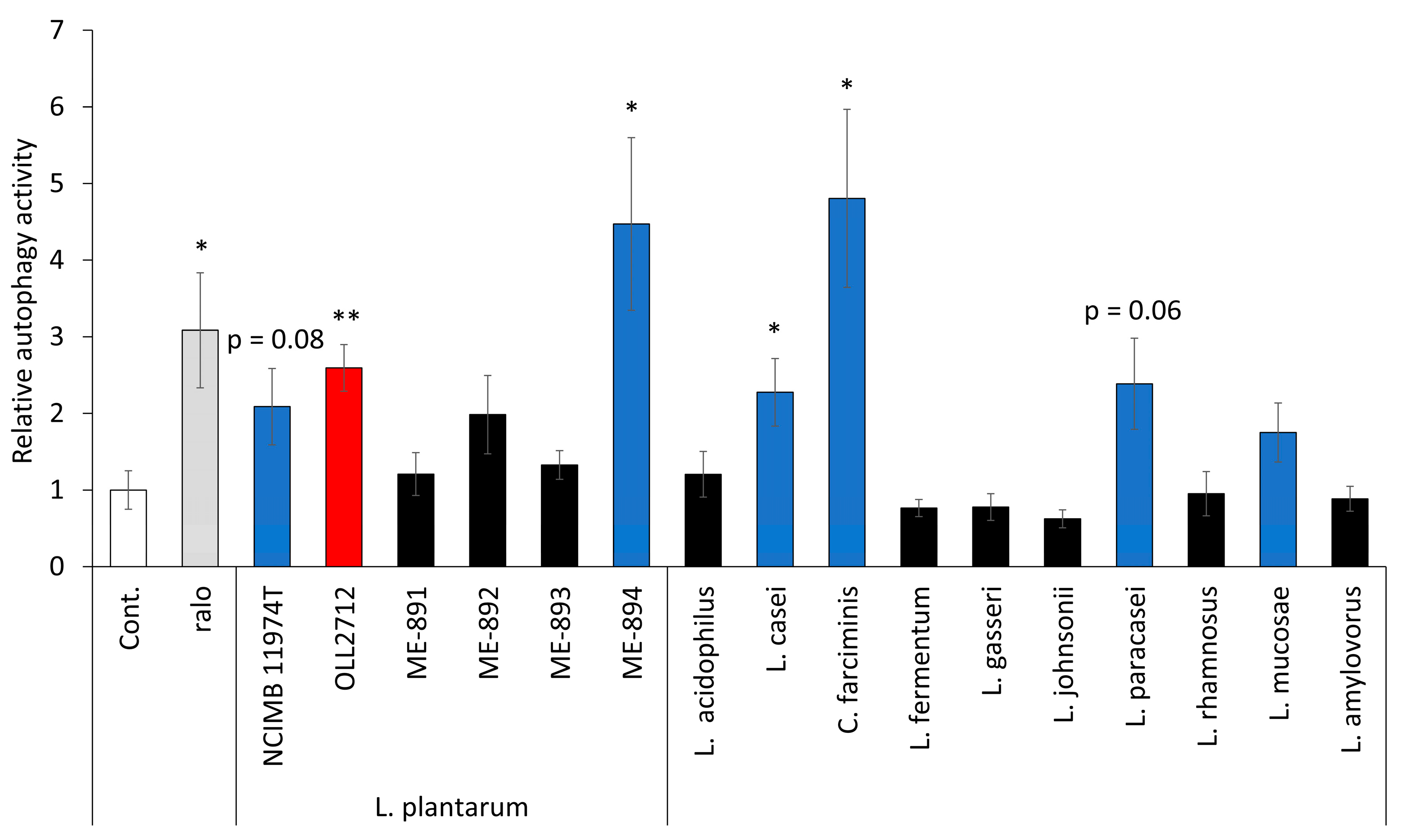
3.5. Some LAB Strains Promote Autophagy in Caco-2 Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, B.; Duden, R.; Rubinsztein, D.C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002, 11, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Ebato, C.; Uchida, T.; Arakawa, M.; Komatsu, M.; Ueno, T.; Komiya, K.; Azuma, K.; Hirose, T.; Tanaka, K.; Kominami, E.; et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008, 8, 325–332. [Google Scholar] [CrossRef]
- Torii, S.; Honda, S.; Murohashi, M.; Yamaguchi, H.; Shimizu, S. Autophagy involvement in oncogenesis. Cancer Sci. 2020, 111, 3993–3999. [Google Scholar] [CrossRef] [PubMed]
- Gardet, A.; Xavier, R.J. Common alleles that influence autophagy and the risk for inflammatory bowel disease. Curr. Opin. Immunol. 2012, 24, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Asano, J.; Sato, T.; Ichinose, S.; Kajita, M.; Onai, N.; Shimizu, S.; Ohteki, T. Intrinsic autophagy is required for the maintenance of intestinal stem cells and for irradiation-induced intestinal regeneration. Cell Rep. 2017, 20, 1050–1060. [Google Scholar] [CrossRef]
- Wlodarska, M.; Thaiss, C.A.; Nowarski, R.; Henao-Mejia, J.; Zhang, J.P.; Brown, E.M.; Frankel, G.; Levy, M.; Katz, M.N.; Philbrick, W.M.; et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell 2014, 156, 1045–1059. [Google Scholar] [CrossRef]
- Cadwell, K.; Liu, J.Y.; Brown, S.L.; Miyoshi, H.; Loh, J.; Lennerz, J.K.; Kishi, C.; Kc, W.; Carrero, J.A.; Hunt, S.; et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008, 456, 259–263. [Google Scholar] [CrossRef]
- Yi, R.; Zhou, X.; Liu, T.; Xue, R.; Yang, Z. Amelioration effect of Lactobacillus plantarum KFY02 on low-fiber diet-induced constipation in mice by regulating gut microbiota. Front. Nutr. 2022, 9, 938869. [Google Scholar] [CrossRef]
- Sashihara, T.; Nagata, M.; Mori, T.; Ikegami, S.; Gotoh, M.; Okubo, K.; Uchida, M.; Itoh, H. Effects of Lactobacillus gasseri OLL2809 and α-lactalbumin on university-student athletes: A randomized, double-blind, placebo-controlled clinical trial. Appl. Physiol. Nutr. Metab. 2013, 38, 1228–1235. [Google Scholar] [CrossRef]
- Kobayashi, K.; Mochizuki, J.; Yamazaki, F.; Sashihara, T. Yogurt starter strains ameliorate intestinal barrier dysfunction via activating AMPK in Caco-2 cells. Tissue Barriers 2023, 2184157. [Google Scholar] [CrossRef]
- Takano, T.; Endo, R.; Wang, Y.; Nakajima-Adachi, H.; Hachimura, S. Lactobacillus plantarum OLL2712 induces IL-10 production by intestinal dendritic cells. Biosci. Microbiota Food Health 2020, 39, 39–44. [Google Scholar] [CrossRef]
- Toshimitsu, T.; Mochizuki, J.; Ikegami, S.; Itou, H. Identification of a Lactobacillus plantarum strain that ameliorates chronic inflammation and metabolic disorders in obese and type 2 diabetic mice. J. Dairy Sci. 2016, 99, 933–946. [Google Scholar] [CrossRef] [PubMed]
