Transcriptomic Analysis of Human Skeletal Muscle in Response to Aerobic Exercise and Protein Intake
Abstract
:1. Introduction
2. Materials and Methods
2.1. Public Datasets
2.2. Data Pre-Processing
2.3. Identification of Differentially Expressed Genes (DEGs)
2.4. Functional Analysis and Network Visualization of co-DEGs
2.5. Gene Set Enrichment Analysis (GSEA)
3. Results
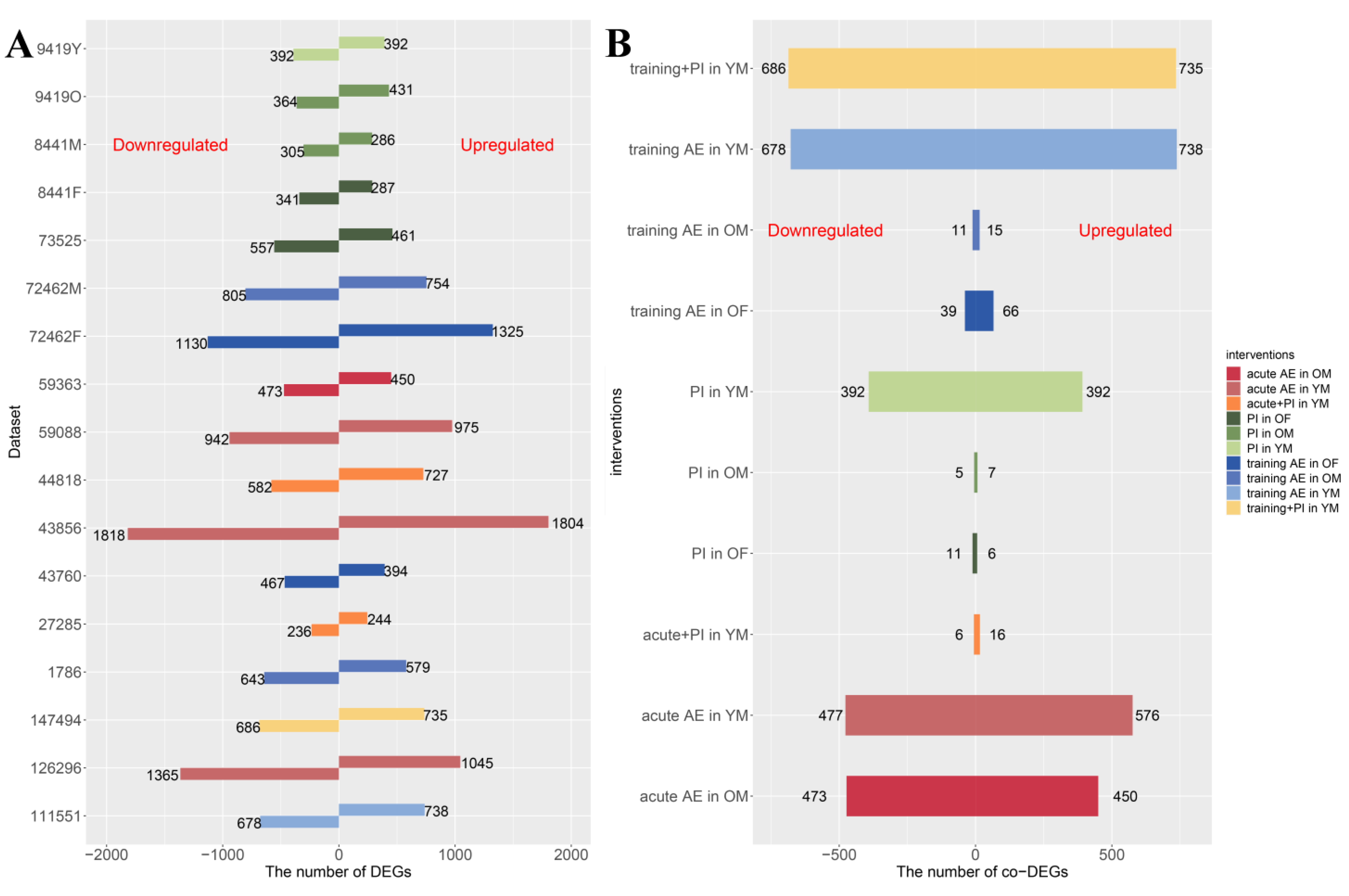
3.1. Overview of Datasets Collection
3.2. DEGs Identification, Hub Gene Screening, and Gene Expression Network Construction
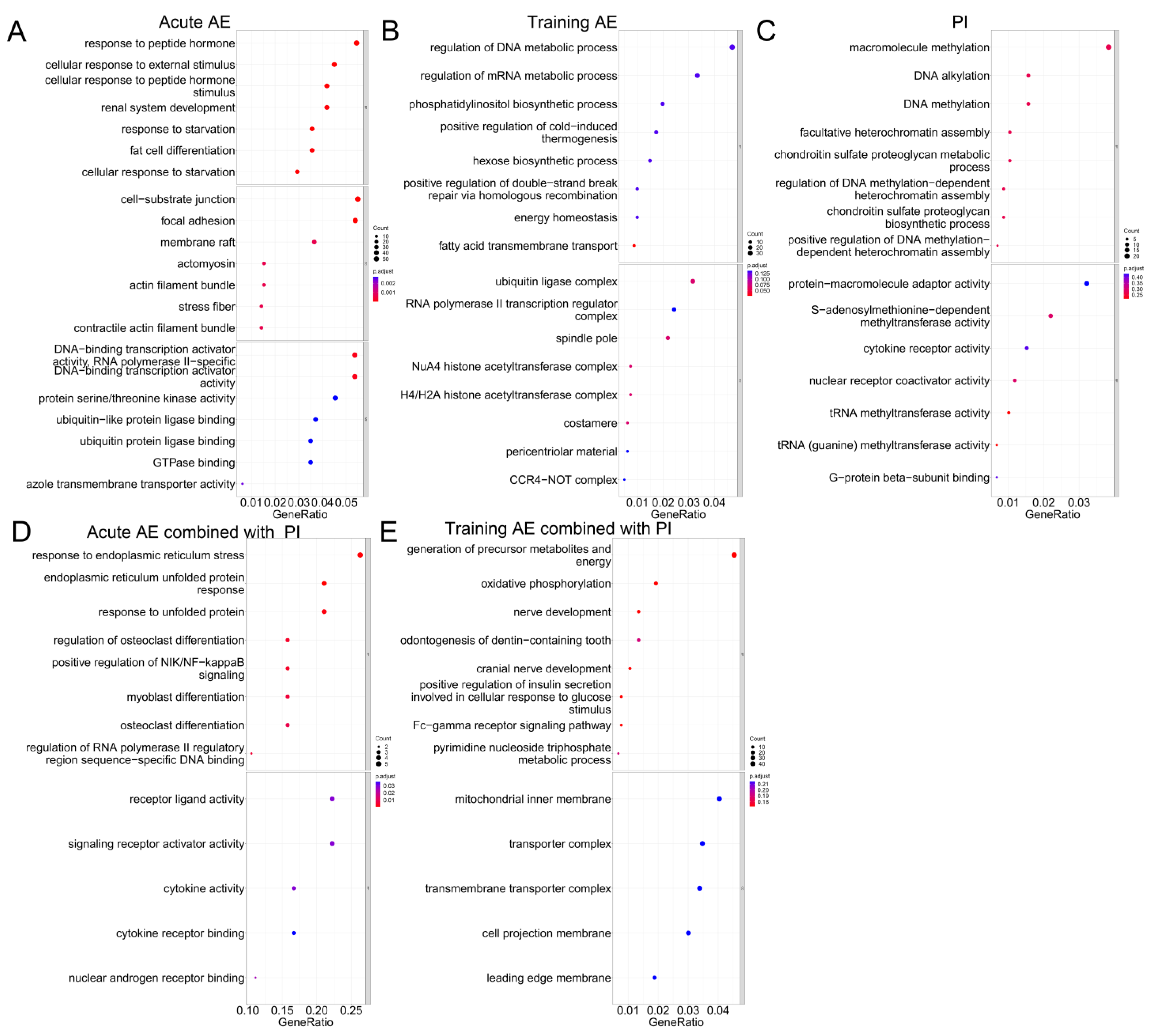
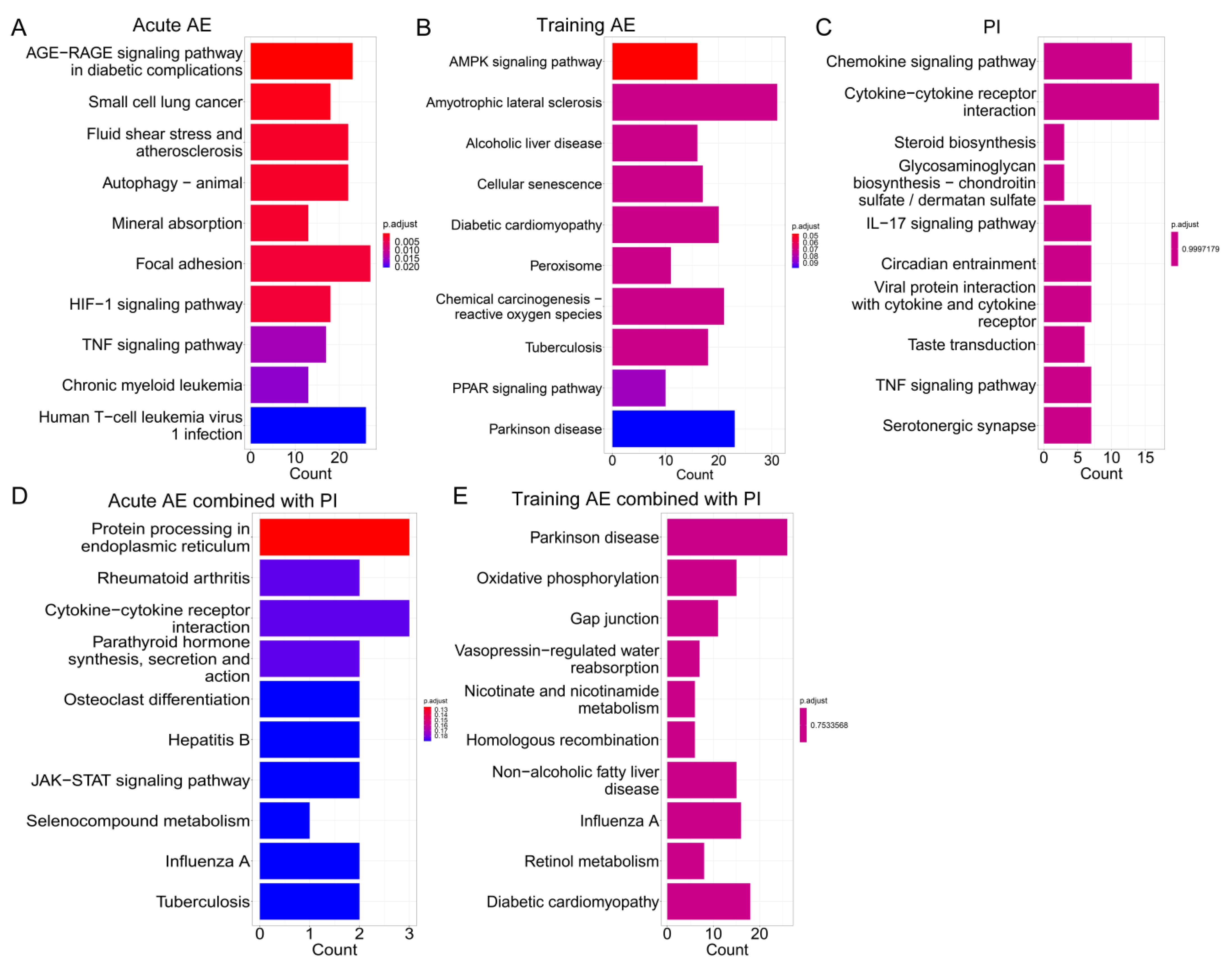
3.3. Functional Enrichment Analysis for co-DEGs
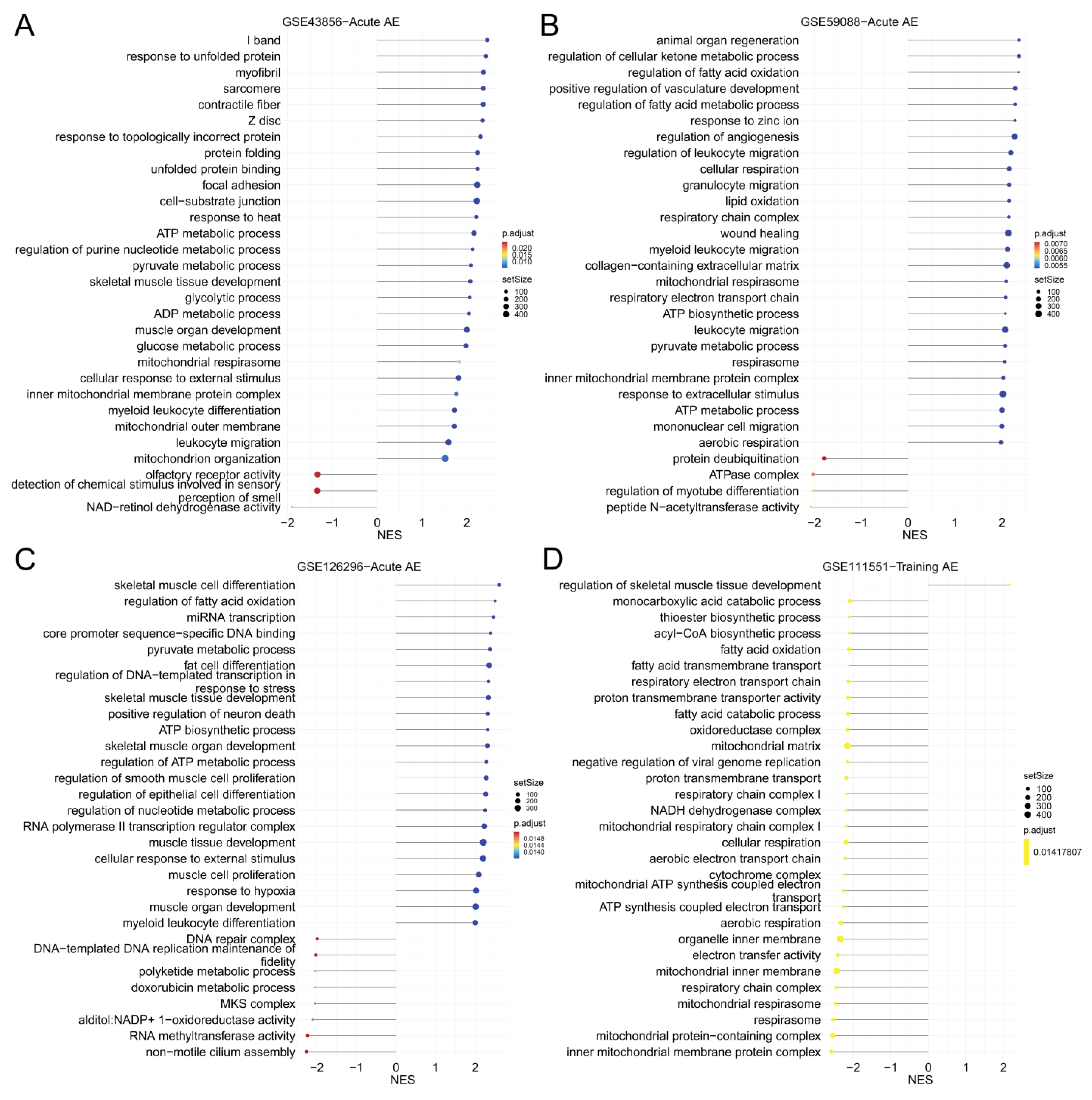
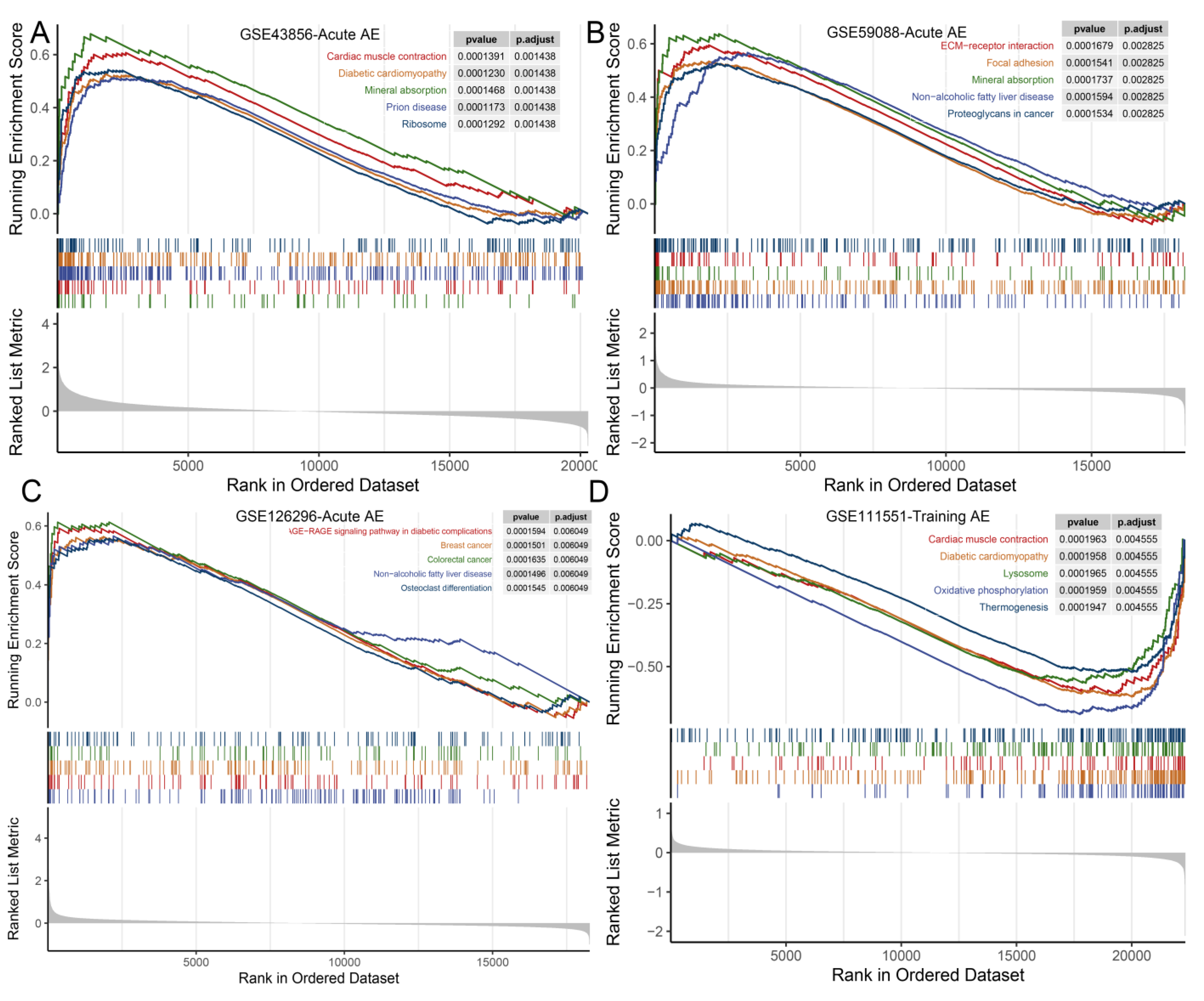
3.4. GSEA
4. Discussion
4.1. Transcriptomic Changes in Skeletal Muscle in Response to Acute AE, PI, and Acute AE Combined with PI
4.2. Transcriptomic Changes in Skeletal Muscle in Response to Training AE, PI, and Training AE Combined with PI
4.3. Strengths and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wolfe, R.R. The underappreciated role of muscle in health and disease. Am. J. Clin. Nutr. 2006, 84, 475–482. [Google Scholar] [CrossRef] [Green Version]
- Bauer, J.; Morley, J.E.; Schols, A.; Ferrucci, L.; Cruz-Jentoft, A.J.; Dent, E.; Baracos, V.E.; Crawford, J.A.; Doehner, W.; Heymsfield, S.B.; et al. Sarcopenia: A Time for Action. An SCWD Position Paper. J. Cachexia Sarcopenia Muscle 2019, 10, 956–961. [Google Scholar] [CrossRef] [Green Version]
- Cruz-Jentoft, A.J.; Sayer, A.A. Sarcopenia. Lancet 2019, 393, 2636–2646. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Jentoft, A.J.; Landi, F.; Schneider, S.M.; Zuniga, C.; Arai, H.; Boirie, Y.; Chen, L.K.; Fielding, R.A.; Martin, F.C.; Michel, J.P.; et al. Prevalence of and interventions for sarcopenia in ageing adults: A systematic review. Report of the International Sarcopenia Initiative (EWGSOP and IWGS). Age Ageing 2014, 43, 748–759. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L.; Degens, H.; Li, M.; Salviati, L.; Lee, Y.I.; Thompson, W.; Kirkland, J.L.; Sandri, M. Sarcopenia: Aging-Related Loss of Muscle Mass and Function. Physiol. Rev. 2019, 99, 427–511. [Google Scholar] [CrossRef]
- Vikberg, S.; Sorlen, N.; Branden, L.; Johansson, J.; Nordstrom, A.; Hult, A.; Nordstrom, P. Effects of Resistance Training on Functional Strength and Muscle Mass in 70-Year-Old Individuals with Pre-sarcopenia: A Randomized Controlled Trial. J. Am. Med. Dir. Assoc. 2019, 20, 28–34. [Google Scholar] [CrossRef] [Green Version]
- Kemmler, W.; Kohl, M.; Frohlich, M.; Jakob, F.; Engelke, K.; von Stengel, S.; Schoene, D. Effects of High-Intensity Resistance Training on Osteopenia and Sarcopenia Parameters in Older Men with Osteosarcopenia-One-Year Results of the Randomized Controlled Franconian Osteopenia and Sarcopenia Trial (FrOST). J. Bone Miner. Res. 2020, 35, 1634–1644. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, Y.; Yamada, Y.; Maeda, A.; Izumo, T.; Rogi, T.; Shibata, H.; Fukuda, M.; Arimitsu, T.; Miyamoto, N.; Hashimoto, T. Effects of resistance training intensity on muscle quantity/quality in middle-aged and older people: A randomized controlled trial. J. Cachexia Sarcopenia Muscle 2022, 13, 894–908. [Google Scholar] [CrossRef]
- Hartono, F.A.; Martin-Arrowsmith, P.W.; Peeters, W.M.; Churchward-Venne, T.A. The Effects of Dietary Protein Supplementation on Acute Changes in Muscle Protein Synthesis and Longer-Term Changes in Muscle Mass, Strength, and Aerobic Capacity in Response to Concurrent Resistance and Endurance Exercise in Healthy Adults: A Systematic Review. Sports Med. 2022, 52, 1295–1328. [Google Scholar]
