Metabolite Profiling of Allium hookeri Leaves Using UHPLC-qTOF-MS/MS and the Senomorphic Activity of Phenolamides
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Plant Material
2.3. UHPLC-qTOF-MS/MS Experiments
2.4. Sample Preparation of A. hookeri for MS/MS Analysis
2.5. Extraction and Isolation
Physicochemical Properties of Isolated Compounds
2.6. Cell Culture
2.7. LPS-Induced NO Production and Cell Viability
2.8. Bleomycin-Induced Senescence Model
Senescence-Associated β-Galactosidase Staining
2.9. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.10. Statistical Analyses
3. Results
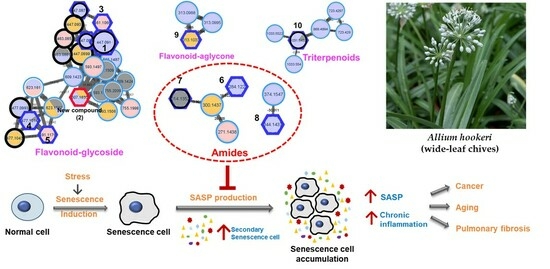
3.1. Feature-Based Molecular Networking of the A. hookeri Leaf Extract
3.2. Isolation and Structural Elucidation of New Compound 2
3.3. Bio-Activity of Isolated Compounds from A. hookeri
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- The World Flora Online Allium L. Available online: https://wfoplantlist.org/plant-list/taxon/wfo-4000001260-2023-06?page=1 (accessed on 24 November 2023).
- Sabiu, S.; Madende, M.; Ajao, A.A.; Aladodo, R.A.; Nurain, I.O.; Ahmad, J.B. The Genus Allium (Amaryllidaceae: Alloideae): Features, Phytoconstituents, and Mechanisms of Antidiabetic Potential of Allium Cepa and Allium Sativum. In Bioactive Food as Dietary Interventions for Diabetes; Elsevier: Amsterdam, The Netherlands, 2019; pp. 137–154. [Google Scholar]
- Ren, F.; Wang, L.; Zhuo, W.; Chen, D.; Huang, H.; Zhang, L. Complete Chloroplast Genome of the Rare Medicinal Vegetable Allium hookeri. Mitochondrial DNA Part B 2022, 7, 6–7. [Google Scholar] [CrossRef] [PubMed]
- Deka, B.; Manna, P.; Borah, J.C.; Talukdar, N.C. A Review on Phytochemical, Pharmacological Attributes and Therapeutic Uses of Allium hookeri. Phytomed. Plus 2022, 2, 100262. [Google Scholar] [CrossRef]
- Ayam, V. Allium hookeri, Thw. Enum. A Lesser Known Terrestrial Perennial Herb Used as Food and Its Ethnobotanical Relevance in Manipur. AJFAND 2011, 11, 5389–5412. [Google Scholar] [CrossRef]
- Bae, G.-C.; Bae, D.-Y. The Anti-Inflammatory Effects of Ethanol Extract of Allium hookeri Cultivated in South Korea. Korea J. Herbol. 2012, 27, 55–61. [Google Scholar] [CrossRef][Green Version]
- Park, E.-K.; Noh, J.-G.; Lee, M.-J.; Nam, S.-Y.; Hong, E.-Y.; Lee, C.-H. Effects of Shading Rates on Growth and Yield of Allium hookeri Cultivation at Greenhouse in Middle Area of Korea. Prot. Hortic. Plant Fact. 2016, 25, 320–327. [Google Scholar] [CrossRef]
- Choi, J.-H.; Lee, E.-B.; Jang, H.-H.; Cha, Y.-S.; Park, Y.-S.; Lee, S.-H. Allium hookeri Extracts Improve Scopolamine-Induced Cognitive Impairment via Activation of the Cholinergic System and Anti-Neuroinflammation in Mice. Nutrients 2021, 13, 2890. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Gohil, R.N.; Kaul, V. Cytological Status of Allium hookeri Thwaites (2n = 22). Genet. Resour. Crop Evol. 2011, 58, 1041–1050. [Google Scholar] [CrossRef]
- Kala, C.P. Ethnomedicinal Botany of the Apatani in the Eastern Himalayan Region of India. J. Ethnobiol. Ethnomed. 2005, 1, 11. [Google Scholar] [CrossRef]
