Totum-070, a Polyphenol-Rich Plant Extract, Prevents Hypercholesterolemia in High-Fat Diet-Fed Hamsters by Inhibiting Intestinal Cholesterol Absorption
Abstract
:1. Introduction
2. Materials and Methods
2.1. Totum-070 (Lipidrive®) Characterization
2.2. Animals
2.3. Serum Lipid Parameters
2.4. Liver Triglycerides
2.5. Histology
2.6. Fecal Neutral Sterols
2.7. Serum Lipopolysaccharide (LPS) Load Quantification
2.8. Intestinal Cholesterol Absorption Test
2.9. Oral Fat Tolerance Test
2.10. Hepatic Triglyceride Production Test
2.11. Lipoprotein Lipase Activity Assay
2.12. Quantitative Reverse Transcription-PCR (RT-qPCR)
2.13. Protein Extraction and Western Blot Analysis
2.14. Cell Cultures
2.15. RNA Sequencing Analysis
2.16. Microscale Thermophoresis (MST) Binding Affinity Assay
2.17. Microbiota Analysis
2.18. Statistical Analysis
3. Results
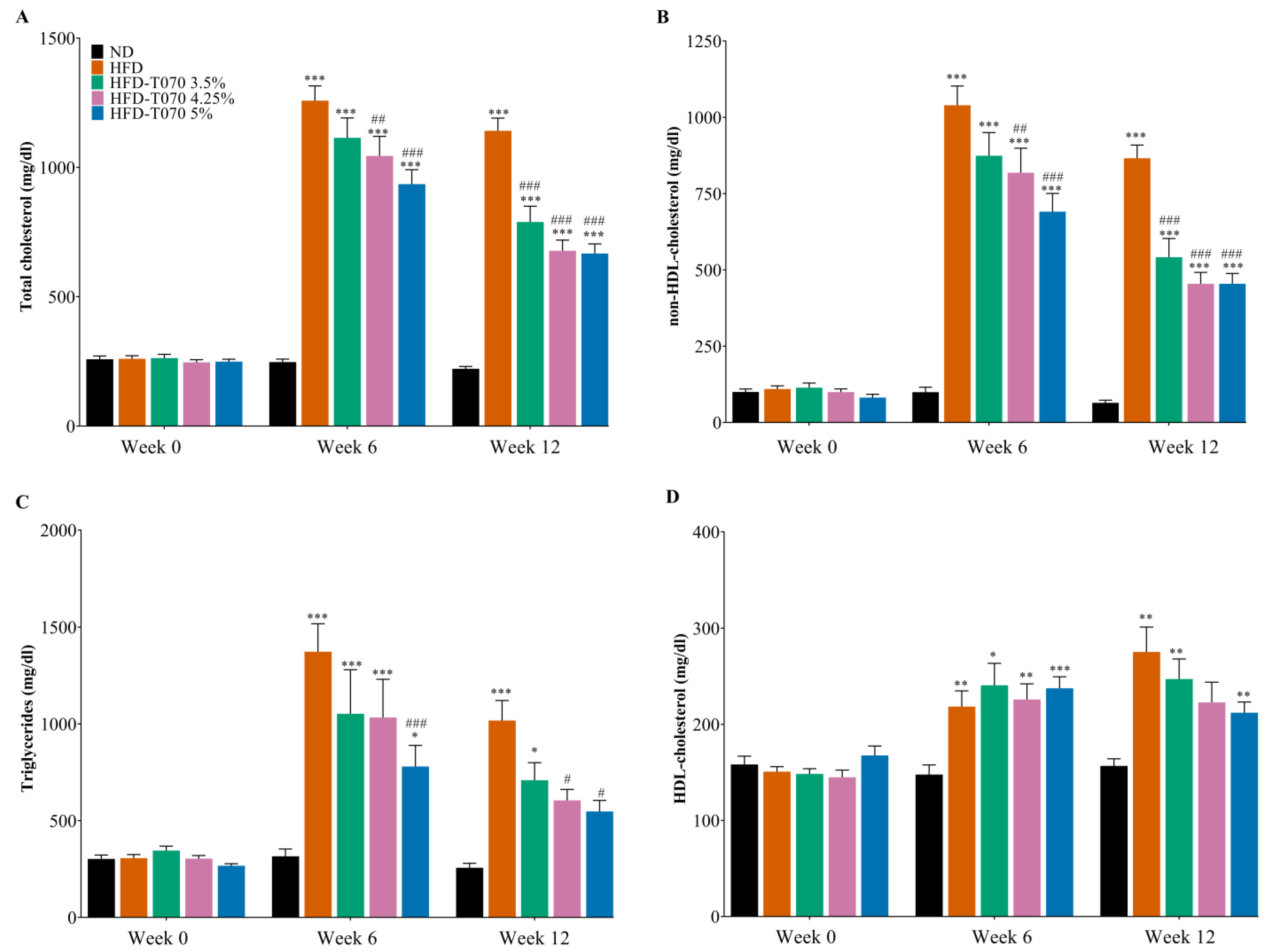
3.1. Supplementation with Totum-070 Reduces Hyperlipidemia and Improves Liver Steatosis and Inflammation in Hamsters without an Effect on Body Weight
3.2. Totum-070 Modulates Intestinal Cholesterol Metabolism in Hamsters
3.3. Totum-070 Inhibits Cholesterol Uptake and Secretion in Enterocytes
3.4. Totum-070 Influences Gut Microbiota in Hamsters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACAT2 | acetyl-CoA acetyltransferase 2 |
| ASCVD | Atherosclerotic cardiovascular disease |
| AUC | Area under the curve |
| FNS | Fecal neutral sterols |
| HDL-C | High-density lipoprotein cholesterol |
| HFD | High-fat diet |
| HTPT | Hepatic triglycerides production test |
| IBAT | Ileal bile acid transporter |
| LDL-C | Low-density lipoprotein cholesterol |
| LPL | Lipoprotein lipase |
| LPS | Lipopolysaccharides |
| MTTP | Microsomal triglyceride transfer protein |
| MST | Microscale thermophoresis |
| ND | Normal diet |
| NTD | N-terminal domain |
| NPC1L1 | Niemann-pick C1-like 1 |
| OFTT | Oral fat tolerance test |
| TG | Triglycerides |
| VLDL | Very low-density lipoprotein |
References
- Dagenais, G.R.; Leong, D.P.; Rangarajan, S.; Lanas, F.; Lopez-Jaramillo, P.; Gupta, R.; Diaz, R.; Avezum, A.; Oliveira, G.B.F.; Wielgosz, A.; et al. Variations in common diseases, hospital admissions, and deaths in middle-aged adults in 21 countries from five continents (PURE): A prospective cohort study. Lancet 2020, 395, 785–794. [Google Scholar] [CrossRef]
- Farzadfar, F. Cardiovascular disease risk prediction models: Challenges and perspectives. Lancet Glob. Health 2019, 7, e1288–e1289. [Google Scholar] [CrossRef]
- Boren, J.; Chapman, M.J.; Krauss, R.M.; Packard, C.J.; Bentzon, J.F.; Binder, C.J.; Daemen, M.J.; Demer, L.L.; Hegele, R.A.; Nicholls, S.J.; et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease: Pathophysiological, genetic, and therapeutic insights: A consensus statement from the European Atherosclerosis Society Consensus Panel. Eur. Heart J. 2020, 41, 2313–2330. [Google Scholar] [CrossRef]
- Prospective Studies, C.; Lewington, S.; Whitlock, G.; Clarke, R.; Sherliker, P.; Emberson, J.; Halsey, J.; Qizilbash, N.; Peto, R.; Collins, R. Blood cholesterol and vascular mortality by age, sex, and blood pressure: A meta-analysis of individual data from 61 prospective studies with 55,000 vascular deaths. Lancet 2007, 370, 1829–1839. [Google Scholar] [CrossRef]
- Zanotti, I.; Dall′Asta, M.; Mena, P.; Mele, L.; Bruni, R.; Ray, S.; Del Rio, D. Atheroprotective effects of (poly)phenols: A focus on cell cholesterol metabolism. Food Funct. 2015, 6, 13–31. [Google Scholar] [CrossRef] [PubMed]
- Morze, J.; Osadnik, T.; Osadnik, K.; Lejawa, M.; Jakubiak, G.; Pawlas, N.; Gasior, M.; Schwingshackl, L.; Banach, M. Comparative effect of nutraceuticals on lipid profile: A protocol for systematic review and network meta-analysis. BMJ Open 2020, 10, e032755. [Google Scholar] [CrossRef] [PubMed]
