Maternal Diet Determines Milk Microbiome Composition and Offspring Gut Colonization in Wistar Rats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. Sample Collection
2.3. Analytical Determinations
2.4. DNA Extraction
2.4.1. Milk
2.4.2. Feces
2.5. DNA Quality, Quantification, and Sequencing
2.6. Metataxonomic Analysis of Bacterial Microbiota
2.7. Immunofluorescence Staining
2.8. Inner Mucus Layer Thickness Analysis
2.9. RNA Extraction and Quantitative RT-PCR
2.10. Statical and Bioinformatic Analyses
3. Results
3.1. Effect of Nutritional Restriction on the Biochemical Profile of Offspring Rats during Lactation
3.2. Effect of Maternal Diet on the Milk and Gut Microbiome of Offspring throughout Lactation
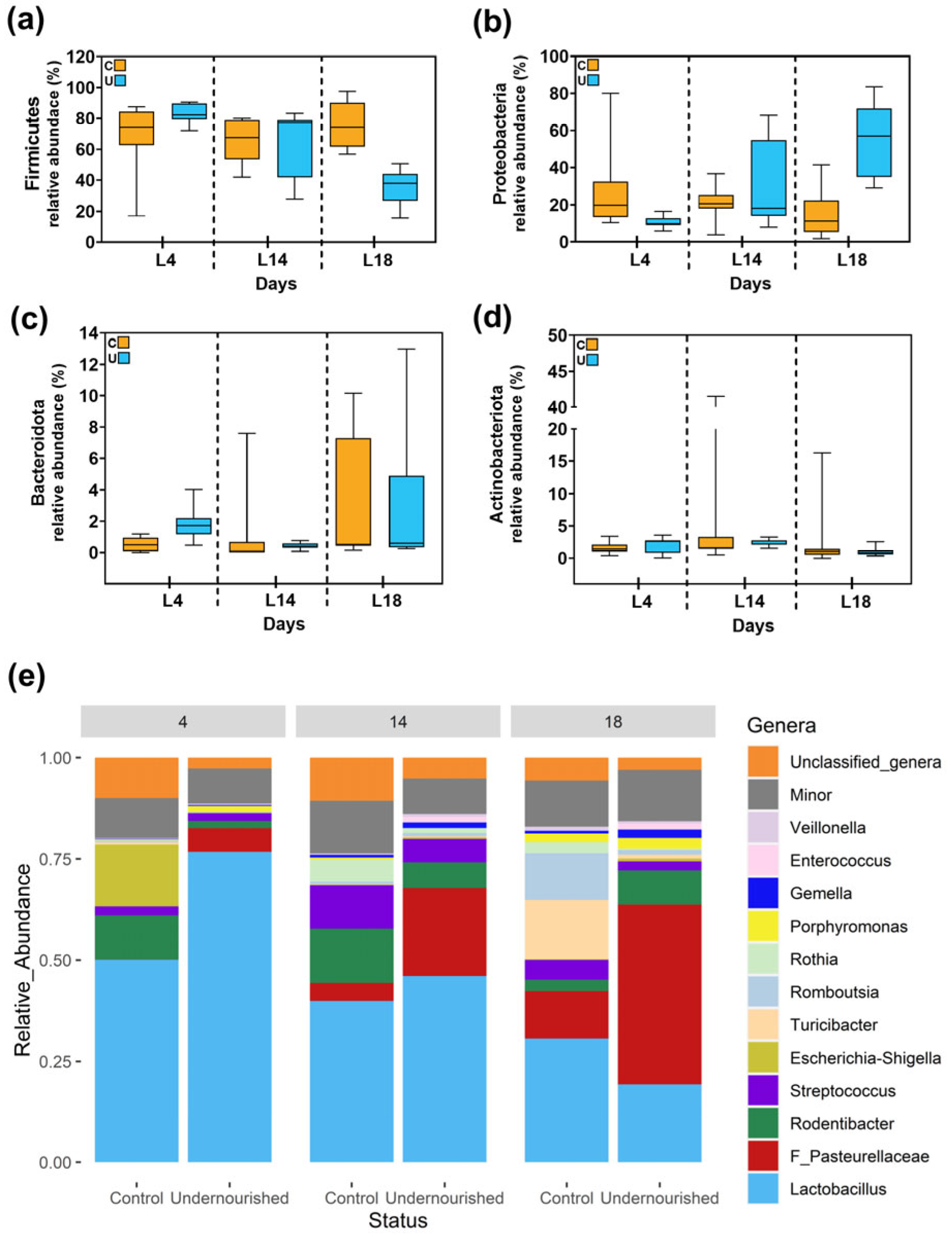
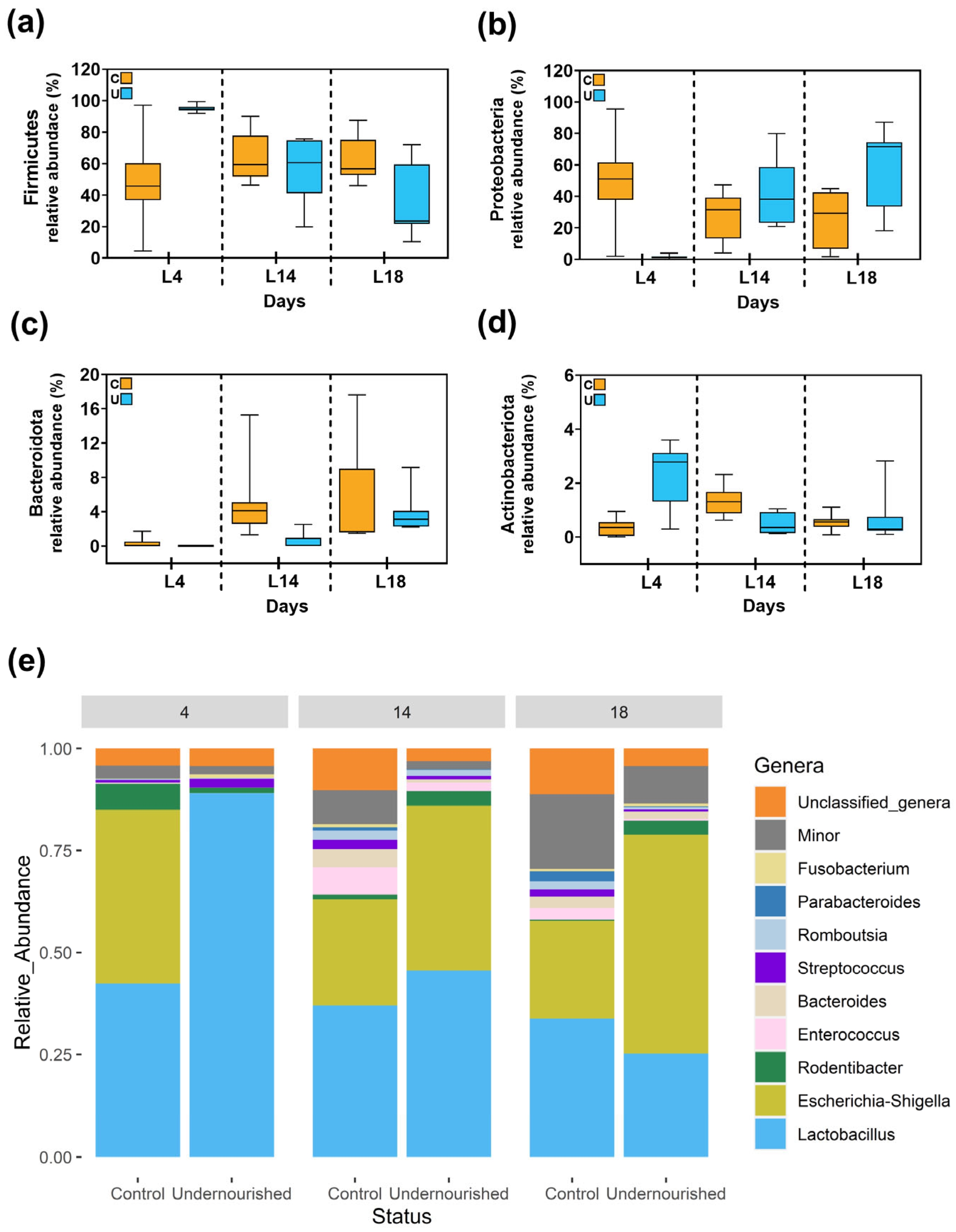
3.2.1. Metataxonomic Analysis of the Milk Samples
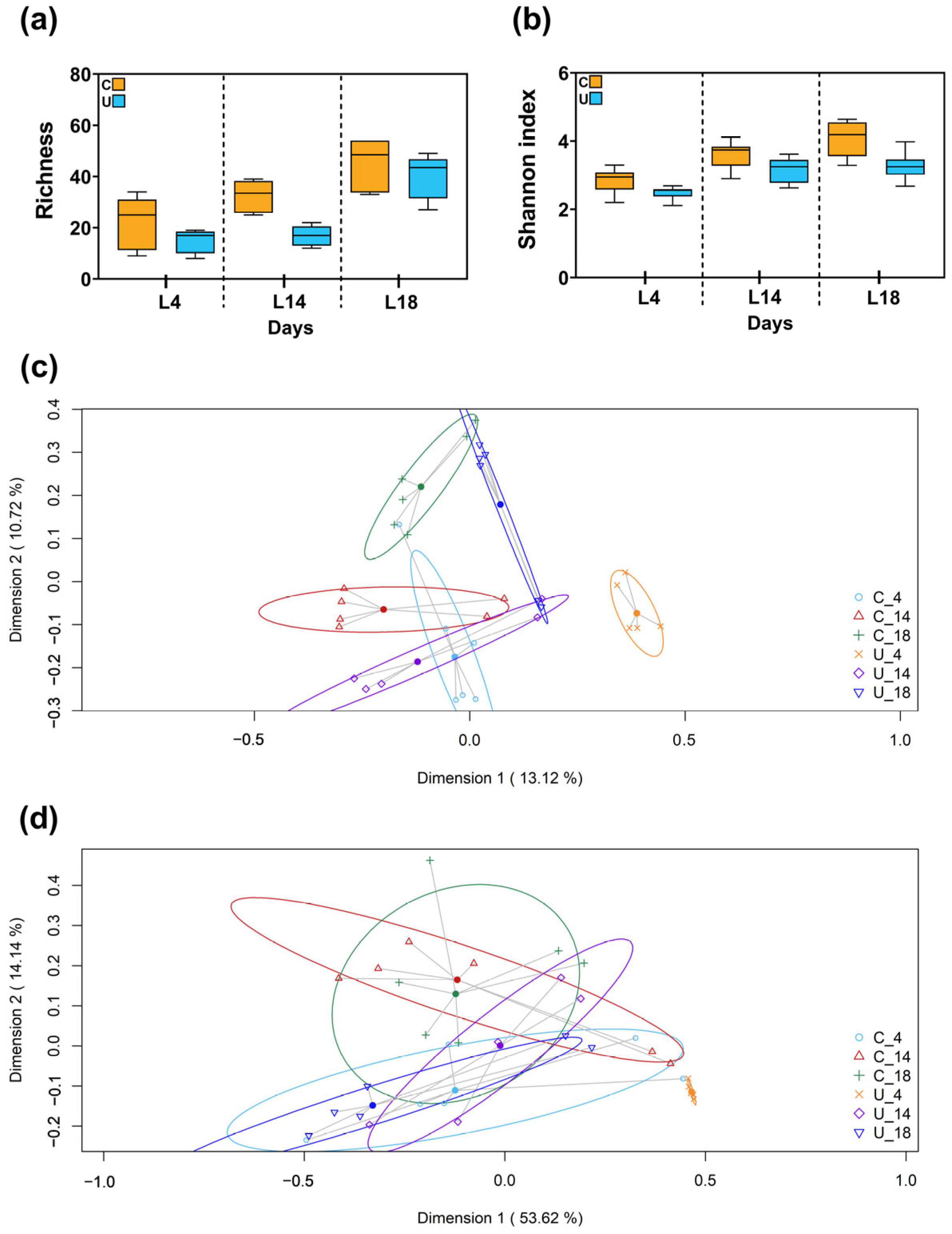
3.2.2. Metataxonomic Analysis of the Fecal Samples
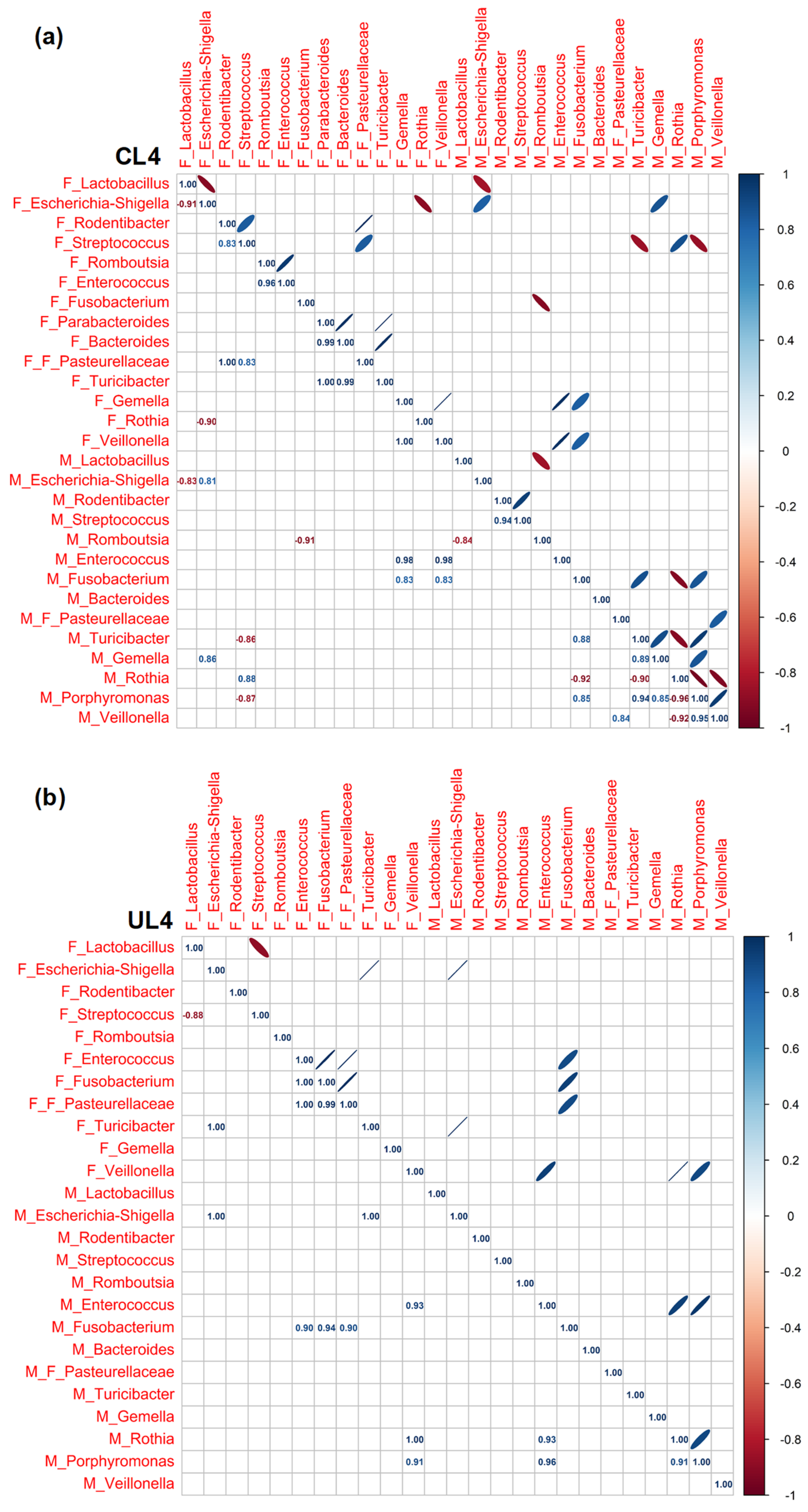
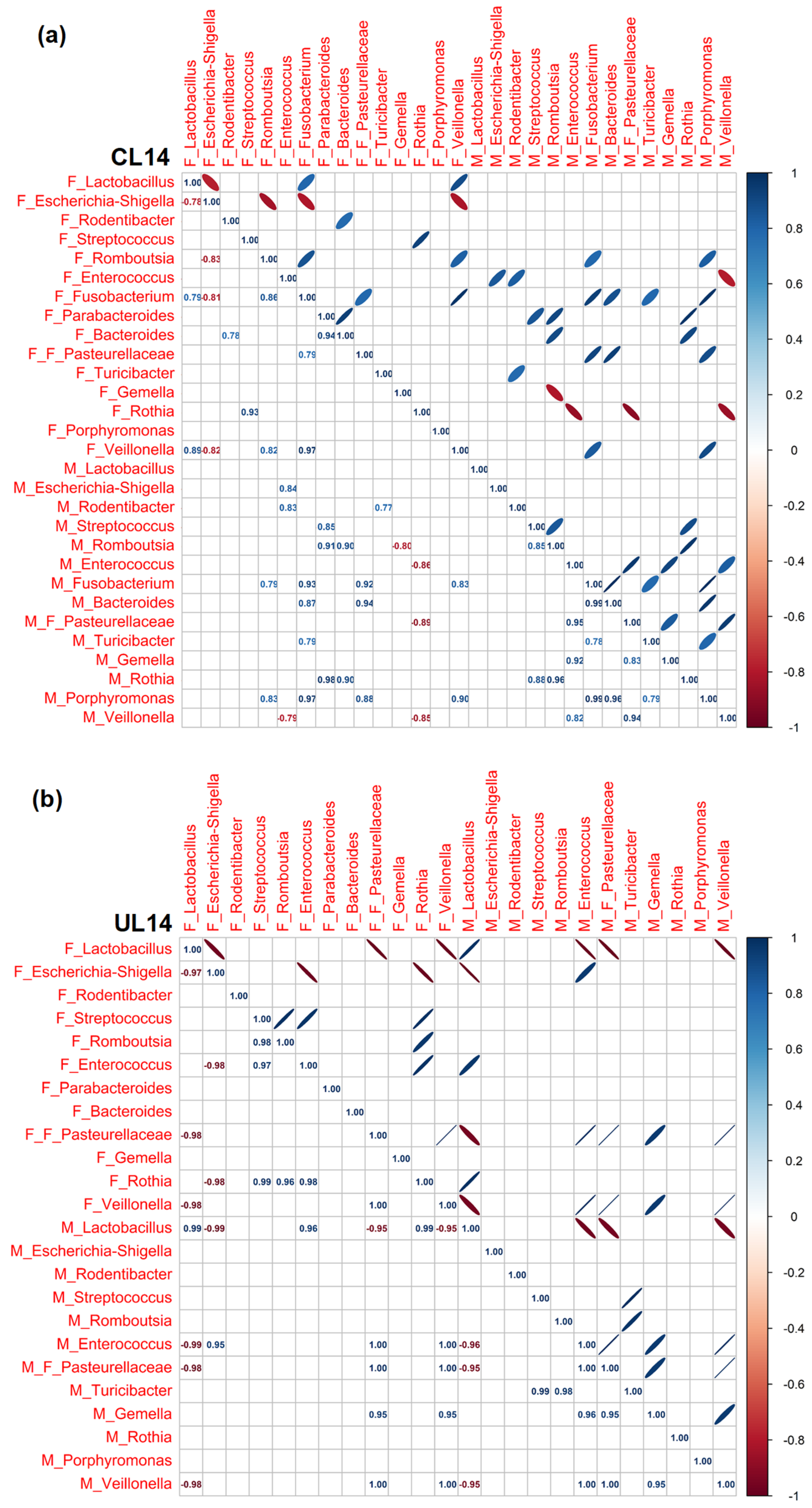
3.3. Pearson Rank Correlation Analysis between Predominant Genera in Milk and Offspring Feces
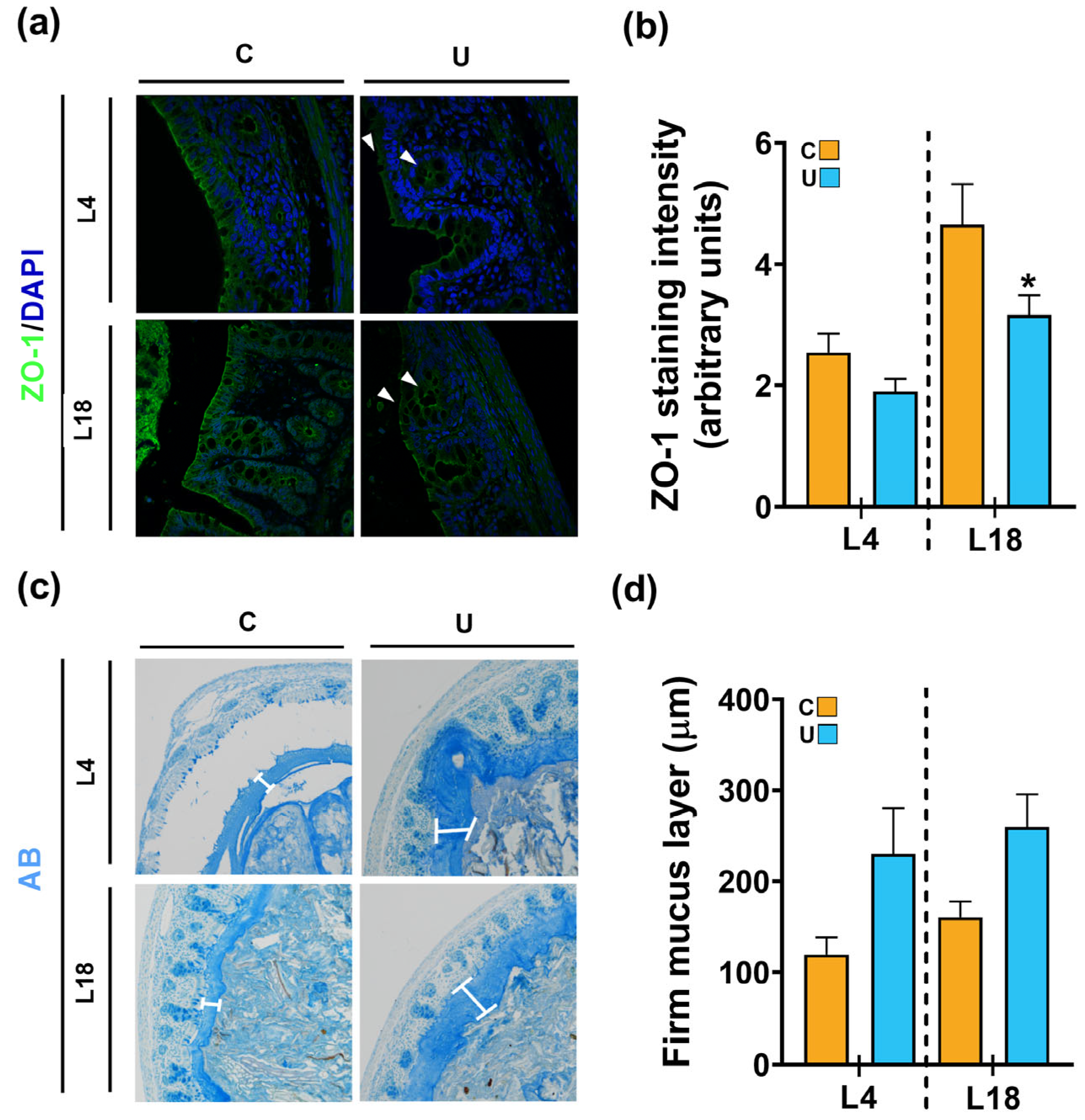
3.4. Nutritional Restriction during Lactation Disrupts the Colonic Barrier Integrity of the Offspring
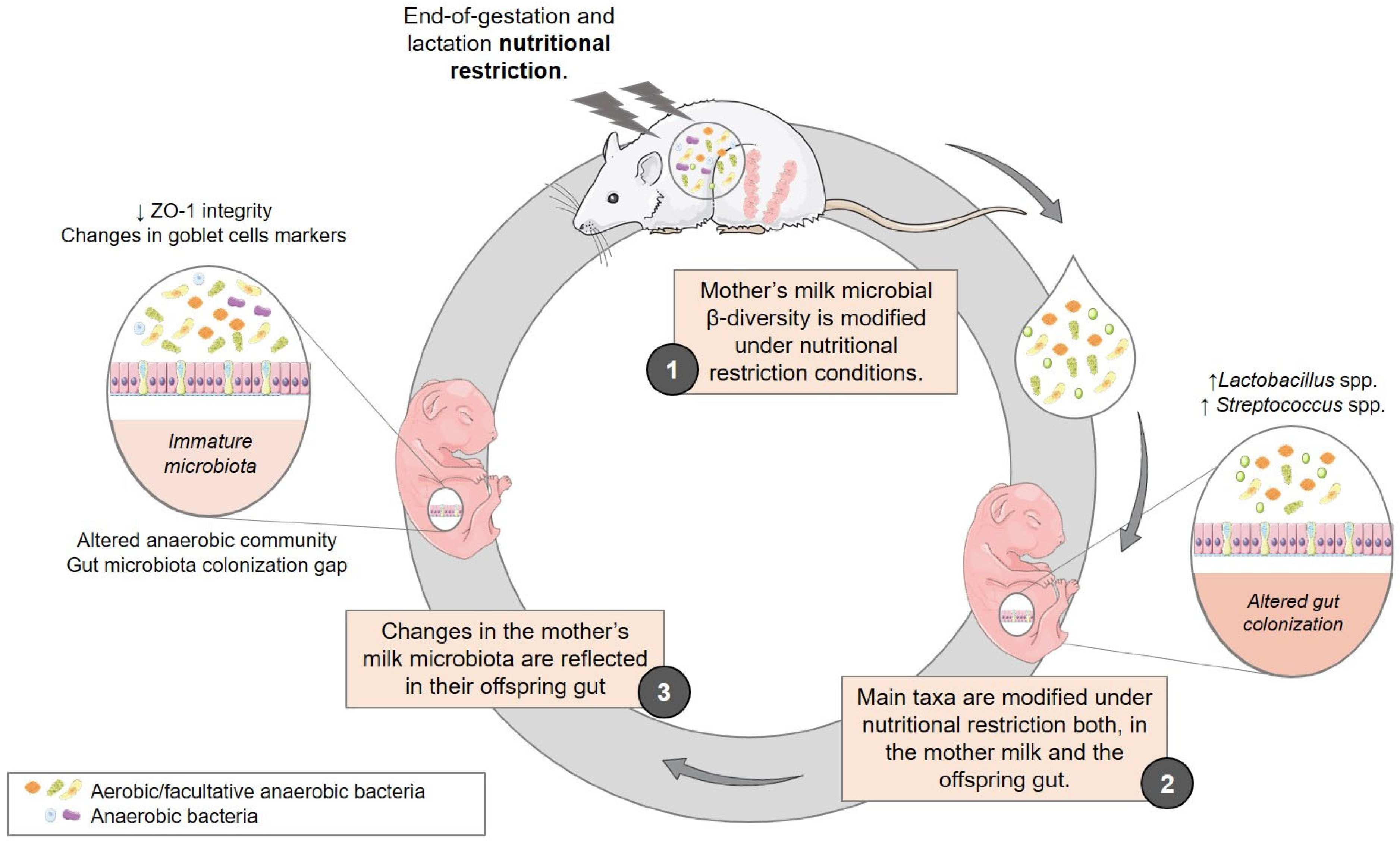
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pena-León, V.; Folgueira, C.; Barja-Fernández, S.; Pérez-Lois, R.; Da Silva Lima, N.; Martin, M.; Heras, V.; Martínez-Martínez, S.; Valero, P.; Iglesias, C.; et al. Prolonged breastfeeding protects from obesity by hypothalamic action of hepatic FGF21. Nat. Metab. 2022, 4, 901–917. [Google Scholar] [CrossRef] [PubMed]
- Picó, C.; Reis, F.; Egas, C.; Mathias, P.; Matafome, P. Lactation as a programming window for metabolic syndrome. Eur. J. Clin. Investig. 2021, 51, e13482. [Google Scholar] [CrossRef]
- Hsu, C.N.; Hou, C.Y.; Hsu, W.H.; Tain, Y.L. Early-Life Origins of Metabolic Syndrome: Mechanisms and Preventive Aspects. Int. J. Mol. Sci. 2021, 22, 11872. [Google Scholar] [CrossRef]
- De Toro-Martín, J.; Fernández-Millán, E.; Lizárraga-Mollinedo, E.; López-Oliva, E.; Serradas, P.; Escrivá, F.; Álvarez, C. Predominant role of GIP in the development of a metabolic syndrome-like phenotype in female Wistar rats submitted to forced catch-up growth. Endocrinology 2014, 155, 3769–3780. [Google Scholar] [CrossRef]