- Bok, S.; Seo, J.; Bae, C.; Kang, B.; Cho, S.; Park, D. Allium hookeri Root Extract Regulates Asthmatic Changes through Immunological Modulation of Th1/Th2-related Factors in an Ovalbumin-induced Asthma Mouse Model. Mol. Med. Rep. 2019, 20, 3215–3223. [Google Scholar] [CrossRef]
- Jun, H.-I.; Jang, H.; Ahn, D.; Kim, D.K.; Yang, J.H.; Yun, B.S.; Kim, Y.S. Isolation and Characterization of Phenolic Compound from Allium hookeri Root for Potential Use as Antioxidant in Foods. Food Sci. Biotechnol. 2015, 24, 2031–2034. [Google Scholar] [CrossRef]
- Rho, S.H.; You, S.; Kim, G.-H.; Park, H.J. Neuroprotective Effect of Allium hookeri against H2O2-Induced PC12 Cell Cytotoxicity by Reducing Oxidative Stress. Food Sci. Biotechnol. 2020, 29, 1519–1530. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Seo, J.H.; Bae, M.S.; Bae, C.S.; Yoo, J.C.; Bang, M.A.; Cho, S.S.; Park, D.H. Antimicrobial Constituents from Allium hookeri Root. Nat. Prod. Commun. 2016, 11, 237–238. [Google Scholar] [CrossRef]
- Roh, S.S.; Kwon, O.J.; Yang, J.H.; Kim, Y.S.; Lee, S.H.; Jin, J.S.; Jeon, Y.D.; Yokozawa, T.; Kim, H.J. Allium hookeri Root Protects Oxidative Stress-Induced Inflammatory Responses and β-Cell Damage in Pancreas of Streptozotocin-Induced Diabetic Rats. BMC Complement. Altern. Med. 2016, 16, 63. [Google Scholar] [CrossRef]
- Park, S.H.; Bae, U.J.; Choi, E.K.; Jung, S.J.; Lee, S.H.; Yang, J.H.; Kim, Y.S.; Jeong, D.Y.; Kim, H.J.; Park, B.H.; et al. A Randomized, Double-Blind, Placebo-Controlled Crossover Clinical Trial to Evaluate the Anti-Diabetic Effects of Allium hookeri Extract in the Subjects with Prediabetes. BMC Complement. Med. Ther. 2020, 20, 211. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Hyen Lee, S.; Sun Jeong, M.; Kim, J.B.; Hee Jang, H.; Choe, J.; Kim, D.W.; Lillehoj, H.S. In Vitro Analysis of the Immunomodulating Effects of Allium hookeri on Lymphocytes, Macrophages, and Tumour Cells. J. Poult. Sci. 2017, 54, 142–148. [Google Scholar] [CrossRef]
- Jang, J.Y.; Lee, M.J.; You, B.R.; Jin, J.S.; Lee, S.H.; Yun, Y.R.; Kim, H.J. Allium hookeri Root Extract Exerts Anti-Inflammatory Effects by Nuclear Factor-ΚB down-Regulation in Lipopolysaccharide-Induced RAW264.7 Cells. BMC Complement. Altern. Med. 2017, 17, 126. [Google Scholar]
- Kim, H.; Rho, S.H.; Lim, J.; Park, H.J.; Jeong, H. Protective Effect of Linoleic Acid against Inflammatory Reactions by Mast Cell via Caspase-1 Cascade Pathways. J. Food Biochem. 2019, 43, e12932. [Google Scholar] [CrossRef]
- Park, H.; Jeong, J.; Hyun, H.; Kim, J.; Kim, H.; Oh, H.; Choi, J.; Hwang, H.; Oh, D.; Kim, J.; et al. Effects of a Hot-Water Extract of Allium hookeri Roots on Bone Formation in Human Osteoblast-Like MG-63 Cells In Vitro and in Rats In Vivo. Planta Med. 2016, 82, 1410–1415. [Google Scholar] [CrossRef]
- Park, S.; No, K.; Lee, J. Anti-Obesity Effect of Allium hookeri Leaf Extract in High-Fat Diet-Fed Mice. J. Med. Food 2018, 21, 254–260. [Google Scholar] [CrossRef]
- Yang, M.H.; Kim, N.H.; Heo, J.D.; Rho, J.R.; Ock, K.J.; Shin, E.C.; Jeong, E.J. Comparative Evaluation of Sulfur Compounds Contents and Antiobesity Properties of Allium hookeri Prepared by Different Drying Methods. Evid.-Based Complement. Altern. Med. 2017, 2017, 2436927. [Google Scholar] [CrossRef]