- Osadnik, T.; Golawski, M.; Lewandowski, P.; Morze, J.; Osadnik, K.; Pawlas, N.; Lejawa, M.; Jakubiak, G.K.; Mazur, A.; Schwingschackl, L.; et al. A network meta-analysis on the comparative effect of nutraceuticals on lipid profile in adults. Pharmacol. Res. 2022, 183, 106402. [Google Scholar] [CrossRef]
- Cicero, A.F.G.; Colletti, A.; Bajraktari, G.; Descamps, O.; Djuric, D.M.; Ezhov, M.; Fras, Z.; Katsiki, N.; Langlois, M.; Latkovskis, G.; et al. Lipid-lowering nutraceuticals in clinical practice: Position paper from an International Lipid Expert Panel. Nutr. Rev. 2017, 75, 731–767. [Google Scholar] [CrossRef]
- Wider, B.; Pittler, M.H.; Thompson-Coon, J.; Ernst, E. Artichoke leaf extract for treating hypercholesterolaemia. Cochrane Database Syst. Rev. 2013, 3, CD003335. [Google Scholar] [CrossRef]
- Sahebkar, A.; Pirro, M.; Banach, M.; Mikhailidis, D.P.; Atkin, S.L.; Cicero, A.F.G. Lipid-lowering activity of artichoke extracts: A systematic review and meta-analysis. Crit. Rev. Food Sci. Nutr. 2018, 58, 2549–2556. [Google Scholar] [CrossRef]
- Acar-Tek, N.; Agagunduz, D. Olive Leaf (Olea europaea L. folium): Potential Effects on Glycemia and Lipidemia. Ann. Nutr. Metab. 2020, 76, 10–15. [Google Scholar] [CrossRef]
- Vogel, P.; Kasper Machado, I.; Garavaglia, J.; Zani, V.T.; de Souza, D.; Morelo Dal Bosco, S. Polyphenols benefits of olive leaf (Olea europaea L) to human health. Nutr. Hosp. 2014, 31, 1427–1433. [Google Scholar] [CrossRef]
- Kulczyński, B.; Gramza-Michałowska, A. Goji Berry (Lycium barbarum): Composition and Health Effects—a Review. Pol. J. Food Nutr. Sci. 2016, 66, 67–75. [Google Scholar] [CrossRef]
- Ben Salem, M.; Affes, H.; Dhouibi, R.; Charfi, S.; Turki, M.; Hammami, S.; Ayedi, F.; Sahnoun, Z.; Zeghal, K.M.; Ksouda, K. Effect of Artichoke (Cynara scolymus) on cardiac markers, lipid profile and antioxidants levels in tissue of HFD-induced obesity. Arch. Physiol. Biochem. 2022, 128, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Ben Salem, M.; Ksouda, K.; Dhouibi, R.; Charfi, S.; Turki, M.; Hammami, S.; Ayedi, F.; Sahnoun, Z.; Zeghal, K.M.; Affes, H. LC-MS/MS Analysis and Hepatoprotective Activity of Artichoke (Cynara scolymus L.) Leaves Extract against High Fat Diet-Induced Obesity in Rats. Biomed. Res. Int. 2019, 2019, 4851279. [Google Scholar] [CrossRef] [PubMed]
- Ben Salem, M.; Ben Abdallah Kolsi, R.; Dhouibi, R.; Ksouda, K.; Charfi, S.; Yaich, M.; Hammami, S.; Sahnoun, Z.; Zeghal, K.M.; Jamoussi, K.; et al. Protective effects of Cynara scolymus leaves extract on metabolic disorders and oxidative stress in alloxan-diabetic rats. BMC Complement. Altern. Med. 2017, 17, 328. [Google Scholar] [CrossRef] [PubMed]
- Qiang, Z.; Lee, S.O.; Ye, Z.; Wu, X.; Hendrich, S. Artichoke extract lowered plasma cholesterol and increased fecal bile acids in Golden Syrian hamsters. Phytother. Res. 2012, 26, 1048–1052. [Google Scholar] [CrossRef] [PubMed]
- Poudyal, H.; Campbell, F.; Brown, L. Olive leaf extract attenuates cardiac, hepatic, and metabolic changes in high carbohydrate-, high fat-fed rats. J. Nutr. 2010, 140, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Yoon, L.; Liu, Y.N.; Park, H.; Kim, H.S. Olive Leaf Extract Elevates Hepatic PPAR alpha mRNA Expression and Improves Serum Lipid Profiles in Ovariectomized Rats. J. Med. Food 2015, 18, 738–744. [Google Scholar] [CrossRef]
- Wang, L.; Geng, C.; Jiang, L.; Gong, D.; Liu, D.; Yoshimura, H.; Zhong, L. The anti-atherosclerotic effect of olive leaf extract is related to suppressed inflammatory response in rabbits with experimental atherosclerosis. Eur. J. Nutr. 2008, 47, 235–243. [Google Scholar] [CrossRef]