- Fyfe, J.J.; Bishop, D.J.; Stepto, N.K. Interference between concurrent resistance and endurance exercise: Molecular bases and the role of individual training variables. Sports Med. 2014, 44, 743–762. [Google Scholar] [CrossRef]
- Hurst, C.; Robinson, S.M.; Witham, M.D.; Dodds, R.M.; Granic, A.; Buckland, C.; De Biase, S.; Finnegan, S.; Rochester, L.; Skelton, D.A.; et al. Resistance exercise as a treatment for sarcopenia: Prescription and delivery. Age Ageing 2022, 51, afac003. [Google Scholar] [CrossRef] [PubMed]
- Petersen, K.F.; Befroy, D.; Dufour, S.; Dziura, J.; Ariyan, C.; Rothman, D.L.; DiPietro, L.; Cline, G.W.; Shulman, G.I. Mitochondrial dysfunction in the elderly: Possible role in insulin resistance. Science 2003, 300, 1140–1142. [Google Scholar] [CrossRef] [Green Version]
- Short, K.R.; Bigelow, M.L.; Kahl, J.; Singh, R.; Coenen-Schimke, J.; Raghavakaimal, S.; Nair, K.S. Decline in skeletal muscle mitochondrial function with aging in humans. Proc. Natl. Acad. Sci. USA 2005, 102, 5618–5623. [Google Scholar] [CrossRef] [PubMed]
- Mitsuishi, M.; Miyashita, K.; Muraki, A.; Tamaki, M.; Tanaka, K.; Itoh, H. Dietary protein decreases exercise endurance through rapamycin-sensitive suppression of muscle mitochondria. Am. J. Physiol. Endocrinol. Metab. 2013, 305, E776–E784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leduc-Gaudet, J.P.; Hussain, S.N.A.; Barreiro, E.; Gouspillou, G. Mitochondrial Dynamics and Mitophagy in Skeletal Muscle Health and Aging. Int. J. Mol. Sci. 2021, 22, 8179. [Google Scholar] [CrossRef]
- Petersen, K.F.; Dufour, S.; Befroy, D.; Garcia, R.; Shulman, G.I. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N. Engl. J. Med. 2004, 350, 664–671. [Google Scholar] [CrossRef] [Green Version]
- Curcio, F.; Testa, G.; Liguori, I.; Papillo, M.; Flocco, V.; Panicara, V.; Galizia, G.; Della-Morte, D.; Gargiulo, G.; Cacciatore, F.; et al. Sarcopenia and Heart Failure. Nutrients 2020, 12, 211. [Google Scholar] [CrossRef] [Green Version]
- Lavine, K.J.; Sierra, O.L. Skeletal muscle inflammation and atrophy in heart failure. Heart Fail. Rev. 2017, 22, 179–189. [Google Scholar] [CrossRef] [Green Version]
- Kalinkovich, A.; Livshits, G. Sarcopenic obesity or obese sarcopenia: A cross talk between age-associated adipose tissue and skeletal muscle inflammation as a main mechanism of the pathogenesis. Ageing Res. Rev. 2017, 35, 200–221. [Google Scholar] [CrossRef]
- Petridou, A.; Siopi, A.; Mougios, V. Exercise in the management of obesity. Metabolism 2019, 92, 163–169. [Google Scholar] [CrossRef] [Green Version]
- Ismail, I.; Keating, S.E.; Baker, M.K.; Johnson, N.A. A systematic review and meta-analysis of the effect of aerobic vs. resistance exercise training on visceral fat. Obes. Rev. 2012, 13, 68–91. [Google Scholar] [CrossRef] [PubMed]
- Beckers, P.J.; Denollet, J.; Possemiers, N.M.; Wuyts, F.L.; Vrints, C.J.; Conraads, V.M. Combined endurance-resistance training vs. endurance training in patients with chronic heart failure: A prospective randomized study. Eur. Heart J. 2008, 29, 1858–1866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, A.; Kitzman, D.W.; Brubaker, P.; Haykowsky, M.J.; Morgan, T.; Becton, J.T.; Berry, J.D. Response to Endurance Exercise Training in Older Adults with Heart Failure with Preserved or Reduced Ejection Fraction. J. Am. Geriatr. Soc. 2017, 65, 1698–1704. [Google Scholar] [CrossRef]
- Joisten, N.; Kummerhoff, F.; Koliamitra, C.; Schenk, A.; Walzik, D.; Hardt, L.; Knoop, A.; Thevis, M.; Kiesl, D.; Metcalfe, A.J.; et al. Exercise and the Kynurenine pathway: Current state of knowledge and results from a randomized cross-over study comparing acute effects of endurance and resistance training. Exerc. Immunol. Rev. 2020, 26, 24–42. [Google Scholar] [PubMed]