- Tsuduki, T.; Kitano, Y.; Honma, T.; Kijima, R.; Ikeda, I. High dietary fat intake during lactation promotes development of diet-induced obesity in male offspring of mice. J. Nutr. Sci. Vitaminol. 2013, 59, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Barker, D.J. The origins of the developmental origins theory. J. Intern. Med. 2007, 261, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Grigor, M.R.; Allan, J.E.; Carrington, J.M.; Carne, A.; Geursen, A.; Young, D.; Thompson, M.P.; Haynes, E.B.; Coleman, R.A. Effect of Dietary Protein and Food Restriction on Milk Production and Composition, Maternal Tissues and Enzymes in Lactating Rats. J. Nutr. 1987, 117, 1247–1258. [Google Scholar] [CrossRef]
- Bautista, C.J.; Bautista, R.J.; Montaño, S.; Reyes-Castro, L.A.; Rodriguez-Peña, O.N.; Ibáñez, C.A.; Nathanielsz, P.W.; Zambrano, E. Effects of maternal protein restriction during pregnancy and lactation on milk composition and offspring development. Br. J. Nutr. 2019, 122, 141–151. [Google Scholar] [CrossRef]
- Wattez, J.S.; Delmont, A.; Bouvet, M.; Beseme, O.; Goers, S.; Delahaye, F.; Laborie, C.; Lesage, J.; Foligné, B.; Breton, C.; et al. Maternal perinatal undernutrition modifies lactose and serotranferrin in milk: Relevance to the programming of metabolic diseases? Am. J. Physiol. Endocrinol. Metab. 2015, 308, E393–E401. [Google Scholar] [CrossRef]
- Fiorotto, M.L.; Burrin, D.G.; Perez, M.; Reeds, P.J. Intake and use of milk nutrients by rat pups suckled in small, medium, or large litters. Am. J. Physiol. 1991, 260, R1104–R1113. [Google Scholar] [CrossRef]
- Palou, M.; Torrens, J.M.; Castillo, P.; Sánchez, J.; Palou, A.; Picó, C. Metabolomic approach in milk from calorie-restricted rats during lactation: A potential link to the programming of a healthy phenotype in offspring. Eur. J. Nutr. 2020, 59, 1191–1204. [Google Scholar] [CrossRef]
- Lizárraga-Mollinedo, E.; Fernández-Millán, E.; de Toro Martín, J.; Martínez-Honduvilla, C.; Escrivá, F.; Álvarez, C. Early undernutrition induces glucagon resistance and insulin hypersensitivity in the liver of suckling rats. Am. J. Physiol. Metab. 2012, 302, E1070–E1077. [Google Scholar] [CrossRef]
- de Toro-Martín, J.; Fernández-Marcelo, T.; González-Rodríguez, Á.; Escrivá, F.; Valverde, Á.M.; Álvarez, C.; Fernández-Millán, E. Defective liver glycogen autophagy related to hyperinsulinemia in intrauterine growth-restricted newborn Wistar rats. Sci. Rep. 2020, 10, 17651. [Google Scholar] [CrossRef]
- Lizárraga-Mollinedo, E.; Fernández-Millán, E.; García-San Frutos, M.; de Toro-Martín, J.; Fernández-Agulló, T.; Ros, M.; Álvarez, C.; Escrivá, F. Early and long-term undernutrition in female rats exacerbates the metabolic risk associated with nutritional rehabilitation. J. Biol. Chem. 2015, 290, 19353–19366. [Google Scholar] [CrossRef] [PubMed]