- Cui, B.; Liu, S.; Lin, X.; Wang, J.; Li, S.; Wang, Q.; Li, S. Effects of Lycium barbarum aqueous and ethanol extracts on high-fat-diet induced oxidative stress in rat liver tissue. Molecules 2011, 16, 9116–9128. [Google Scholar] [CrossRef]
- Pai, P.G.; Habeeba, P.U.; Ullal, S.D.; Shoeb, P.A.; Pradeepti, M.S.; Ramya, K. Evaluation of Hypolipidemic Effects of Lyciumbarbarum (goji Berry) in a Murine Model. J. Nat. Remedies 2013, 13, 4–8. [Google Scholar]
- Crepaldi, L.D.; Mariano, I.R.; Trondoli, A.J.P.C.; Moreno, F.N.; Piovan, S.; Formigoni, M.; Salgueiro-Pagadigorria, C.L.; Godoi, V.A.F.; Brito, M.d.N.; Garcia, R.F. Goji Berry (Lycium barbarum) Extract Improves Biometric, Plasmatic and Hepatic Parameters of Rats Fed a High-Carbohydrate Diet. J. Pharm. Pharmacol. 2018, 6, 877–889. [Google Scholar]
- Luo, Q.; Cai, Y.; Yan, J.; Sun, M.; Corke, H. Hypoglycemic and hypolipidemic effects and antioxidant activity of fruit extracts from Lycium barbarum. Life Sci. 2004, 76, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Parim, B.; Harishankar, N.; Balaji, M.; Pothana, S.; Sajjalaguddam, R.R. Effects of Piper nigrum extracts: Restorative perspectives of high-fat diet-induced changes on lipid profile, body composition, and hormones in Sprague-Dawley rats. Pharm. Biol. 2015, 53, 1318–1328. [Google Scholar] [CrossRef]
- Vijayakumar, R.S.; Surya, D.; Senthilkumar, R.; Nalini, N. Hypolipidemic Effect of Black Pepper (Piper nigrum Linn.) in Rats Fed High Fat Diet. J. Clin. Biochem. Nutr. 2002, 32, 31–42. [Google Scholar] [CrossRef]
- Cottet, J.; Etienne, J. Determination of serum lipids by the sulfo-phospho-vanillic method of E. Chabrol and R. Charonnat. Bull. Acad. Natl. Med. 1965, 149, 331–338. [Google Scholar] [PubMed]
- Cheng, Y.S.; Zheng, Y.; VanderGheynst, J.S. Rapid quantitative analysis of lipids using a colorimetric method in a microplate format. Lipids 2011, 46, 95–103. [Google Scholar] [CrossRef]
- Benson, J.R.; Hare, P.E. O-phthalaldehyde: Fluorogenic detection of primary amines in the picomole range. Comparison with fluorescamine and ninhydrin. Proc. Natl. Acad. Sci. USA 1975, 72, 619–622. [Google Scholar] [CrossRef] [PubMed]
- Giera, M.; Plossl, F.; Bracher, F. Fast and easy in vitro screening assay for cholesterol biosynthesis inhibitors in the post-squalene pathway. Steroids 2007, 72, 633–642. [Google Scholar] [CrossRef]
- Maillard, F.; Vazeille, E.; Sauvanet, P.; Sirvent, P.; Bonnet, R.; Combaret, L.; Chausse, P.; Chevarin, C.; Otero, Y.F.; Delcros, G.; et al. Preventive Effect of Spontaneous Physical Activity on the Gut-Adipose Tissue in a Mouse Model That Mimics Crohn′s Disease Susceptibility. Cells 2019, 8, 33. [Google Scholar] [CrossRef]
- Turley, S.D.; Daggy, B.P.; Dietschy, J.M. Psyllium augments the cholesterol-lowering action of cholestyramine in hamsters by enhancing sterol loss from the liver. Gastroenterology 1994, 107, 444–452. [Google Scholar] [CrossRef]
- Turley, S.D.; Herndon, M.W.; Dietschy, J.M. Reevaluation and application of the dual-isotope plasma ratio method for the measurement of intestinal cholesterol absorption in the hamster. J. Lipid Res. 1994, 35, 328–339. [Google Scholar] [CrossRef] [PubMed]
- Ono-Moore, K.D.; Ferguson, M.; Blackburn, M.L.; Issafras, H.; Adams, S.H. Application of an In Vivo Hepatic Triacylglycerol Production Method in the Setting of a High-Fat Diet in Mice. Nutrients 2016, 9, 16. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.F.; Uffelman, K.; Naples, M.; Szeto, L.; Haidari, M.; Adeli, K. Intestinal lipoprotein overproduction, a newly recognized component of insulin resistance, is ameliorated by the insulin sensitizer rosiglitazone: Studies in the fructose-fed Syrian golden hamster. Endocrinology 2005, 146, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Taghibiglou, C.; Carpentier, A.; Van Iderstine, S.C.; Chen, B.; Rudy, D.; Aiton, A.; Lewis, G.F.; Adeli, K. Mechanisms of hepatic very low density lipoprotein overproduction in insulin resistance. Evidence for enhanced lipoprotein assembly, reduced intracellular ApoB degradation, and increased microsomal triglyceride transfer protein in a fructose-fed hamster model. J. Biol. Chem. 2000, 275, 8416–8425. [Google Scholar] [CrossRef] [PubMed]