- Schlagheck, M.L.; Walzik, D.; Joisten, N.; Koliamitra, C.; Hardt, L.; Metcalfe, A.J.; Wahl, P.; Bloch, W.; Schenk, A.; Zimmer, P. Cellular immune response to acute exercise: Comparison of endurance and resistance exercise. Eur. J. Haematol. 2020, 105, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Mao, L.; Feng, Y.; Ainsworth, B.E.; Liu, Y.; Chen, N. Effects of different exercise training modes on muscle strength and physical performance in older people with sarcopenia: A systematic review and meta-analysis. BMC Geriatr. 2021, 21, 708. [Google Scholar] [CrossRef]
- Strasser, B.; Keinrad, M.; Haber, P.; Schobersberger, W. Efficacy of systematic endurance and resistance training on muscle strength and endurance performance in elderly adults—A randomized controlled trial. Wien. Klin. Wochenschr. 2009, 121, 757–764. [Google Scholar] [CrossRef]
- Rondanelli, M.; Cereda, E.; Klersy, C.; Faliva, M.A.; Peroni, G.; Nichetti, M.; Gasparri, C.; Iannello, G.; Spadaccini, D.; Infantino, V.; et al. Improving rehabilitation in sarcopenia: A randomized-controlled trial utilizing a muscle-targeted food for special medical purposes. J. Cachexia Sarcopenia Muscle 2020, 11, 1535–1547. [Google Scholar] [CrossRef]
- Park, S.J.; Park, J.; Won, C.W.; Lee, H.J. The Inverse Association of Sarcopenia and Protein-Source Food and Vegetable Intakes in the Korean Elderly: The Korean Frailty and Aging Cohort Study. Nutrients 2022, 14, 1375. [Google Scholar] [CrossRef]
- Verlaan, S.; Maier, A.B.; Bauer, J.M.; Bautmans, I.; Brandt, K.; Donini, L.M.; Maggio, M.; McMurdo, M.E.T.; Mets, T.; Seal, C.; et al. Sufficient levels of 25-hydroxyvitamin D and protein intake required to increase muscle mass in sarcopenic older adults—The PROVIDE study. Clin. Nutr. 2018, 37, 551–557. [Google Scholar] [CrossRef] [Green Version]
- Nunes, E.A.; Colenso-Semple, L.; McKellar, S.R.; Yau, T.; Ali, M.U.; Fitzpatrick-Lewis, D.; Sherifali, D.; Gaudichon, C.; Tome, D.; Atherton, P.J.; et al. Systematic review and meta-analysis of protein intake to support muscle mass and function in healthy adults. J Cachexia Sarcopenia Muscle 2022, 13, 795–810. [Google Scholar] [CrossRef]
- Park, Y.; Choi, J.E.; Hwang, H.S. Protein supplementation improves muscle mass and physical performance in undernourished prefrail and frail elderly subjects: A randomized, double-blind, placebo-controlled trial. Am. J. Clin. Nutr. 2018, 108, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.Y.; Huang, K.S.; Chen, K.M.; Chou, C.P.; Tu, Y.K. Exercise, Nutrition, and Combined Exercise and Nutrition in Older Adults with Sarcopenia: A Systematic Review and Network Meta-analysis. Maturitas 2021, 145, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Deldicque, L. Protein Intake and Exercise-Induced Skeletal Muscle Hypertrophy: An Update. Nutrients 2020, 12, 2023. [Google Scholar] [CrossRef]
- Longland, T.M.; Oikawa, S.Y.; Mitchell, C.J.; Devries, M.C.; Phillips, S.M. Higher compared with lower dietary protein during an energy deficit combined with intense exercise promotes greater lean mass gain and fat mass loss: A randomized trial. Am. J. Clin. Nutr. 2016, 103, 738–746. [Google Scholar] [CrossRef] [Green Version]
- Kemmler, W.; Kohl, M.; Jakob, F.; Engelke, K.; von Stengel, S. Effects of High Intensity Dynamic Resistance Exercise and Whey Protein Supplements on Osteosarcopenia in Older Men with Low Bone and Muscle Mass. Final Results of the Randomized Controlled FrOST Study. Nutrients 2020, 12, 2341. [Google Scholar] [CrossRef] [PubMed]
- Breen, L.; Philp, A.; Witard, O.C.; Jackman, S.R.; Selby, A.; Smith, K.; Baar, K.; Tipton, K.D. The influence of carbohydrate-protein co-ingestion following endurance exercise on myofibrillar and mitochondrial protein synthesis. J. Physiol. 2011, 589, 4011–4025. [Google Scholar] [CrossRef]