- López, M.; Seoane, L.M.; Tovar, S.; García, M.C.; Nogueiras, R.; Diéguez, C.; Señarís, R.M. A possible role of neuropeptide Y, agouti-related protein and leptin receptor isoforms in hypothalamic programming by perinatal feeding in the rat. Diabetologia 2005, 48, 140–148. [Google Scholar] [CrossRef]
- Martínez-Oca, P.; Robles-Vera, I.; Sánchez-Roncero, A.; Escrivá, F.; Pérez-Vizcaíno, F.; Duarte, J.; Álvarez, C.; Fernández-Millán, E. Gut DYSBIOSIS and altered barrier function precedes the appearance of metabolic syndrome in a rat model of nutrient-induced catch-up growth. J. Nutr. Biochem. 2020, 81, 08383. [Google Scholar] [CrossRef]
- Fança-Berthon, P.; Michel, C.; Pagniez, A.; Rival, M.; Van Seuningen, I.; Darmaun, D.; Hoebler, C. Intrauterine growth restriction alters postnatal colonic barrier maturation in rats. Pediatr. Res. 2009, 66, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Dogra, S.K.; Kwong Chung, C.; Wang, D.; Sakwinska, O.; Colombo Mottaz, S.; Sprenger, N. Nurturing the Early Life Gut Microbiome and Immune Maturation for Long Term Health. Microorganisms 2021, 9, 2110. [Google Scholar] [CrossRef]
- Xiao, L.; Zhao, F. Microbial transmission, colonisation and succession: From pregnancy to infancy. Gut 2023, 72, 772–786. [Google Scholar] [CrossRef] [PubMed]
- Lackey, K.A.; Williams, J.E.; Meehan, C.L.; Zachek, J.A.; Benda, E.D.; Price, W.J.; Foster, J.A.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W.; et al. What’s normal? microbiomes in human milk and infant feces are related to each other but vary geographically: The INSPIRE study. Front. Nutr. 2019, 6, 45. [Google Scholar] [CrossRef]
- Togo, A.; Dufour, J.-C.; Lagier, J.-C.; Dubourg, G.; Raoult, D.; Million, M. Repertoire of human breast and milk microbiota: A systematic review. Future Microbiol. 2019, 14, 623–641. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Cabrera-Rubio, R.; Mira-Pascual, L.; Mira, A.; Collado, M.C. Impact of mode of delivery on the milk microbiota composition of healthy women. J. Dev. Orig. Health Dis. 2015, 7, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Rubio, R.; Collado, M.C.; Laitinen, K.; Salminen, S.; Isolauri, E.; Mira, A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am. J. Clin. Nutr. 2012, 96, 544–551. [Google Scholar] [CrossRef]
- Collado, M.C.; Laitinen, K.; Salminen, S.; Isolauri, E. Maternal weight and excessive weight gain during pregnancy modify the immunomodulatory potential of breast milk. Pediatr. Res. 2012, 72, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Kalliomäki, M.; Collado, M.C.; Salminen, S.; Isolauri, E. Early differences in fecal microbiota composition in children may predict overweight. Am. J. Clin. Nutr. 2008, 87, 534–538. [Google Scholar] [CrossRef]