- Cheema, S.K.; Cornish, M.L. Bio F1B hamster: A unique animal model with reduced lipoprotein lipase activity to investigate nutrient mediated regulation of lipoprotein metabolism. Nutr. Metab. 2007, 4, 27. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.F.; Murdoch, S.; Uffelman, K.; Naples, M.; Szeto, L.; Albers, A.; Adeli, K.; Brunzell, J.D. Hepatic lipase mRNA, protein, and plasma enzyme activity is increased in the insulin-resistant, fructose-fed Syrian golden hamster and is partially normalized by the insulin sensitizer rosiglitazone. Diabetes 2004, 53, 2893–2900. [Google Scholar] [CrossRef]
- Yamazaki, T.; Kishimoto, K.; Ezaki, O. The ddY mouse: A model of postprandial hypertriglyceridemia in response to dietary fat. J. Lipid Res. 2012, 53, 2024–2037. [Google Scholar] [CrossRef]
- Chavanelle, V.; Otero, Y.F.; Le Joubioux, F.; Ripoche, D.; Bargetto, M.; Vluggens, A.; Montaurier, C.; Pickering, G.; Ducheix, G.; Dubray, C.; et al. Effects of Totum-63 on glucose homeostasis and postprandial glycemia: A translational study. Am. J. Physiol. Endocrinol. Metab. 2021, 320, E1119–E1137. [Google Scholar] [CrossRef]
- Gilda, J.E.; Gomes, A.V. Western blotting using in-gel protein labeling as a normalization control: Stain-free technology. Methods Mol. Biol. 2015, 1295, 381–391. [Google Scholar] [CrossRef]
- Chateau, D.; Pauquai, T.; Delers, F.; Rousset, M.; Chambaz, J.; Demignot, S. Lipid micelles stimulate the secretion of triglyceride-enriched apolipoprotein B48-containing lipoproteins by Caco-2 cells. J. Cell Physiol. 2005, 202, 767–776. [Google Scholar] [CrossRef]
- Nakano, T.; Inoue, I.; Katayama, S.; Seo, M.; Takahashi, S.; Hokari, S.; Shinozaki, R.; Hatayama, K.; Komoda, T. Lysophosphatidylcholine for efficient intestinal lipid absorption and lipoprotein secretion in caco-2 cells. J. Clin. Biochem. Nutr. 2009, 45, 227–234. [Google Scholar] [CrossRef]
- Vallier, M.; Suwandi, A.; Ehrhardt, K.; Belheouane, M.; Berry, D.; Cepic, A.; Galeev, A.; Johnsen, J.M.; Grassl, G.A.; Baines, J.F. Pathometagenomics reveals susceptibility to intestinal infection by Morganella to be mediated by the blood group-related B4galnt2 gene in wild mice. Gut Microbes 2023, 15, 2164448. [Google Scholar] [CrossRef] [PubMed]
- Betters, J.L.; Yu, L. NPC1L1 and cholesterol transport. FEBS Lett. 2010, 584, 2740–2747. [Google Scholar] [CrossRef] [PubMed]
- Jerabek-Willemsen, M.; André, T.; Wanner, R.; Roth, H.M.; Duhr, S.; Baaske, P.; Breitsprecher, D. MicroScale Thermophoresis: Interaction analysis and beyond. J. Mol. Struct. 2014, 1077, 101–113. [Google Scholar] [CrossRef]
- Haguet, Q.; Le Joubioux, F.; Chavanelle, V.; Groult, H.; Schoonjans, N.; Langhi, C.; Michaux, A.; Otero, Y.F.; Boisseau, N.; Peltier, S.L.; et al. Inhibitory Potential of alpha-Amylase, alpha-Glucosidase, and Pancreatic Lipase by a Formulation of Five Plant Extracts: TOTUM-63. Int. J. Mol. Sci. 2023, 24, 3652. [Google Scholar] [CrossRef]
- Verbeek, R.; Hovingh, G.K.; Boekholdt, S.M. Non-high-density lipoprotein cholesterol: Current status as cardiovascular marker. Curr. Opin. Lipidol. 2015, 26, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Arsenault, B.J.; Rana, J.S.; Stroes, E.S.; Despres, J.P.; Shah, P.K.; Kastelein, J.J.; Wareham, N.J.; Boekholdt, S.M.; Khaw, K.T. Beyond low-density lipoprotein cholesterol: Respective contributions of non-high-density lipoprotein cholesterol levels, triglycerides, and the total cholesterol/high-density lipoprotein cholesterol ratio to coronary heart disease risk in apparently healthy men and women. J. Am. Coll. Cardiol. 2009, 55, 35–41. [Google Scholar] [CrossRef]