- Toshimitsu, T.; Gotou, A.; Sashihara, T.; Furuichi, K.; Hachimura, S.; Shioya, N.; Suzuki, S.; Asami, Y. Ingesting yogurt containing Lactobacillus plantarum OLL2712 reduces abdominal fat accumulation and chronic inflammation in overweight adults in a randomized placebo-controlled trial. Curr. Dev. Nutr. 2021, 5, nzab006. [Google Scholar] [CrossRef]
- Toshimitsu, T.; Gotou, A.; Sashihara, T.; Hachimura, S.; Shioya, N.; Suzuki, S.; Asami, Y. Effects of 12-week ingestion of yogurt containing Lactobacillus plantarum OLL2712 on glucose metabolism and chronic inflammation in prediabetic adults: A randomized placebo-controlled trial. Nutrients 2020, 12, 374. [Google Scholar] [CrossRef]
- Lumeng, C.N.; Bodzin, J.L.; Saltiel, A.R. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Investig. 2007, 117, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Kawano, Y.; Nakae, J.; Watanabe, N.; Kikuchi, T.; Tateya, S.; Tamori, Y.; Kaneko, M.; Abe, T.; Onodera, M.; Itoh, H. Colonic pro-inflammatory macrophages cause insulin resistance in an intestinal Ccl2/Ccr2-dependent manner. Cell Metab. 2016, 24, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Yasueda, A.; Kayama, H.; Murohashi, M.; Nishimura, J.; Wakame, K.; Komatsu, K.I.; Ogino, T.; Miyoshi, N.; Takahashi, H.; Uemura, M.; et al. Sanguisorba officinalis L. derived from herbal medicine prevents intestinal inflammation by inducing autophagy in macrophages. Sci. Rep. 2020, 10, 9972. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Tang, X.; Wang, W.; Qian, W.; Fu, X.; Deng, X.; Zhou, S.; Han, C.; Hou, X. Lactobacillus rhamnosus attenuates intestinal inflammation induced by Fusobacterium nucleatum infection by restoring the autophagic flux. Int. J. Mol. Med. 2021, 47, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Chen, G.; Zhang, J.; Pei, C.; Chen, Y.; Gong, J.; Deng, S.; Cai, K.; Li, H.; Wang, D.; et al. Live Lactobacillus acidophilus alleviates ulcerative colitis via the SCFAs/mitophagy/NLRP3 inflammasome axis. Food Funct. 2022, 13, 2985–2997. [Google Scholar] [CrossRef]
- Wang, Y.; Takano, T.; Zhou, Y.; Wang, R.; Toshimitsu, T.; Sashihara, T.; Tanokura, M.; Miyakawa, T.; Nakajima-Adachi, H.; Hachimura, S. Orally administered Lactiplantibacillus plantarum OLL2712 decreased intestinal permeability, especially in the ileum: Ingested lactic acid bacteria alleviated obesity-induced inflammation by collaborating with gut microbiota. Front. Immunol. 2023, 14, 1123052. [Google Scholar] [CrossRef]
- Iwashita, H.; Sakurai, H.T.; Nagahora, N.; Ishiyama, M.; Shioji, K.; Sasamoto, K.; Okuma, K.; Shimizu, S.; Ueno, Y. Small fluorescent molecules for monitoring autophagic flux. FEBS Lett. 2018, 592, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Kim, J.; Lee, W. Raloxifene prevents intracellular invasion of pathogenic bacteria through modulation of cell metabolic pathways. J. Antimicrob. Chemother. 2022, 77, 1617–1624. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Kabeya, Y.; Mizushima, N.; Ueno, T.; Yamamoto, A.; Kirisako, T.; Noda, T.; Kominami, E.; Ohsumi, Y.; Yoshimori, T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000, 19, 5720–5728. [Google Scholar] [CrossRef] [PubMed]