- Hawley, J.A.; Burke, L.M.; Phillips, S.M.; Spriet, L.L. Nutritional modulation of training-induced skeletal muscle adaptations. J Appl. Physiol. 2011, 110, 834–845. [Google Scholar] [CrossRef] [Green Version]
- Pillon, N.J.; Gabriel, B.M.; Dollet, L.; Smith, J.A.B.; Sardon Puig, L.; Botella, J.; Bishop, D.J.; Krook, A.; Zierath, J.R. Transcriptomic profiling of skeletal muscle adaptations to exercise and inactivity. Nat. Commun. 2020, 11, 470. [Google Scholar] [CrossRef] [Green Version]
- Deane, C.S.; Willis, C.R.G.; Phillips, B.E.; Atherton, P.J.; Harries, L.W.; Ames, R.M.; Szewczyk, N.J.; Etheridge, T. Transcriptomic meta-analysis of disuse muscle atrophy vs. resistance exercise-induced hypertrophy in young and older humans. J. Cachexia Sarcopenia Muscle 2021, 12, 629–645. [Google Scholar] [CrossRef]
- Amar, D.; Lindholm, M.E.; Norrbom, J.; Wheeler, M.T.; Rivas, M.A.; Ashley, E.A. Time trajectories in the transcriptomic response to exercise-a meta-analysis. Nat. Commun. 2021, 12, 3471. [Google Scholar] [CrossRef] [PubMed]
- Knuiman, P.; van Loon, L.J.C.; Wouters, J.; Hopman, M.; Mensink, M. Protein supplementation elicits greater gains in maximal oxygen uptake capacity and stimulates lean mass accretion during prolonged endurance training: A double-blind randomized controlled trial. Am. J. Clin. Nutr. 2019, 110, 508–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonvik, K.L.; Paulussen, K.J.M.; Danen, S.L.; Ceelen, I.J.M.; Horstman, A.M.; Wardenaar, F.C.; VAN Loon, L.J.C.; VAN Dijk, J.-W. Protein Supplementation Does Not Augment Adaptations to Endurance Exercise Training. Med. Sci. Sports Exerc. 2019, 51, 2041–2049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williamson, E.; Kato, H.; Volterman, K.A.; Suzuki, K.; Moore, D.R. The Effect of Dietary Protein on Protein Metabolism and Performance in Endurance-trained Males. Med. Sci. Sports Exerc. 2019, 51, 352–360. [Google Scholar] [CrossRef]
- Churchward-Venne, T.A.; Pinckaers, P.J.M.; Smeets, J.S.J.; Betz, M.W.; Senden, J.M.; Goessens, J.P.B.; Gijsen, A.P.; Rollo, I.; Verdijk, L.B.; van Loon, L.J.C. Dose-response effects of dietary protein on muscle protein synthesis during recovery from endurance exercise in young men: A double-blind randomized trial. Am. J. Clin. Nutr. 2020, 112, 303–317. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. S4), S11. [Google Scholar] [CrossRef] [Green Version]
- Neubauer, O.; Sabapathy, S.; Lazarus, R.; Jowett, J.B.; Desbrow, B.; Peake, J.M.; Cameron-Smith, D.; Haseler, L.J.; Wagner, K.H.; Bulmer, A.C. Transcriptome analysis of neutrophils after endurance exercise reveals novel signaling mechanisms in the immune response to physiological stress. J. Appl. Physiol. 2013, 114, 1677–1688. [Google Scholar] [CrossRef] [Green Version]
- Vissing, K.; Schjerling, P. Simplified data access on human skeletal muscle transcriptome responses to differentiated exercise. Sci. Data 2014, 1, 140041. [Google Scholar] [CrossRef] [Green Version]
- Rundqvist, H.C.; Montelius, A.; Osterlund, T.; Norman, B.; Esbjornsson, M.; Jansson, E. Acute sprint exercise transcriptome in human skeletal muscle. PLoS ONE 2019, 14, e0223024. [Google Scholar] [CrossRef] [Green Version]
- Hansen, J.S.; Zhao, X.; Irmler, M.; Liu, X.; Hoene, M.; Scheler, M.; Li, Y.; Beckers, J.; Hrabe de Angelis, M.; Haring, H.U.; et al. Type 2 diabetes alters metabolic and transcriptional signatures of glucose and amino acid metabolism during exercise and recovery. Diabetologia 2015, 58, 1845–1854. [Google Scholar] [CrossRef] [Green Version]