- Hwang, J.S.; Lee, B.H.; An, X.; Jeong, H.R.; Kim, Y.E.; Lee, I.; Lee, H.; Kim, D.O. Total Phenolics, Total Flavonoids, and Antioxidant Capacity in the Leaves, Bulbs, and Roots of Allium hookeri. Korean J. Food Sci. Technol. 2015, 47, 261–266. [Google Scholar] [CrossRef]
- Park, S.Y.; Je, J.Y.; Ahn, C.B. Phenolic Composition and Hepatoprotective Activities of Allium hookeri Against Hydrogen-Peroxide-Induced Oxidative Stress in Cultured Hepatocytes. J. Food Biochem. 2016, 40, 284–293. [Google Scholar] [CrossRef]
- Li, R.; Wang, Y.F.; Sun, Q.; Hu, H. Bin Chemical Composition and Antimicrobial Activity of the Essential Oil from Allium hookeri Consumed in Xishuangbanna, Southwest China. Nat. Prod. Commun. 2014, 9, 863–864. [Google Scholar] [PubMed]
- Thiyagarajan, J.A.; Mikton, C.; Harwood, R.H.; Gichu, M.; Gaigbe-Togbe, V.; Jhamba, T.; Pokorna, D.; Stoevska, V.; Hada, R.; Steffan, G.S.; et al. The UN Decade of Healthy Ageing: Strengthening Measurement for Monitoring Health and Wellbeing of Older People. Age Ageing 2022, 51, afac147. [Google Scholar] [CrossRef] [PubMed]
- Beard, J.R.; Officer, A.; de Carvalho, I.A.; Sadana, R.; Pot, A.M.; Michel, J.-P.; Lloyd-Sherlock, P.; Epping-Jordan, J.E.; Peeters, G.M.E.E.G.; Mahanani, W.R.; et al. The World Report on Ageing and Health: A Policy Framework for Healthy Ageing. Lancet 2016, 387, 2145–2154. [Google Scholar] [CrossRef] [PubMed]
- Boccardi, V.; Mecocci, P. Senotherapeutics: Targeting Senescent Cells for the Main Age-Related Diseases. Mech. Ageing Dev. 2021, 197, 111526. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Coppé, J.P.; Lam, E.W.F. Cellular Senescence: The Sought or the Unwanted? Trends Mol. Med. 2018, 24, 871–885. [Google Scholar] [CrossRef] [PubMed]
- van Deursen, J.M. The Role of Senescent Cells in Ageing. Nature 2014, 509, 439–446. [Google Scholar] [CrossRef]
- Huang, W.; Hickson, L.J.; Eirin, A.; Kirkland, J.L.; Lerman, L.O. Cellular Senescence: The Good, the Bad and the Unknown. Nat. Rev. Nephrol. 2022, 18, 611–627. [Google Scholar] [CrossRef]
- Jun, J.-I.; Lau, L.F. The Matricellular Protein CCN1 Induces Fibroblast Senescence and Restricts Fibrosis in Cutaneous Wound Healing. Nat. Cell Biol. 2010, 12, 676–685. [Google Scholar] [CrossRef]
- Thompson, E.L.; Pitcher, L.E.; Niedernhofer, L.J.; Robbins, P.D. Targeting Cellular Senescence with Senotherapeutics: Development of New Approaches for Skin Care. Plast. Reconstr. Surg. 2022, 150, 12S–19S. [Google Scholar] [CrossRef] [PubMed]
- Vande Berg, J.S.; Rose, M.A.; Haywood-Reid, P.L.; Rudolph, R.; Payne, W.G.; Robson, M.C. Cultured Pressure Ulcer Fibroblasts Show Replicative Senescence with Elevated Production of Plasmin, Plasminogen Activator Inhibitor-1, and Transforming Growth Factor-β1. Wound Repair Regen. 2005, 13, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Qin, L.; Feng, R.; Hu, G.; Sun, H.; He, Y.; Zhang, R. Emerging Senolytic Agents Derived from Natural Products. Mech. Ageing Dev. 2019, 181, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Luís, C.; Maduro, A.T.; Pereira, P.; Mendes, J.J.; Soares, R.; Ramalho, R. Nutritional Senolytics and Senomorphics: Implications to Immune Cells Metabolism and Aging—From Theory to Practice. Front. Nutr. 2022, 9, 958563. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, H.M.; Kim, J.H.; Lee, J.-H.; Zhang, K.X.; Gao, E.M.; Jeon, J.-S.; Syed, A.S.; Son, R.H.; Kim, J.-Y.; et al. Chemical Constituents of the Ajuga Multiflora Bunge and Their Protective Effects on Dexamethasone-Induced Muscle Atrophy in C2C12 Myotubes. Nat. Prod. Res. 2023, 37, 1978–1985. [Google Scholar] [CrossRef]