- Eckel, R.H.; Grundy, S.M.; Zimmet, P.Z. The metabolic syndrome. Lancet 2005, 365, 1415–1428. [Google Scholar] [CrossRef]
- Kolovou, G.D.; Anagnostopoulou, K.K.; Cokkinos, D.V. Pathophysiology of dyslipidaemia in the metabolic syndrome. Postgrad. Med. J. 2005, 81, 358–366. [Google Scholar] [CrossRef]
- Kusku-Kiraz, Z.; Mehmetcik, G.; Dogru-Abbasoglu, S.; Uysal, M. Artichoke leaf extract reduces oxidative stress and lipoprotein dyshomeostasis in rats fed on high cholesterol diet. Phytother. Res. 2010, 24, 565–570. [Google Scholar] [CrossRef]
- Kucukgergin, C.; Aydin, A.F.; Ozdemirler-Erata, G.; Mehmetcik, G.; Kocak-Toker, N.; Uysal, M. Effect of artichoke leaf extract on hepatic and cardiac oxidative stress in rats fed on high cholesterol diet. Biol. Trace Elem. Res. 2010, 135, 264–274. [Google Scholar] [CrossRef]
- Cheurfa, M.; Abdallah, H.H.; Allem, R.; Noui, A.; Picot-Allain, C.M.N.; Mahomoodally, F. Hypocholesterolaemic and antioxidant properties of Olea europaea L. leaves from Chlef province, Algeria using in vitro, in vivo and in silico approaches. Food Chem. Toxicol. 2019, 123, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Olmez, E.; Vural, K.; Gok, S.; Ozturk, Z.; Kayalar, H.; Ayhan, S.; Var, A. Olive Leaf Extract Improves the Atherogenic Lipid Profile in Rats Fed a High Cholesterol Diet. Phytother. Res. 2015, 29, 1652–1657. [Google Scholar] [CrossRef] [PubMed]
- Dillard, A.; Matthan, N.R.; Lichtenstein, A.H. Use of hamster as a model to study diet-induced atherosclerosis. Nutr Metab (Lond) 2010, 7, 89. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, G.; Rizvi, F.; Saxena, R.; Puri, A.; Khanna, A.K.; Chander, R.; Wulff, E.M.; Rastogi, A.K. In vivo model for dyslipidemia with diabetes mellitus in hamster. Indian. J. Exp. Biol. 2003, 41, 1456–1459. [Google Scholar] [PubMed]
- Blacklow, S.C. Versatility in ligand recognition by LDL receptor family proteins: Advances and frontiers. Curr. Opin. Struct. Biol. 2007, 17, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, A.; Miyata, S.; Shimizu, M.; Inoue, J.; Sato, R. Piperine Induces Hepatic Low-Density Lipoprotein Receptor Expression through Proteolytic Activation of Sterol Regulatory Element-Binding Proteins. PLoS ONE 2015, 10, e0139799. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Qin, Y.; Wan, X.; Liu, H.; Iv, C.; Ruan, W.; Lu, L.; He, L.; Guo, X. Hydroxytyrosol Plays Antiatherosclerotic Effects through Regulating Lipid Metabolism via Inhibiting the p38 Signal Pathway. Biomed. Res. Int. 2020, 2020, 5036572. [Google Scholar] [CrossRef]
- Twisk, J.; Gillian-Daniel, D.L.; Tebon, A.; Wang, L.; Barrett, P.H.; Attie, A.D. The role of the LDL receptor in apolipoprotein B secretion. J. Clin. Investig. 2000, 105, 521–532. [Google Scholar] [CrossRef]
- Blasiole, D.A.; Oler, A.T.; Attie, A.D. Regulation of ApoB secretion by the low density lipoprotein receptor requires exit from the endoplasmic reticulum and interaction with ApoE or ApoB. J. Biol. Chem. 2008, 283, 11374–11381. [Google Scholar] [CrossRef]
- Nakamura, K.; Miyoshi, T.; Yunoki, K.; Ito, H. Postprandial hyperlipidemia as a potential residual risk factor. J. Cardiol. 2016, 67, 335–339. [Google Scholar] [CrossRef]
- Duangjai, A.; Ingkaninan, K.; Praputbut, S.; Limpeanchob, N. Black pepper and piperine reduce cholesterol uptake and enhance translocation of cholesterol transporter proteins. J. Nat. Med. 2013, 67, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S. The Effect of Polyphenols on Hypercholesterolemia through Inhibiting the Transport and Expression of Niemann-Pick C1-Like 1. Int. J. Mol. Sci. 2019, 20, 4939. [Google Scholar] [CrossRef] [PubMed]