- Warren, M.F.; Hallowell, H.A.; Higgins, K.V.; Liles, M.R.; Hood, W.R. Maternal Dietary Protein Intake Influences Milk and Offspring Gut Microbial Diversity in a Rat (Rattus norvegicus) Model. Nutrients 2019, 11, 2257. [Google Scholar] [CrossRef]
- Cabinian, A.; Sinsimer, D.; Tang, M.; Zumba, O.; Mehta, H.; Toma, A.; Sant’Angelo, D.; Laouar, Y.; Laouar, A. Transfer of Maternal Immune Cells by Breastfeeding: Maternal Cytotoxic T Lymphocytes Present in Breast Milk Localize in the Peyer’s Patches of the Nursed Infant. PLoS ONE 2016, 11, e0156762. [Google Scholar] [CrossRef]
- Rodríguez-Cruz, M.; Alba, C.; Aparicio, M.; Checa, M.Á.; Fernández, L.; Rodríguez, J.M. Effect of Sample Collection (Manual Expression vs. Pumping) and Skimming on the Microbial Profile of Human Milk Using Culture Techniques and Metataxonomic Analysis. Microorganisms 2020, 8, 1278. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Odenwald, M.A.; Turner, J.R. The intestinal epithelial barrier: A therapeutic target? Nat. Rev. Gastroenterol. Hepatol. 2012, 14, 9–21. [Google Scholar] [CrossRef]
- Crosnier, C.; Stamataki, D.; Lewis, J. Organizing cell renewal in the intestine: Stem cells, signals and combinatorial control. Nat. Rev. Genet. 2006, 7, 349–359. [Google Scholar] [CrossRef]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef]
- Fernández, L.; Pannaraj, P.S.; Rautava, S.; Rodríguez, J.M. The Microbiota of the Human Mammary Ecosystem. Front. Cell. Infect. Microbiol. 2020, 10, 586667. [Google Scholar] [CrossRef]
- Pannaraj, P.S.; Li, F.; Cerini, C.; Bender, J.M.; Yang, S.; Rollie, A.; Adisetiyo, H.; Zabih, S.; Lincez, P.J.; Bittinger, K.; et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017, 171, 647–654. [Google Scholar] [CrossRef]
- Murphy, K.; Curley, D.; O’Callaghan, T.F.; O’Shea, C.A.; Dempsey, E.M.; O’Toole, P.W.; Ross, R.P.; Ryan, C.A.; Stanton, C. The composition of human milk and infant fecal microbiota over the first three months of life: A pilot study. Sci. Rep. 2017, 7, 40597. [Google Scholar] [CrossRef] [PubMed]
- Yelverton, C.A.; Killeen, S.L.; Feehily, C.; Moore, R.L.; Callaghan, S.L.; Geraghty, A.A.; Byrne, D.F.; Walsh, C.J.; Lawton, E.M.; Murphy, E.F.; et al. Maternal breastfeeding is associated with offspring microbiome diversity; a secondary analysis of the MicrobeMom randomized control trial. Front. Microbiol. 2023, 14, 1154114. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Huq, S.; Yatsunenko, T.; Haque, R.; Mahfuz, M.; Alam, M.A.; Benezra, A.; Destefano, J.; Meier, M.F.; Muegge, B.D.; et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014, 510, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.I.; Yatsunenko, T.; Manary, M.J.; Trehan, I.; Mkakosya, R.; Cheng, J.; Kau, A.L.; Rich, S.S.; Concannon, P.; Mychaleckyj, J.C.; et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 2013, 339, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Olivares, M.; Marín, M.; Xaus, J.; Fernández, L.; Rodríguez, J. Characterization of a reuterin-producing Lactobacillus coryniformis strain isolated from a goat’s milk cheese. Int. J. Food Microbiol. 