- Böhm, A.; Hoffmann, C.; Irmler, M.; Schneeweiss, P.; Schnauder, G.; Sailer, C.; Schmid, V.; Hudemann, J.; Machann, J.; Schick, F.; et al. TGF-b Contributes to Impaired Exercise Response by Suppression of Mitochondrial Key Regulators in Skeletal Muscle. Diabetes 2016, 65, 2846–2861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radom-Aizik, S.; Hayek, S.; Shahar, I.; Rechavi, G.; Kaminski, N.; Ben-Dov, I. Effects of aerobic training on gene expression in skeletal muscle of elderly men. Med. Sci. Sports Exerc. 2005, 37, 1680–1696. [Google Scholar] [CrossRef]
- Poelkens, F.; Lammers, G.; Pardoel, E.M.; Tack, C.J.; Hopman, M.T. Upregulation of skeletal muscle inflammatory genes links inflammation with insulin resistance in women with the metabolic syndrome. Exp. Physiol. 2013, 98, 1485–1494. [Google Scholar] [CrossRef]
- Thalacker-Mercer, A.E.; Fleet, J.C.; Craig, B.A.; Carnell, N.S.; Campbell, W.W. Inadequate protein intake affects skeletal muscle transcript profiles in older humans. Am. J. Clin. Nutr. 2007, 85, 1344–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thalacker-Mercer, A.E.; Fleet, J.C.; Craig, B.A.; Campbell, W.W. The skeletal muscle transcript profile reflects accommodative responses to inadequate protein intake in younger and older males. J. Nutr. Biochem. 2010, 21, 1076–1082. [Google Scholar] [CrossRef]
- Smith, G.I.; Yoshino, J.; Kelly, S.C.; Reeds, D.N.; Okunade, A.; Patterson, B.W.; Klein, S.; Mittendorfer, B. High-Protein Intake during Weight Loss Therapy Eliminates the Weight-Loss-Induced Improvement in Insulin Action in Obese Postmenopausal Women. Cell Rep. 2016, 17, 849–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rowlands, D.S.; Thomson, J.S.; Timmons, B.W.; Raymond, F.; Fuerholz, A.; Mansourian, R.; Zwahlen, M.C.; Metairon, S.; Glover, E.; Stellingwerff, T.; et al. Transcriptome and translational signaling following endurance exercise in trained skeletal muscle: Impact of dietary protein. Physiol. Genom. 2011, 43, 1004–1020. [Google Scholar] [CrossRef] [Green Version]
- Rowlands, D.S.; Nelson, A.R.; Raymond, F.; Metairon, S.; Mansourian, R.; Clarke, J.; Stellingwerff, T.; Phillips, S.M. Protein-leucine ingestion activates a regenerative inflammo-myogenic transcriptome in skeletal muscle following intense endurance exercise. Physiol. Genom. 2016, 48, 21–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knuiman, P.; Hangelbroek, R.; Boekschoten, M.; Hopman, M.; Mensink, M. Impact of protein supplementation during endurance training on changes in skeletal muscle transcriptome. BMC Genom. 2020, 21, 397. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Gil, A.M.; Elizondo-Montemayor, L. The Role of Exercise in the Interplay between Myokines, Hepatokines, Osteokines, Adipokines, and Modulation of Inflammation for Energy Substrate Redistribution and Fat Mass Loss: A Review. Nutrients 2020, 12, 1899. [Google Scholar] [CrossRef]
- Murphy, R.M.; Watt, M.J.; Febbraio, M.A. Metabolic communication during exercise. Nat. Metab. 2020, 2, 805–816. [Google Scholar] [CrossRef]
- Smeets, A.J.; Soenen, S.; Luscombe-Marsh, N.D.; Ueland, O.; Westerterp-Plantenga, M.S. Energy expenditure, satiety, and plasma ghrelin, glucagon-like peptide 1, and peptide tyrosine-tyrosine concentrations following a single high-protein lunch. J. Nutr. 2008, 138, 698–702. [Google Scholar] [CrossRef] [Green Version]
- Gaffney, K.; Lucero, A.; Macartney-Coxson, D.; Clapham, J.; Whitfield, P.; Palmer, B.R.; Wakefield, S.; Faulkner, J.; Stoner, L.; Rowlands, D.S. Effects of whey protein on skeletal muscle microvascular and mitochondrial plasticity following 10 weeks of exercise training in men with type 2 diabetes. Appl. Physiol. Nutr. Metab. 2021, 46, 915–924. [Google Scholar] [CrossRef] [PubMed]
- Peake, J.M.; Neubauer, O.; Della Gatta, P.A.; Nosaka, K. Muscle damage and inflammation during recovery from exercise. J. Appl. Physiol. 2017, 122, 559–570. [Google Scholar] [CrossRef]