- Nekohashi, M.; Ogawa, M.; Ogihara, T.; Nakazawa, K.; Kato, H.; Misaka, T.; Abe, K.; Kobayashi, S. Luteolin and quercetin affect the cholesterol absorption mediated by epithelial cholesterol transporter niemann-pick c1-like 1 in caco-2 cells and rats. PLoS ONE 2014, 9, e97901. [Google Scholar] [CrossRef]
- Pirillo, A.; Catapano, A.L.; Norata, G.D. Niemann-Pick C1-Like 1 (NPC1L1) Inhibition and Cardiovascular Diseases. Curr. Med. Chem. 2016, 23, 983–999. [Google Scholar] [CrossRef]
- Huang, C.S.; Yu, X.; Fordstrom, P.; Choi, K.; Chung, B.C.; Roh, S.H.; Chiu, W.; Zhou, M.; Min, X.; Wang, Z. Cryo-EM structures of NPC1L1 reveal mechanisms of cholesterol transport and ezetimibe inhibition. Sci. Adv. 2020, 6, eabb1989. [Google Scholar] [CrossRef]
- Ghazalpour, A.; Cespedes, I.; Bennett, B.J.; Allayee, H. Expanding role of gut microbiota in lipid metabolism. Curr. Opin. Lipidol. 2016, 27, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Kriaa, A.; Bourgin, M.; Potiron, A.; Mkaouar, H.; Jablaoui, A.; Gerard, P.; Maguin, E.; Rhimi, M. Microbial impact on cholesterol and bile acid metabolism: Current status and future prospects. J. Lipid Res. 2019, 60, 323–332. [Google Scholar] [CrossRef]
- Yu, Y.; Raka, F.; Adeli, K. The Role of the Gut Microbiota in Lipid and Lipoprotein Metabolism. J. Clin. Med. 2019, 8, 2227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gerard, P. Diet-gut microbiota interactions on cardiovascular disease. Comput. Struct. Biotechnol. J. 2022, 20, 1528–1540. [Google Scholar] [CrossRef] [PubMed]
- Bisanz, J.E.; Upadhyay, V.; Turnbaugh, J.A.; Ly, K.; Turnbaugh, P.J. Meta-Analysis Reveals Reproducible Gut Microbiome Alterations in Response to a High-Fat Diet. Cell Host Microbe 2019, 26, 265–272.e264. [Google Scholar] [CrossRef] [PubMed]
- Kunstner, A.; Aherrahrou, R.; Hirose, M.; Bruse, P.; Ibrahim, S.M.; Busch, H.; Erdmann, J.; Aherrahrou, Z. Effect of Differences in the Microbiome of Cyp17a1-Deficient Mice on Atherosclerotic Background. Cells 2021, 10, 1292. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.T.; Xiao, T.; Wang, L.; Lu, C.; Liu, L.; Zhou, X.; Wang, A.; Qin, W.; Wang, F. Plant protein reduces serum cholesterol levels in hypercholesterolemia hamsters by modulating the compositions of gut microbiota and metabolites. iScience 2021, 24, 103435. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.; Li, Q.; Liu, L.; Wang, S.; Wu, Z.; Tao, Y.; Huang, P.; Wang, P. Crocetin Prolongs Recovery Period of DSS-Induced Colitis via Altering Intestinal Microbiome and Increasing Intestinal Permeability. Int. J. Mol. Sci. 2022, 23, 3832. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yi, C.; Han, J.; Ming, T.; Zhou, J.; Lu, C.; Li, Y.; Su, X. Gut microbiome and metabolome analyses reveal the protective effect of special high-docosahexaenoic acid tuna oil on d-galactose-induced aging in mice. Food Sci. Nutr. 2022, 10, 3814–3827. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, H.; Xia, H.; Yang, X.; Yang, L.; Wang, S.; Wen, J.; Sun, G. Different effects of high-fat diets rich in different oils on lipids metabolism, oxidative stress and gut microbiota. Food Res. Int. 2021, 141, 110078. [Google Scholar] [CrossRef]
- Li, Y.; Chen, M.; Ma, Y.; Yang, Y.; Cheng, Y.; Ma, H.; Ren, D.; Chen, P. Regulation of viable/inactivated/lysed probiotic Lactobacillus plantarum H6 on intestinal microbiota and metabolites in hypercholesterolemic mice. NPJ Sci. Food 2022, 6, 50. [Google Scholar] [CrossRef]