2005, 104, 267–277. [Google Scholar] [CrossRef]
- Martino, C.; Dilmore, A.H.; Burcham, Z.M.; Metcalf, J.L.; Jeste, D.; Knight, R. Microbiota succession throughout life from the cradle to the grave. Nat. Rev. Microbiol. 2022, 20, 707–720. [Google Scholar] [CrossRef] [PubMed]
- Kozyrskyj, A.L.; Kalu, R.; Koleva, P.T.; Bridgman, S.L. Fetal programming of overweight through the microbiome: Boys are disproportionately affected. J. Dev. Orig. Health Dis. 2016, 7, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Jašarević, E.; Bale, T.L. Prenatal and postnatal contributions of the maternal microbiome on offspring programming. Front. Neuroendocrinol. 2019, 55, 100797. [Google Scholar] [CrossRef] [PubMed]
- Milano, W.; Ambrosio, P.; Carizzone, F.; De Biasio, V.; Foia, M.G.; Saetta, B.; Milano, M.F.; Capasso, A. Menstrual Disorders Related to Eating Disorders. Endocr. Metab. Immune Disord. Drug Targets 2022, 22, 471–480. [Google Scholar] [CrossRef]
- Ribet, D.; Cossart, P. How Bacterial Pathogens Colonize Their Hosts and Invade Deeper Tissues. Microbes Infect. 2015, 17, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Song, H.; Zhang, X.; Song, G.; Wang, Y.; Ding, X.; Duan, X.; Li, L.; Sun, T.; Kan, Q. Metformin attenuated sepsis-related liver injury by modulating gut microbiota. Emerg. Microbes Infect. 2022, 11, 815–828. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, C.; Li, N.; Ding, L.; Chen, H.; Wan, J.; Yang, X.; Xia, L.; He, W.; Xiong, H.; et al. The interplay between the gut microbiota and NLRP3 activation affects the severity of acute pancreatitis in mice. Gut Microbes 2020, 11, 1774–1789. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, A.; Cattane, N.; Galluzzi, S.; Provasi, S.; Lopizzo, N.; Festari, C.; Ferrari, C.; Guerra, U.P.; Paghera, B.; Muscio, C.; et al. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol. Aging 2017, 49, 60–68. [Google Scholar] [CrossRef]
- Moossavi, M.; Sepehri, S.; Robertson, B.; Bode, L.; Goruk, S.; Field, C.J.; Lix, L.M.; de Souza, R.J.; Becker, A.B.; Mandhane, P.J.; et al. Composition and Variation of the Human Milk Microbiota Are Influenced by Maternal and Early-Life Factors. Cell Host Microbe 2019, 25, 324–335. [Google Scholar] [CrossRef]