- Kabeya, Y.; Mizushima, N.; Yamamoto, A.; Oshitani-Okamoto, S.; Ohsumi, Y.; Yoshimori, T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004, 117, 2805–2812. [Google Scholar] [CrossRef]
- Yamamoto, A.; Tagawa, Y.; Yoshimori, T.; Moriyama, Y.; Masaki, R.; Tashiro, Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 1998, 23, 33–42. [Google Scholar] [CrossRef]
- Yu, Q.; Wang, Z.; Yang, Q. Lactobacillus amylophilus D14 protects tight junction from enteropathogenic bacteria damage in Caco-2 cells. J. Dairy Sci. 2012, 95, 5580–5587. [Google Scholar] [CrossRef]
- Shukla, P.K.; Meena, A.S.; Manda, B.; Gomes-Solecki, M.; Dietrich, P.; Dragatsis, I.; Rao, R. Lactobacillus plantarum prevents and mitigates alcohol-induced disruption of colonic epithelial tight junctions, endotoxemia, and liver damage by an EGF receptor-dependent mechanism. FASEB J. 2018, 32, fj201800351R. [Google Scholar] [CrossRef]
- Nighot, P.K.; Hu, C.A.; Ma, T.Y. Autophagy enhances intestinal epithelial tight junction barrier function by targeting claudin-2 protein degradation. J. Biol. Chem. 2015, 290, 7234–7246. [Google Scholar] [CrossRef]
- Johansson, M.E.; Hansson, G.C. Mucus and the goblet cell. Dig. Dis. 2013, 31, 305–309. [Google Scholar] [CrossRef]
- Martínez-Maqueda, D.; Miralles, B.; Recio, I. HT29 Cell Line. In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer: Cham, Switzerland, 2015; pp. 113–124. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef]
- Benjamin, J.L.; Sumpter, R., Jr.; Levine, B.; Hooper, L.V. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe 2013, 13, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Yi, J.; Gubas, A.; Wang, Y.T.; Wu, Y.; Ren, Y.; Wu, M.; Shi, Y.; Ouyang, C.; Tan, H.W.S.; et al. Suppression of autophagy during mitosis via CUL4-RING ubiquitin ligases-mediated WIPI2 polyubiquitination and proteasomal degradation. Autophagy 2019, 15, 1917–1934. [Google Scholar] [CrossRef] [PubMed]
- Powell, D.W. Barrier function of epithelia. Am. J. Physiol. 1981, 241, G275–G288. [Google Scholar] [CrossRef]
- Yee, S. In vitro permeability across Caco-2 cells (colonic) can predict in vivo (small intestinal) absorption in man--fact or myth. Pharm. Res. 1997, 14, 763–766. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Lv, Q.; Qiao, S.; Yang, L.; Tao, Y.; Yan, W.; Wang, P.; Cao, N.; Dai, Y.; Wei, Z. Alpinetin improves intestinal barrier homeostasis via regulating AhR/suv39h1/TSC2/mTORC1/autophagy pathway. Toxicol. Appl. Pharmacol. 2019, 384, 114772. [Google Scholar] [CrossRef]
- Heitman, J.; Movva, N.R.; Hall, M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991, 253, 905–909. [Google Scholar] [CrossRef]
- Ren, H.; Shao, Y.; Wu, C.; Ma, X.; Lv, C.; Wang, Q. Metformin alleviates oxidative stress and enhances autophagy in diabetic kidney disease via AMPK/SIRT1-FoxO1 pathway. Mol. Cell. Endocrinol. 2020, 500, 110628. [Google Scholar] [CrossRef]
- Song, Y.M.; Lee, Y.H.; Kim, J.W.; Ham, D.S.; Kang, E.S.; Cha, B.S.; Lee, H.C.; Lee, B.W. Metformin alleviates hepatosteatosis by restoring SIRT1-mediated autophagy induction via an AMP-activated protein kinase-independent pathway. Autophagy 2015, 11, 46–59. [Google Scholar] [CrossRef]