- Lehbili, M.; Alabdul Magid, A.; Kabouche, A.; Voutquenne-Nazabadioko, L.; Abedini, A.; Morjani, H.; Gangloff, S.C.; Kabouche, Z. Antibacterial, Antioxidant and Cytotoxic Activities of Triterpenes and Flavonoids from the Aerial Parts of Salvia Barrelieri Etl. Nat. Prod. Res. 2018, 32, 2683–2691. [Google Scholar] [CrossRef]
- Myagchilov, A.V.; Gorovoi, P.G.; Sokolova, L.I. Flavonoids from Serratula Komarovii Iljin (the Asteraceae Family). Russ. J. Bioorg. Chem. 2021, 47, 1418–1423. [Google Scholar] [CrossRef]
- Yanting, L.R.J.B.X.Z. Chrysoeriol-7-O-Beta-D Glucuronic Acid Methyl Ester and Extraction Method and Application Thereof. Patent CN107446009A, 8 December 2017. [Google Scholar]
- Sutthanut, K.; Sripanidkulchai, B.; Yenjai, C.; Jay, M. Simultaneous Identification and Quantitation of 11 Flavonoid Constituents in Kaempferia Parviflora by Gas Chromatography. J. Chromatogr. A 2007, 1143, 227–233. [Google Scholar] [CrossRef]
- van Zadelhoff, A.; Vincken, J.P.; de Bruijn, W.J.C. Facile Amidation of Non-Protected Hydroxycinnamic Acids for the Synthesis of Natural Phenol Amides. Molecules 2022, 27, 2203. [Google Scholar] [CrossRef]
- Zheng, Y.; Su, B.; Wang, Y.; Wang, H.; Liao, H.; Liang, D. New Tyramine- and Aporphine-Type Alkamides with NO Release Inhibitory Activities from Piper puberulum. J. Nat. Prod. 2021, 84, 1316–1325. [Google Scholar] [CrossRef]
- Chen, S.; Snyder, J.K. Diosgenin-Bearing, Molluscicidal Saponins from Allium Vineale: An NMR Approach for the Structural Assignment of Oligosaccharide Units. J. Org. Chem. 1989, 54, 3679–3689. [Google Scholar] [CrossRef]
- Thangnipon, W.; Ngampramuan, S.; Suthprasertporn, N.; Jantrachotechatchawan, C.; Tuchinda, P.; Nobsathian, S. Protective Roles of N-Trans-Feruloyltyramine Against Scopolamine-Induced Cholinergic Dysfunction on Cortex and Hippocampus of Rat Brains. Siriraj Med. J. 2021, 73, 413–422. [Google Scholar] [CrossRef]
- Thangnipon, W.; Suwanna, N.; Kitiyanant, N.; Soi-ampornkul, R.; Tuchinda, P.; Munyoo, B.; Nobsathian, S. Protective Role of N-Trans-Feruloyltyramine against β-Amyloid Peptide-Induced Neurotoxicity in Rat Cultured Cortical Neurons. Neurosci. Lett. 2012, 513, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Efdi, M.; Ohguchi, K.; Akao, Y.; Nozawa, Y.; Koketsu, M.; Ishihara, H. N-Trans-Feruloyltyramine as a Melanin Biosynthesis Inhibitor. Biol. Pharm. Bull. 2007, 30, 1972–1974. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wang, C.; Chen, Z.; Chen, Y.; Santhanam, R.K.; Xue, Z.; Ma, Q.; Guo, Q.; Liu, W.; Zhang, M.; et al. Effects of N-Trans-Feruloyltyramine Isolated from Laba Garlic on Antioxidant, Cytotoxic Activities and H2O2-Induced Oxidative Damage in HepG2 and L02 cells. Food Chem. Toxicol. 