- Zenobia, C.; Hajishengallis, G. Porphyromonas gingivalis virulence factors involved in subversion of leukocytes and microbial dysbiosis. Virulence 2015, 6, 236–243. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, J.; Zhang, C.; Yu, N.; Lu, Z.; Zhang, S.; Li, Y.; Li, Q.; Liu, J.; Liu, D.; et al. Porphyromonas gingivalis exacerbates ulcerative colitis via Porphyromonas gingivalis peptidylarginine deiminase. Int. J. Oral Sci. 2021, 3, 31. [Google Scholar] [CrossRef]
- Koliarakis, I.; Messaritakis, I.; Nikolouzakis, T.K.; Hamilos, G.; Souglakos, J.; Tsiaoussis, J. Oral Bacteria and Intestinal Dysbiosis in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 4146. [Google Scholar] [CrossRef]
- Ohtsu, A.; Takeuchi, Y.; Katagiri, S.; Suda, W.; Maekawa, S.; Shiba, T.; Komazaki, R.; Udagawa, S.; Sasaki, N.; Hattori, M.; et al. Influence of Porphyromonas gingivalis in gut microbiota of streptozotocin-induced diabetic mice. Oral Dis. 2019, 25, 868–880. [Google Scholar] [CrossRef] [PubMed]
- Kerr, C.A.; Grice, D.M.; Tran, C.D.; Bauer, D.C.; Li, D.; Hendry, P.; Hannan, G.N. Early life events influence whole-of-life metabolic health via gut microflora and gut permeability. Crit. Rev. Microbiol. 2015, 41, 326–340. [Google Scholar] [CrossRef]
- Taylor, S.N.; Basile, L.A.; Ebeling, M.; Wagner, C.L. Intestinal permeability in preterm infants by feeding type: Mother’s milk versus formula. Breastfeed Med. 2009, 4, 11–15. [Google Scholar] [CrossRef]
- Catassi, C.; Bonucci, A.; Coppa, G.V.; Carlucci, A.; Giorgi, P.L. Intestinal permeability changes during the first month—Effect of natural versus artificial feeding. J. Pediatr. Gastroenterol. Nutr. 1995, 21, 383–386. [Google Scholar] [CrossRef] [PubMed]
- Edwinson, A.L.; Yang, L.; Peters, S.; Hanning, N.; Jeraldo, P.; Jagtap, P.; Simpson, J.B.; Yang, T.Y.; Kumar, P.; Mehta, S.; et al. Gut microbial β-glucuronidases regulate host luminal proteases and are depleted in irritable bowel syndrome. Nat. Microbiol. 2022, 7, 680–694. [Google Scholar] [CrossRef]
- Ling, C.; Versloot, C.J.; Arvidsson Kvissberg, M.E.; Hu, G.; Swain, N.; Horcas-Nieto, J.M.; Miraglia, E.; Thind, M.K.; Farooqui, A.; Gerding, A.; et al. Rebalancing of mitochondrial homeostasis through an NAD+-SIRT1 pathway preserves intestinal barrier function in severe malnutrition. EBioMedicine 2023, 20, 104809. [Google Scholar] [CrossRef] [PubMed]
- Amadi, B.; Besa, E.; Zyambo, K.; Kaonga, P.; Louis-Auguste, J.; Chandwe, K.; Tarr, P.I.; Denno, D.M.; Nataro, J.P.; Faubion, W.; et al. Impaired barrier function and autoantibody generation in malnutrition enteropathy in Zambia. EBioMedicine 2017, 22, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Patterson, G.T.; Osorio, E.Y.; Peniche, A.; Dann, S.M.; Cordova, E.; Preidis, G.A.; Suh, J.H.; Ito, I.; Saldarriaga, O.A.; Loeffelholz, M.; et al. Pathologic Inflammation in Malnutrition Is Driven by Proinflammatory Intestinal Microbiota, Large Intestine Barrier Dysfunction, and Translocation of Bacterial Lipopolysaccharide. Front. Immunol. 2022, 26, 846155. [Google Scholar] [CrossRef]