- Lang, J.M.; Sedgeman, L.R.; Cai, L.; Layne, J.D.; Wang, Z.; Pan, C.; Lee, R.; Temel, R.E.; Lusis, A.J. Dietary and Pharmacologic Manipulations of Host Lipids and Their Interaction With the Gut Microbiome in Non-human Primates. Front. Med. 2021, 8, 646710. [Google Scholar] [CrossRef]
- Just, S.; Mondot, S.; Ecker, J.; Wegner, K.; Rath, E.; Gau, L.; Streidl, T.; Hery-Arnaud, G.; Schmidt, S.; Lesker, T.R.; et al. The gut microbiota drives the impact of bile acids and fat source in diet on mouse metabolism. Microbiome 2018, 6, 134. [Google Scholar] [CrossRef]
- Palmisano, B.T.; Zhu, L.; Eckel, R.H.; Stafford, J.M. Sex differences in lipid and lipoprotein metabolism. Mol. Metab. 2018, 15, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Russo, G.T.; Giandalia, A.; Romeo, E.L.; Cucinotta, D. Gender differences in lipoprotein metabolism. Ital. J. Gend.-Specif. Med. 2015, 1, 58–65. [Google Scholar] [CrossRef]
- Davies, J.P.; Scott, C.; Oishi, K.; Liapis, A.; Ioannou, Y.A. Inactivation of NPC1L1 causes multiple lipid transport defects and protects against diet-induced hypercholesterolemia. J. Biol. Chem. 2005, 280, 12710–12720. [Google Scholar] [CrossRef] [PubMed]
- Davis, H.R., Jr.; Zhu, L.J.; Hoos, L.M.; Tetzloff, G.; Maguire, M.; Liu, J.; Yao, X.; Iyer, S.P.; Lam, M.H.; Lund, E.G.; et al. Niemann-Pick C1 Like 1 (NPC1L1) is the intestinal phytosterol and cholesterol transporter and a key modulator of whole-body cholesterol homeostasis. J. Biol. Chem. 2004, 279, 33586–33592. [Google Scholar] [CrossRef]
- Kamato, D.; Ilyas, I.; Xu, S.; Little, P.J. Non-Mouse Models of Atherosclerosis: Approaches to Exploring the Translational Potential of New Therapies. Int. J. Mol. Sci. 2022, 23, 12964. [Google Scholar] [CrossRef]
- Hermans, M.P.; Lempereur, P.; Salembier, J.P.; Maes, N.; Albert, A.; Jansen, O.; Pincemail, J. Supplementation Effect of a Combination of Olive (Olea europea L.) Leaf and Fruit Extracts in the Clinical Management of Hypertension and Metabolic Syndrome. Antioxidants 2020, 9, 872. [Google Scholar] [CrossRef]
- Rondanelli, M.; Riva, A.; Petrangolini, G.; Allegrini, P.; Bernardinelli, L.; Fazia, T.; Peroni, G.; Gasparri, C.; Nichetti, M.; Faliva, M.A.; et al. The Metabolic Effects of Cynara Supplementation in Overweight and Obese Class I Subjects with Newly Detected Impaired Fasting Glycemia: A Double-Blind, Placebo-Controlled, Randomized Clinical Trial. Nutrients 2020, 12, 3298. [Google Scholar] [CrossRef]




| Compound Type (Sorted by Families) | Totum-070 Content (w/w) |
|---|---|
| Total sugars | 41.35 |
| Total lipids | 7.30 |
| Total proteins | 2.05 |
| Betaine | 0.23 |
| Total phenolic compounds | 15.42 |
| Mono-caffeoylquinic acids | |
| Chlorogenic acid | 0.81 |
| Cryptochlorogenic acid | 0.01 |
| Other mono-caffeoylquinic acids | 0.25 |
| Dicaffeoylquinic acids | |
| Cynarin | 0.16 |
| 3-5 dicaffeoylquinic acid | 0.18 |
| 4-5 dicaffeoylquinic acid | 0.14 |
| Other dicaffeoylquinic acids | 0.11 |
| Caffeic acid | 0.01 |
| Oleuropein | 4.6 |
| Oleuropein isomers | 0.47 |
| Hydroxytyrosol | 0.08 |
| Luteolin | 0.01 |
| Luteolin-7-O-glucoside | 0.27 |
| Luteolin-7-O-glucoside isomer | 0.02 |
| Luteolin-4-O-glucoside | 0.06 |
| Luteolin-7-O-glucuronide | 0.29 |
| Apigenin-7-O-glucoside | 0.03 |
| Apigenin-7-O-glucuronide | 0.17 |
| Apigenin-6-C-glucoside-8-C-arabinoside (Shaftoside) | 0.01 |
| Apigenin-6,8-C-diglucoside (Vicenin 2) | 0.02 |
| Apigenin-7-O-rutinoside | 0.01 |
| Eriodictyol-7-O-glucoside | 0.04 |
| Flavanomarein | 0.14 |
| Marein | 0.05 |
| Maritimein | 0.05 |
| Rutin | 0.02 |
| Verbascoside | 0.08 |
| Terpenes and terpenoids | |
| Oleanolic acid | 1.64 |
| Saponins | |
| Chrysanthellin A | 0.28 |
| Chrysanthellin B | 0.27 |
| Iridoids | |
| Oleoside | 0.01 |
| Cynaropicrin | 0.07 |
| Alkaloids | |
| Piperine | 0.06 |