2019, 130, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Pu, Z.; Shen, C.; Zhang, W.; Xie, H.; Wang, W. Avenanthramide C from Oats Protects Pyroptosis through Dependent ROS-Induced Mitochondrial Damage by PI3K Ubiquitination and Phosphorylation in Pediatric Pneumonia. J. Agric. Food Chem. 2022, 70, 2339–2353. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, H.; Yang, E.-J.; Lee, S.; Kim, M.-J.; Baek, M.-C.; Lee, B.; Park, P.-H.; Kwon, T.K.; Khang, D.; Song, K.-S.; et al. Avenanthramide C from Germinated Oats Exhibits Anti-Allergic Inflammatory Effects in Mast Cells. Sci. Rep. 2019, 9, 6884. [Google Scholar] [CrossRef]
- Lim, J.S.; Lee, D.Y.; Kim, H.S.; Park, S.C.; Park, J.T.; Kim, H.S.; Oh, W.K.; Cho, K.A. Identification of a Novel Senomorphic Agent, Avenanthramide C, via the Suppression of the Senescence-Associated Secretory Phenotype. Mech. Ageing Dev. 2020, 192, 111355. [Google Scholar] [CrossRef]
- Ramasamy, V.S.; Samidurai, M.; Park, H.J.; Wang, M.; Park, R.Y.; Yu, S.Y.; Kang, H.K.; Hong, S.; Choi, W.-S.; Lee, Y.Y.; et al. Avenanthramide-C Restores Impaired Plasticity and Cognition in Alzheimer’s Disease Model Mice. Mol. Neurobiol. 2020, 57, 315–330. [Google Scholar] [CrossRef]








| Pos. | 2 | |
|---|---|---|
| δC a | δH (J in Hz) b | |
| 2 | 165.5 | |
| 3 | 104.5 | 6.93, s |
| 4 | 183.3 | |
| 5 | 158.3 | |
| 6 | 95.6 | 7.20, overlap. |
| 7 | 163.7 | |
| 8 | 100.8 | 7.04, s |
| 9 | 163.4 | |
| 10 | 107.4 | |
| 1′ | 122.4 | |
| 2′ | 129.5 | 7.85, d (5.1) |
| 3′ | 117.4 | 7.19, overlap. |
| 4′ | 163.3 | |
| 5′ | 117.4 | |
| 6′ | 129.5 | |
| 1″ | 100.0 | 6.03, d (7.3) |
| 2″ | 77.8 | 4.65, t (7.8) |
| 3″ | 78.7 | 4.47, t (8.8) |
| 4″ | 73.3 | 4.52, t (8.8) |
| 5″ | 77.5 | 4.88, d (8.5) |
| -C=O | 170.4 | |
| -COOCH3 | 52.6 | 3.62, s |
| 4′-OCH3 | ||
| 1‴ | 103.1 | 6.45, s |
| 2‴ | 72.9 | 4.84, br s |
| 3‴ | 73.2 | 4.58, d (7.7) |
| 4‴ | 74.5 | 4.38, t (9.1) |
| 5‴ | 70.6 | 4.80, m |
| 6‴ | 19.4 | 1.85, d (6.0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doan, T.-P.; Zhang, M.; An, J.-P.; Ponce-Zea, J.-E.; Mai, V.-H.; Ryu, B.; Park, E.-J.; Oh, W.-K. Metabolite Profiling of Allium hookeri Leaves Using UHPLC-qTOF-MS/MS and the Senomorphic Activity of Phenolamides. Nutrients 2023, 15, 5109. https://doi.org/10.3390/nu15245109
Doan T-P, Zhang M, An J-P, Ponce-Zea J-E, Mai V-H, Ryu B, Park E-J, Oh W-K. Metabolite Profiling of Allium hookeri Leaves Using UHPLC-qTOF-MS/MS and the Senomorphic Activity of Phenolamides. Nutrients. 2023; 15(24):5109. https://doi.org/10.3390/nu15245109
Chicago/Turabian StyleDoan, Thi-Phuong, Mi Zhang, Jin-Pyo An, Jorge-Eduardo Ponce-Zea, Van-Hieu Mai, Byeol Ryu, Eun-Jin Park, and Won-Keun Oh. 2023. "Metabolite Profiling of Allium hookeri Leaves Using UHPLC-qTOF-MS/MS and the Senomorphic Activity of Phenolamides" Nutrients 15, no. 24: 5109. https://doi.org/10.3390/nu15245109
APA StyleDoan, T.-P., Zhang, M., An, J.-P., Ponce-Zea, J.-E., Mai, V.-H., Ryu, B., Park, E.-J., & Oh, W.-K. (2023). Metabolite Profiling of Allium hookeri Leaves Using UHPLC-qTOF-MS/MS and the Senomorphic Activity of Phenolamides. Nutrients, 15(24), 5109. https://doi.org/10.3390/nu15245109