- Louis, E.; Raue, U.; Yang, Y.; Jemiolo, B.; Trappe, S. Time course of proteolytic, cytokine, and myostatin gene expression after acute exercise in human skeletal muscle. J. Appl. Physiol. 2007, 103, 1744–1751. [Google Scholar] [CrossRef] [Green Version]
- Draganidis, D.; Karagounis, L.G.; Athanailidis, I.; Chatzinikolaou, A.; Jamurtas, A.Z.; Fatouros, I.G. Inflammaging and Skeletal Muscle: Can Protein Intake Make a Difference? J. Nutr. 2016, 146, 1940–1952. [Google Scholar] [CrossRef] [Green Version]
- Koliaraki, V.; Prados, A.; Armaka, M.; Kollias, G. The mesenchymal context in inflammation, immunity and cancer. Nat. Immunol. 2020, 21, 974–982. [Google Scholar] [CrossRef]
- Rowlands, D.S.; Nelson, A.R.; Phillips, S.M.; Faulkner, J.A.; Clarke, J.; Burd, N.A.; Moore, D.; Stellingwerff, T. Protein-leucine fed dose effects on muscle protein synthesis after endurance exercise. Med. Sci. Sports Exerc. 2015, 47, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Burd, N.A.; Tang, J.E.; Moore, D.R.; Phillips, S.M. Exercise training and protein metabolism: Influences of contraction, protein intake, and sex-based differences. J. Appl. Physiol. 2009, 106, 1692–1701. [Google Scholar] [CrossRef]
- Kilroe, S.P.; Fulford, J.; Jackman, S.; Holwerda, A.; Gijsen, A.; van Loon, L.; Wall, B.T. Dietary protein intake does not modulate daily myofibrillar protein synthesis rates or loss of muscle mass and function during short-term immobilization in young men: A randomized controlled trial. Am. J. Clin. Nutr. 2021, 113, 548–561. [Google Scholar] [CrossRef] [PubMed]
- Trommelen, J.; van Lieshout, G.A.A.; Pabla, P.; Nyakayiru, J.; Hendriks, F.K.; Senden, J.M.; Goessens, J.P.B.; van Kranenburg, J.M.X.; Gijsen, A.P.; Verdijk, L.B.; et al. Pre-sleep Protein Ingestion Increases Mitochondrial Protein Synthesis Rates during Overnight Recovery from Endurance Exercise: A Randomized Controlled Trial. Sports Med. 2023, 53, 1445–1455. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, J.; Tomita, T.; Arimitsu, T.; Fujita, S. Evenly Distributed Protein Intake over 3 Meals Augments Resistance Exercise-Induced Muscle Hypertrophy in Healthy Young Men. J. Nutr. 2020, 150, 1845–1851. [Google Scholar] [CrossRef] [Green Version]
- Dirks, M.L.; Wall, B.T.; Nilwik, R.; Weerts, D.H.; Verdijk, L.B.; van Loon, L.J. Skeletal muscle disuse atrophy is not attenuated by dietary protein supplementation in healthy older men. J. Nutr. 2014, 144, 1196–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franzke, B.; Neubauer, O.; Cameron-Smith, D.; Wagner, K.H. Dietary Protein, Muscle and Physical Function in the Very Old. Nutrients 2018, 10, 935. [Google Scholar] [CrossRef] [Green Version]
- Lim, M.T.; Pan, B.J.; Toh, D.W.K.; Sutanto, C.N.; Kim, J.E. Animal Protein versus Plant Protein in Supporting Lean Mass and Muscle Strength: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Nutrients 2021, 13, 661. [Google Scholar] [CrossRef]
- Chen, G.; Zou, Y.; Zhang, X.; Xu, L.; Hu, Q.; Li, T.; Yao, C.; Yu, S.; Wang, X.; Wang, C. Beta-Actin protein expression differs in the submandibular glands of male and female mice. Cell Biol. Int. 2016, 40, 779–786. [Google Scholar] [CrossRef]
- Yang, S.; Zhao, Y.; Chen, X.; Lu, X.; Chen, Y.; Zhao, X.; Zhu, L.; Fang, Z.; Zhao, H.; Yao, Y.; et al. The ACTB Variants and Alcohol Drinking Confer Joint Effect to Ischemic Stroke in Chinese Han Population. J. Atheroscler. Thromb. 2020, 27, 226–244. [Google Scholar] [CrossRef]
- Ha, A.T.; Cho, J.Y.; Kim, D. MLK3 Regulates Inflammatory Response via Activation of AP-1 Pathway in HEK293 and RAW264.7 Cells. Int. J. Mol. Sci. 2022, 23, 10874. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Hou, P.; Pan, W.; He, W.; He, D.C.; Wang, H.; He, H. DDIT3 Targets Innate Immunity via the DDIT3-OTUD1-MAVS Pathway to Promote Bovine Viral Diarrhea Virus Replication. J. Virol. 2021, 95, e02351-20. [Google Scholar] [CrossRef] [PubMed]