- Kim, D.E.; Kim, Y.; Cho, D.H.; Jeong, S.Y.; Kim, S.B.; Suh, N.; Lee, J.S.; Choi, E.K.; Koh, J.Y.; Hwang, J.J.; et al. Raloxifene induces autophagy-dependent cell death in breast cancer cells via the activation of AMP-activated protein kinase. Mol. Cells 2015, 38, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.; Phillipson, M.; Petersson, J.; Velcich, A.; Holm, L.; Hansson, G.C. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 15064–15069. [Google Scholar] [CrossRef]
- Wang, S.; Ahmadi, S.; Nagpal, R.; Jain, S.; Mishra, S.P.; Kavanagh, K.; Zhu, X.; Wang, Z.; McClain, D.A.; Kritchevsky, S.B.; et al. Lipoteichoic acid from the cell wall of a heat killed Lactobacillus paracasei D3-5 ameliorates aging-related leaky gut, inflammation and improves physical and cognitive functions: From C. elegans to mice. GeroScience 2020, 42, 333–352. [Google Scholar] [CrossRef]
- Fischer, W.; Mannsfeld, T.; Hagen, G. On the basic structure of poly(glycerophosphate) lipoteichoic acids. Biochem. Cell Biol. 1990, 68, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Toll-like receptors; their physiological role and signal transduction system. Int. Immunopharmacol. 2001, 1, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Sanjuan, M.A.; Dillon, C.P.; Tait, S.W.; Moshiach, S.; Dorsey, F.; Connell, S.; Komatsu, M.; Tanaka, K.; Cleveland, J.L.; Withoff, S.; et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 2007, 450, 1253–1257. [Google Scholar] [CrossRef]
- Shi, C.S.; Kehrl, J.H. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J. Biol. Chem. 2008, 283, 33175–33182. [Google Scholar] [CrossRef]
- Jia, Y.P.; Wang, K.; Zhang, Z.J.; Tong, Y.N.; Han, D.; Hu, C.Y.; Li, Q.; Xiang, Y.; Mao, X.H.; Tang, B. TLR2/TLR4 activation induces Tregs and suppresses intestinal inflammation caused by Fusobacterium nucleatum in vivo. PLoS ONE 2017, 12, e0186179. [Google Scholar] [CrossRef]
- Sun, D.; Ding, A. MyD88-mediated stabilization of interferon-gamma-induced cytokine and chemokine mRNA. Nat. Immunol. 2006, 7, 375–381. [Google Scholar] [CrossRef]
- Luo, L.; Jiang, S.; Huang, D.; Lu, N.; Luo, Z. MLK3 phophorylates AMPK independently of LKB1. PLoS ONE 2015, 10, e0123927. [Google Scholar] [CrossRef]
- Alers, S.; Löffler, A.S.; Wesselborg, S.; Stork, B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: Cross talk, shortcuts, and feedbacks. Mol. Cell. Biol. 2012, 32, 2–11. [Google Scholar] [CrossRef]
- Shi, M.; Yue, Y.; Ma, C.; Dong, L.; Chen, F. Pasteurized Akkermansia muciniphila ameliorate the LPS-induced intestinal barrier dysfunction via modulating AMPK and NF-κB through TLR2 in Caco-2 cells. Nutrients 2022, 14, 764. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Yang, Q.; Rogers, C.J.; Du, M.; Zhu, M.J. AMPK improves gut epithelial differentiation and barrier function via regulating Cdx2 expression. Cell Death Differ. 2017, 24, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Honme, Y.; Sashihara, T. Lactobacillus delbrueckii subsp. bulgaricus 2038 and Streptococcus thermophilus 1131 Induce the Expression of the REG3 Family in the Small Intestine of Mice via the Stimulation of Dendritic Cells and Type 3 Innate Lymphoid Cells. Nutrients 2019, 11, 2998. [Google Scholar] [CrossRef] [PubMed]