- Lieleg, O.; Lieleg, C.; Bloom, J.; Buck, C.B.; Ribbeck, K. Mucin Biopolymers As Broad-Spectrum Antiviral Agents. Biomacromolecules 2012, 13, 1724–1732. [Google Scholar] [CrossRef] [PubMed]
- Teng, X.; Yang, Y.; Liu, L.; Yang, L.; Wu, J.; Sun, M.; Xu, L. Evaluation of inflammatory bowel disease activity in children using serum trefoil factor peptide. Pediatr. Res. 2020, 88, 792–795. [Google Scholar] [CrossRef]
- Bureau, C.; Hanoun, N.; Torrisani, J.; Vinel, J.-P.; Buscail, L.; Cordelier, P. Expression and Function of Kruppel Like-Factors (KLF) in Carcinogenesis. Curr. Genom. 2009, 10, 10353–10360. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jiang, Z.; Zhang, Y.; Wang, X.; Liu, L.; Fan, Z. RNA sequencing analysis reveals protective role of kruppel-like factor 3 in colorectal cancer. Oncotarget 2017, 8, 21984–21993. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Cang, S.; Jiao, J.; Rong, W.; Xu, H.; Bi, K.; Li, Q.; Liu, R. Integrated study of metabolomics and gut metabolic activity from ulcerative colitis to colorectal cancer: The combined action of disordered gut microbiota and linoleic acid metabolic pathway might fuel cancer. J. Chromatogr. A 2020, 1629, 461503. [Google Scholar] [CrossRef] [PubMed]



| Gene | Symbol | Sequence (5′–3′) |
|---|---|---|
| MUC2 | Muc2 | AAGCCAGATCCCGAAACCAT |
| ATGGCCCCATTCACAACTGCC | ||
| TFF1 | Tff1 | CAAGGTGACCTGTGTCCTC |
| CTTGCTGGTTCTCAATGACC | ||
| TFF3 | Tff3 | GACTCCAGCATCCCAAATGT |
| GCAGATCAGGGGTGAGTGTT | ||
| KLF3 | Klf3 | TCATGTACACCAGCCACCTG |
| TAGTCAGTCCTCTGTGGTTC | ||
| 18S ARNr | Rps18 | GTAACCCGTTGAACCCCATT |
| CCATCCAATCGGTAGTAGCG |
| Control | Undernourished | |
|---|---|---|
| Newborns weight (g) | 6.16 ± 0.14 | 5.69 ± 0.09 ** |
| Litter size | 11.5 ± 1.30 | 9.86 ± 0.72 |
| Number of male newborns | 6.25 ± 11.11 | 5.71 ± 0.65 |
| Number of female newborns | 5.25 ± 0.63 | 4.00 ± 0.67 |
| Ratio males/females | 1.23 ± 0.24 | 1.88 ± 0.25 |
| Control | Undernourished | |||||
|---|---|---|---|---|---|---|
| L4 | L14 | L18 | L4 | L14 | L18 | |
| Body weight (g) | 11.36 ± 0.23 | 27.94 ± 0.4 | 34.65 ± 1.02 | 8.35 ± 0.15 *** | 16.5 ± 0.41 *** | 16.62 ± 0.36 *** |
| Glycemia (mg/dL) | 119.1 ± 1.43 | 107.8 ± 1.03 | 146.19 ± 1.19 | 85.9 ± 4.02 *** | 85.3 ± 5.29 ** | 93.15 ± 4.94 *** |
| Insulinemia (ng/mL) | 0.67 ± 0.06 | 0.70 ± 0.08 | 0.78 ± 0.17 | 0.29 ± 0.04 *** | 0.25 ± 0.06 ** | 0.36 ± 0.09 * |