- Granata, C.; Jamnick, N.A.; Bishop, D.J. Training-Induced Changes in Mitochondrial Content and Respiratory Function in Human Skeletal Muscle. Sports Med. 2018, 48, 1809–1828. [Google Scholar] [CrossRef]
- Jones, A.M.; Carter, H. The effect of endurance training on parameters of aerobic fitness. Sports Med. 2000, 29, 373–386. [Google Scholar] [CrossRef]
- Rodriguez, N.R.; Vislocky, L.M.; Gaine, P.C. Dietary protein, endurance exercise, and human skeletal-muscle protein turnover. Curr. Opin. Clin. Nutr. Metab. Care 2007, 10, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Bozaykut, P.; Ozer, N.K.; Karademir, B. Regulation of protein turnover by heat shock proteins. Free Radic. Biol. Med. 2014, 77, 195–209. [Google Scholar] [CrossRef]
- Cunha, T.F.; Bacurau, A.V.; Moreira, J.B.; Paixao, N.A.; Campos, J.C.; Ferreira, J.C.; Leal, M.L.; Negrao, C.E.; Moriscot, A.S.; Wisloff, U.; et al. Exercise training prevents oxidative stress and ubiquitin-proteasome system overactivity and reverse skeletal muscle atrophy in heart failure. PLoS ONE 2012, 7, e41701. [Google Scholar] [CrossRef]
- Huertas, J.R.; Rodríguez Lara, A.; González Acevedo, O.; Mesa-García, M.D. Milk and dairy products as vehicle for calcium and vitamin D: Role of calcium enriched milks. Nutr. Hosp. 2019, 36, 962–973. [Google Scholar]
- Dang, C.V.; O’Donnell, K.A.; Zeller, K.I.; Nguyen, T.; Osthus, R.C.; Li, F. The c-Myc target gene network. Semin. Cancer Biol. 2006, 16, 253–264. [Google Scholar] [CrossRef]
- McLean, C.S.; Mielke, C.; Cordova, J.M.; Langlais, P.R.; Bowen, B.; Miranda, D.; Coletta, D.K.; Mandarino, L.J. Gene and MicroRNA Expression Responses to Exercise; Relationship with Insulin Sensitivity. PLoS ONE 2015, 10, e0127089. [Google Scholar] [CrossRef]
| Datasets | Groups | Intervention Details | Age (Year) | Sex (n) | Muscle Biopsy | Platform |
|---|---|---|---|---|---|---|
| Acute aerobic exercise (n = 28, included 28 males) | ||||||
| GSE43856 Neubauer O 2013, Austria [50] | Pre- and post-exercise | 1 h of cycling followed by 1 h of running. | 25 ± 4.1 | M (8) | NR (3 h, 48 h, 96 h) | GPL10558; Illumina Human HT-12 V4.0 expression beadchip |
| GSE59088 Kristian Vissing 2014, Denmark [51] | Endurance group and control pre-, 2.5 h, 5 h | 2 h of bicycle exercise at 60% post-training VO2 peak. | 23.4 ± 0.8 | M (6) | Vastus lateralis biopsy | GPL6244; [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version] |
| GSE126296 Rundqvist HC 2019, Sweden [52] | Pre- and post-exercise | Three all-out cycle sprints interspersed by 1/3-h recovery. | 26 ± 4 | M (7) | Vastus lateralis biopsy (7/3 h after the third sprint) | GPL6244; [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version] |
| GSE59363 Hansen JS 2015, Germany [53] | Pre- and post-exercise | A 1-h bicycle ergometer exercise | 57.3 ± 1.7 | M (7) | NR (0 h, 1 h, 3 h) | GPL6244; [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version] |
| Training aerobic exercise (n = 45, included 26 males and 19 females) | ||||||
| GSE111551 Michelle Silva, 2018, Brazil | Pre- and post-training | 18 weeks of running (3 times/week; 1 h every time). | Two groups group1, 26 ± 4; group2, 27 ± 3 | M (13) | NR | GPL17586; [HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
| GSE72462 Böhm A, 2016, Germany [54] | Pre- and post-training | The training program lasted 8 weeks and consisted of three supervised exercise sessions per week. Each training session consisted of 0.5 h of bicycle ergometer exercise and 0.5 h walking on a treadmill. | 46.4 ± 11 | M (7) F (13) | Muscle biopsies were taken from the lateral portion of the vastus lateralis of the quadriceps femoris after local anesthesia | GPL17586; [HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version] |