- Holczbauer, Á.; Gyöngyösi, B.; Lotz, G.; Szijártó, A.; Kupcsulik, P.; Schaff, Z.; Kiss, A. Distinct claudin expression profiles of hepatocellular carcinoma and metastatic colorectal and pancreatic carcinomas. J. Histochem. Cytochem. 2013, 61, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Braniste, V.; Leveque, M.; Buisson-Brenac, C.; Bueno, L.; Fioramonti, J.; Houdeau, E. Oestradiol decreases colonic permeability through oestrogen receptor beta-mediated up-regulation of occludin and junctional adhesion molecule-A in epithelial cells. J. Physiol. 2009, 587, 3317–3328. [Google Scholar] [CrossRef] [PubMed]
- Spandidos, A.; Wang, X.; Wang, H.; Seed, B. PrimerBank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010, 38, D792–D799. [Google Scholar] [CrossRef]

| Species | Strains |
|---|---|
| Lactiplantibacillus plantarum subsp. Plantarum * | NCIMB 11974T |
| Lactiplantibacillus plantarum * | OLL2712 |
| Lactiplantibacillus plantarum * | ME-891 ** |
| Lactiplantibacillus plantarum * | ME-892 ** |
| Lactiplantibacillus plantarum * | ME-893 ** |
| Lactiplantibacillus plantarum * | ME-894 ** |
| Lactobacillus acidophilus | JCM 1132T |
| Lacticaseibacillus casei * | JCM 1134T |
| Companilactobacillus farciminis * | ATCC 29644T |
| Limosilactobacillus fermentum * | NRIC 1752T |
| Lactobacillus gasseri | JCM 1131T |
| Lactobacillus johnsonii | JCM 2012T |
| Lacticaseibacillus paracasei * | NBRC 15889T |
| Lacticaseibacillus rhamnosus * | JCM 1136T |
| Limosilactobacillus mucosae * | NCIMB 13705T |
| Lactobacillus amylovorus | JCM 1126T |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watanabe-Yasuoka, Y.; Gotou, A.; Shimizu, S.; Sashihara, T. Lactiplantibacillus plantarum OLL2712 Induces Autophagy via MYD88 and Strengthens Tight Junction Integrity to Promote the Barrier Function in Intestinal Epithelial Cells. Nutrients 2023, 15, 2655. https://doi.org/10.3390/nu15122655
Watanabe-Yasuoka Y, Gotou A, Shimizu S, Sashihara T. Lactiplantibacillus plantarum OLL2712 Induces Autophagy via MYD88 and Strengthens Tight Junction Integrity to Promote the Barrier Function in Intestinal Epithelial Cells. Nutrients. 2023; 15(12):2655. https://doi.org/10.3390/nu15122655
Chicago/Turabian StyleWatanabe-Yasuoka, Yumiko, Ayako Gotou, Shigeomi Shimizu, and Toshihiro Sashihara. 2023. "Lactiplantibacillus plantarum OLL2712 Induces Autophagy via MYD88 and Strengthens Tight Junction Integrity to Promote the Barrier Function in Intestinal Epithelial Cells" Nutrients 15, no. 12: 2655. https://doi.org/10.3390/nu15122655
APA StyleWatanabe-Yasuoka, Y., Gotou, A., Shimizu, S., & Sashihara, T. (2023). Lactiplantibacillus plantarum OLL2712 Induces Autophagy via MYD88 and Strengthens Tight Junction Integrity to Promote the Barrier Function in Intestinal Epithelial Cells. Nutrients, 15(12), 2655. https://doi.org/10.3390/nu15122655