| Diet Supplementation | ND - | HFD - | HFD T070 3.5% | HFD T070 4.25% | HFD T070 5% |
|---|---|---|---|---|---|
| Final body weight (g) | 129.2 ± 2.6 | 124.4 ± 2.7 | 124.8 ± 2.6 | 126.5 ± 3.3 | 123.4 ± 1.2 |
| Final fat mass (g) | 18.12 ± 1.3 | 20.14 ± 0.6 | 22.79 ± 1 | 23.36 ± 1 * | 22.26 ± 1 |
| Final lean mass (g) | 106.3 ± 2.2 | 98.61 ± 2.3 | 97.17 ± 1.9 * | 98.74 ± 2.7 | 96.78 ± 1.8 * |
| Cumulative food intake (g) | 436 ± 7.1 | 375 ± 11.5 ** | 386 ± 10.5 | 401.5 ± 8 | 397 ± 15.4 |
| Cumulative caloric intake (Kcal) | 1680 ± 27.3 | 1688 ± 52 | 1700 ± 46.3 | 1766 ± 35.2 | 1748 ± 68 |
| Log Fold-Change | p-Values HFD | Golden Hamster Gene ID | Gene Name | Protein |
|---|---|---|---|---|
| 2.43 | 0.001509 | ENSMAUG00000000351 | Lipg | lipase G, endothelial type |
| 2.39 | 0.017545 | ENSMAUG00000000465 | Acly | ATP-citrate synthase |
| 2.24 | 0.000089 | ENSMAUG00000001919 | Atp6v0d2 | V-type proton ATPase subunit d 2 |
| 2.08 | 0.000001 | ENSMAUG00000008803 | Alpl | alkaline phosphatase |
| 2.06 | 0.000125 | ENSMAUG00000004155 | Asns | asparagine synthetase |
| 1.51 | 0.000422 | ENSMAUG00000009435 | Bst1 | ADP-ribosyl cyclase 2 |
| 1.47 | 0.001238 | ENSMAUG00000005505 | Mboat2 | lysophospholipid acyltransferase 2 |
| 1.35 | 0.002228 | ENSMAUG00000018983 | Alox5 | polyunsaturated fatty acid 5-lipoxygenase |
| 1.21 | 0.000716 | ENSMAUG00000001135 | B3galnt1 | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 |
| 1.18 | 0.000011 | ENSMAUG00000000598 | N/A | N/A |
| 1.14 | 0.000212 | ENSMAUG00000009261 | Mthfd2 | NAD-dependent methylene tetrahydrofolate dehydrogenase cyclohydrolase |
| 1.14 | 0.000757 | ENSMAUG00000017913 | Mboat1 | lysophospholipid acyltransferase 1 |
| 1.07 | 0.003228 | ENSMAUG00000020920 | Hk2 | hexokinase-2 |
| 1.07 | 0.001393 | ENSMAUG00000007951 | Elovl7 | elongation of very long chain fatty acids protein 7 |
| 1.02 | 0.000411 | ENSMAUG00000016079 | Acss1 | acetyl-coenzyme A synthetase 2-like, mitochondrial |
| 1.01 | 0.002529 | ENSMAUG00000004073 | Sqle | squalene monooxygenase |
| 1.00 | 0.001829 | ENSMAUG00000019624 | Pla2g7 | platelet-activating factor acetylhydrolase |
| −1.02 | 0.005813 | ENSMAUG00000003438 | Gck | hexokinase-4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Langhi, C.; Vallier, M.; Otero, Y.F.; Maura, M.; Le Joubioux, F.; Groult, H.; Achour, O.; Pebriana, R.B.; Giera, M.; Guigas, B.; et al. Totum-070, a Polyphenol-Rich Plant Extract, Prevents Hypercholesterolemia in High-Fat Diet-Fed Hamsters by Inhibiting Intestinal Cholesterol Absorption. Nutrients 2023, 15, 5056. https://doi.org/10.3390/nu15245056
Langhi C, Vallier M, Otero YF, Maura M, Le Joubioux F, Groult H, Achour O, Pebriana RB, Giera M, Guigas B, et al. Totum-070, a Polyphenol-Rich Plant Extract, Prevents Hypercholesterolemia in High-Fat Diet-Fed Hamsters by Inhibiting Intestinal Cholesterol Absorption. Nutrients. 2023; 15(24):5056. https://doi.org/10.3390/nu15245056
Chicago/Turabian StyleLanghi, Cédric, Marie Vallier, Yolanda F. Otero, Maheva Maura, Florian Le Joubioux, Hugo Groult, Oussama Achour, Ratna Budhi Pebriana, Martin Giera, Bruno Guigas, and et al. 2023. "Totum-070, a Polyphenol-Rich Plant Extract, Prevents Hypercholesterolemia in High-Fat Diet-Fed Hamsters by Inhibiting Intestinal Cholesterol Absorption" Nutrients 15, no. 24: 5056. https://doi.org/10.3390/nu15245056
APA StyleLanghi, C., Vallier, M., Otero, Y. F., Maura, M., Le Joubioux, F., Groult, H., Achour, O., Pebriana, R. B., Giera, M., Guigas, B., Maugard, T., Chassaing, B., Peltier, S., Bard, J. -M., & Sirvent, P. (2023). Totum-070, a Polyphenol-Rich Plant Extract, Prevents Hypercholesterolemia in High-Fat Diet-Fed Hamsters by Inhibiting Intestinal Cholesterol Absorption. Nutrients, 15(24), 5056. https://doi.org/10.3390/nu15245056