| GSE1786 Radom-Aizik S, 2005, USA [55] | Pre- and post-training | 6 healthy sedentary men were trained on a cycle ergometer 3/wk for 12 wk at 80% of the predetermined maximal heart rate. | 68 ± 2.7 | M (6) | Vastus Lateralis muscle | GPL96; [HG-U133A] Affymetrix Human Genome U133A Array |
| GSE43760 Poelkens F, 2013, The Netherlands [56] | Pre- and post-training | 6 months guided endurance exercise training program | 49 ± 11 | F (6) | Vastus lateralis muscle | GPL6244; [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version] |
| Protein intake (n = 48, included 27 males and 21 females) | ||||||
| GSE8441 Thalacker-Mercer AE, 2007, USA [57] | Adequate protein vs. inadequate protein group | 11 subjects consumed 1.2 g/kg/d protein for a week first, then consumed 0.5 g/kg/d protein for the second week. | 55–80 (67 ± 7) | M (5) F (6) | Vastus lateralis biopsy | GPL96; [HG-U133A] Affymetrix Human Genome U133A Array |
| GSE9419 Thalacker-Mercer AE, 2008, USA [58] | Three groups with protein intakes of 0.50 g/kg/d, 0.75 g/kg/d, and 1.00 g/kg/d. | 22 healthy young and old males treated with protein intakes of 0.50 g/kg/d, 0.75 g/kg/d, and 1.00 g/kg/d. | Young 21–43 Elderly 63–79 | M (22) | Vastus lateralis biopsy | GPL570; [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array |
| GSE73525 Smith GI, 2016, Japan [59] | High-protein group vs. low-protein group | 9 subjects in a high protein (1.2 g/kg/d) group and 6 subjects in a normal protein (0.8 g/kg/d) group | 50–65 (58 ± 1) | F (15) | NR | GPL6244; [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version] |
| Acute aerobic exercise + protein intake (n = 20, included 20 males) | ||||||
| GSE27285 Rowlands DS, 2011, Switzerland [60] | Acute aerobic exercise with or without protein intake (iso-calorie) | Group1: 1 h intense cycling + 0.2/1.2/0.4 g/kg (fat/carbohydrate/protein). Group2: 1 h intense cycling + 0.2/1.6 g/kg (fat/carbohydrate). | 32.8 ± 6.4 | M (8) | Quadriceps at rest, at 3 h and at 48 h after exercise training | GPL6104; Illumina humanRef-8 v2.0 expression beadchip |
| GSE44818 Rowlands DS, 2015, Switzerland [61] | High-intensity cycling without protein intake; high-intensity cycling + 70 g protein; high-intensity cycling + 23 g protein | 5/3 h of high-intensity cycling with 70/15/180/30 g (protein/leucine/carbohydrate/fat); or with 23/5/180/30 g; or with 0/0/274/30 g | Mean age 30 years | M (12) | Vastus lateralis biopsy | GPL6947; Illumina HumanHT-12 V3.0 expression beadchip |
| Training aerobic exercise + protein intake (n = 40, included 40 males) | ||||||
| GSE147494 Knuiman P, 2020, UK [62] | Protein-treated group vs. control group | 12 weeks endurance training program with 30 g protein drink; or with 30 g carbohydrate drink (isocaloric) | 18–30 Protein group 21.5; Controls group 22.5 | M (40) | Vastus lateralis biopsy | GPL28236; [HuGene-2_1-st] Affymetrix Human Gene 2.1 ST Array |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, X.; Li, L.; Xia, Z.; Zou, L.; Kwok, T.; Su, Y. Transcriptomic Analysis of Human Skeletal Muscle in Response to Aerobic Exercise and Protein Intake. Nutrients 2023, 15, 3485. https://doi.org/10.3390/nu15153485
Zeng X, Li L, Xia Z, Zou L, Kwok T, Su Y. Transcriptomic Analysis of Human Skeletal Muscle in Response to Aerobic Exercise and Protein Intake. Nutrients. 2023; 15(15):3485. https://doi.org/10.3390/nu15153485
Chicago/Turabian StyleZeng, Xueqing, Linghong Li, Zhilin Xia, Lianhong Zou, Timothy Kwok, and Yi Su. 2023. "Transcriptomic Analysis of Human Skeletal Muscle in Response to Aerobic Exercise and Protein Intake" Nutrients 15, no. 15: 3485. https://doi.org/10.3390/nu15153485