| Glucagonemia (pg/mL) | 252.3 ± 10.6 | 298 ± 11.2 | 129.1 ± 6.9 | 103.9 ± 4.4 *** | 97 ± 1.3 *** | 83.2 ± 4.3 *** |
| Cholesterol (mg/dL) | 154.66 ± 4.11 | 127.48 ± 5.81 | 119.95 ± 4.5 | 144.38 ± 3.28 | 119.62 ± 4.51 | 116.86 ± 5.2 |
| Triglycerides (mg/dL) | 157.08 ± 5.26 | 79.94 ± 4.75 | 63.41 ± 5.42 | 160.17 ± 8.57 | 74.02 ± 3.07 | 68.42 ± 3.51 |
| Control | Undernourished | |||||
|---|---|---|---|---|---|---|
| L4 | L14 | L18 | L4 | L14 | L18 | |
| Body length (cm) | 6.23 ± 0.07 | 8.89 ± 0.09 | 9.36 ± 0.114 | 6.28 ± 0.17 | 7.4 ± 0.09 *** | 8.01 ± 0.13 *** |
| Total intestine length (cm) | 33.47 ± 0.37 | 48.0 ± 0.90 | 48.43 ± 1.53 | 29.54 ± 0.67 | 41.0 ± 2.30 | 39.41 ± 0.72 *** |
| Small intestine length (cm) Colon length (cm) Cecum length (cm) | 29.15 ± 0.38 | 40.6 ± 0.81 | 41.47 ± 1.34 | 25.27 ± 0.64 ** | 35.0 ± 2.20 | 33.14 ± 0.70 *** |
| 3.64 ± 0.13 | 5.83 ± 0.14 | 5.90 ± 0.24 | 3.16 ± 0.12 * | 5.17 ± 0.19 | 5.52 ± 0.14 | |
| 0.68 ± 0.04 | 1.35 ± 0.06 | 1.38 ± 0.05 | 0.65 ± 0.03 ** | 1.43 ± 0.08 | 1.26 ± 0.05 | |
| Small Intes. Length/Body Length | 4.68 ± 0.14 | 4.57 ± 0.12 | 0.48 ± 0.19 | 4.64 ± 0.10 | 5.23 ± 0.32 | 4.49 ± 0.20 |
| Colon Length/Body Length | 0.56 ± 0.03 | 0.67 ± 0.01 | 0.63 ± 0.03 | 0.57 ± 0.02 | 0.76 ± 0.03 * | 0.75 ± 0.03 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Oca, P.; Alba, C.; Sánchez-Roncero, A.; Fernández-Marcelo, T.; Martín, M.Á.; Escrivá, F.; Rodríguez, J.M.; Álvarez, C.; Fernández-Millán, E. Maternal Diet Determines Milk Microbiome Composition and Offspring Gut Colonization in Wistar Rats. Nutrients 2023, 15, 4322. https://doi.org/10.3390/nu15204322
Martínez-Oca P, Alba C, Sánchez-Roncero A, Fernández-Marcelo T, Martín MÁ, Escrivá F, Rodríguez JM, Álvarez C, Fernández-Millán E. Maternal Diet Determines Milk Microbiome Composition and Offspring Gut Colonization in Wistar Rats. Nutrients. 2023; 15(20):4322. https://doi.org/10.3390/nu15204322
Chicago/Turabian StyleMartínez-Oca, Paula, Claudio Alba, Alicia Sánchez-Roncero, Tamara Fernández-Marcelo, María Ángeles Martín, Fernando Escrivá, Juan Miguel Rodríguez, Carmen Álvarez, and Elisa Fernández-Millán. 2023. "Maternal Diet Determines Milk Microbiome Composition and Offspring Gut Colonization in Wistar Rats" Nutrients 15, no. 20: 4322. https://doi.org/10.3390/nu15204322
APA StyleMartínez-Oca, P., Alba, C., Sánchez-Roncero, A., Fernández-Marcelo, T., Martín, M. Á., Escrivá, F., Rodríguez, J. M., Álvarez, C., & Fernández-Millán, E. (2023). Maternal Diet Determines Milk Microbiome Composition and Offspring Gut Colonization in Wistar Rats. Nutrients, 15(20), 4322. https://doi.org/10.3390/nu15204322






