High-Throughput CAMP Assay (HiTCA): A Novel Tool for Evaluating the Vitamin D-Dependent Antimicrobial Response
Abstract
:1. Introduction
2. Materials and Methods
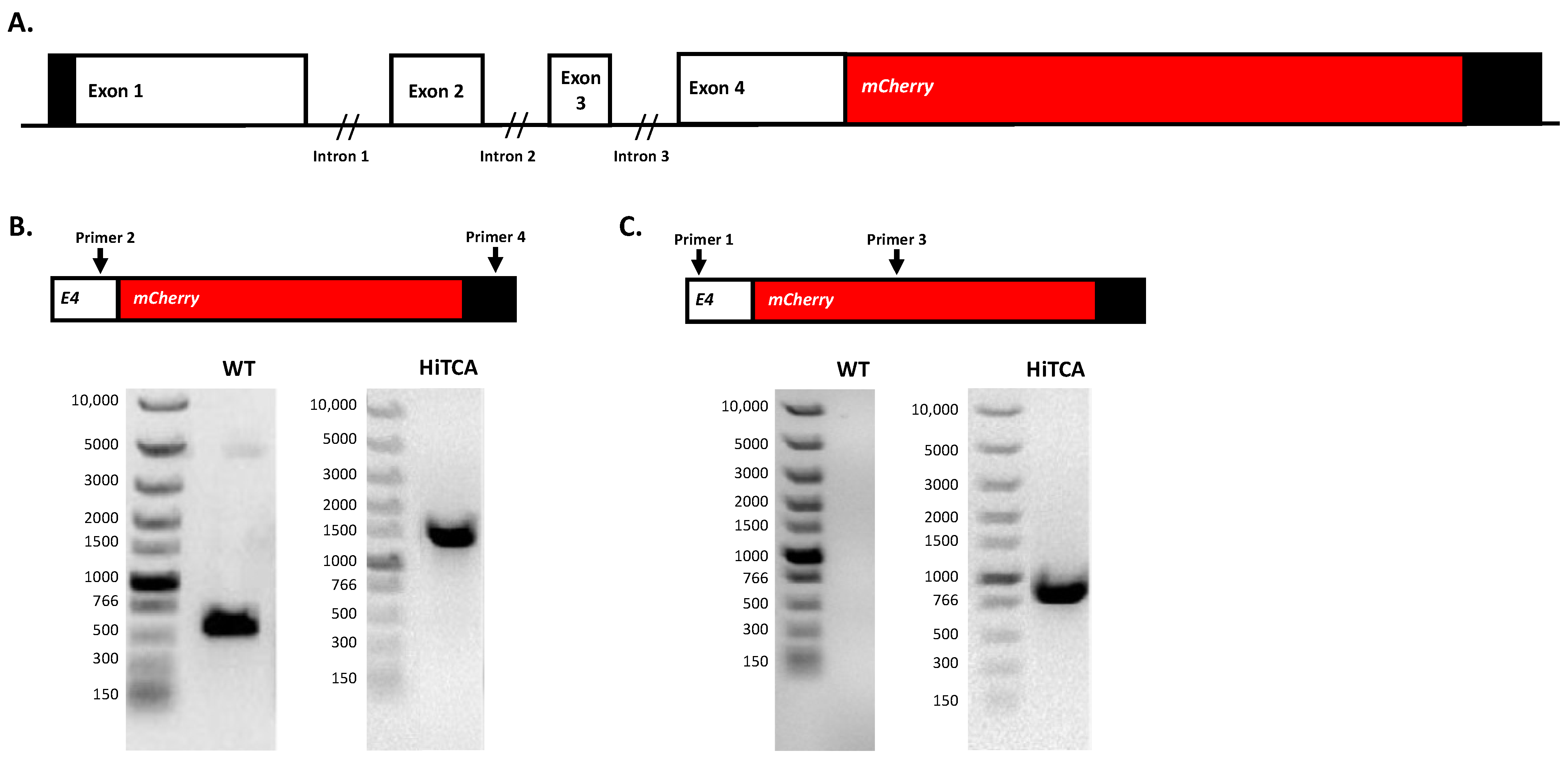
2.1. Insertion of mCherry at 3′ End of CAMP Gene in Human SC Cells
2.2. Cell Culture and Maintenance
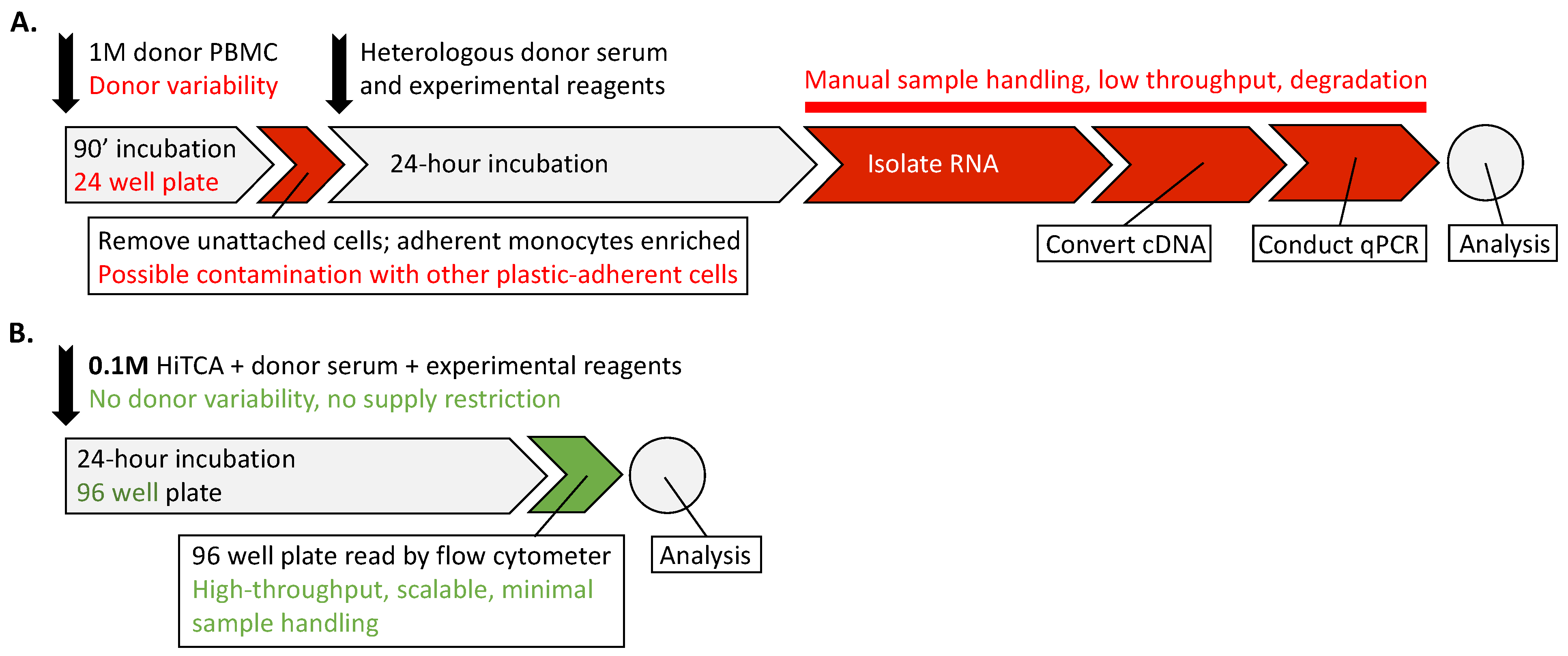
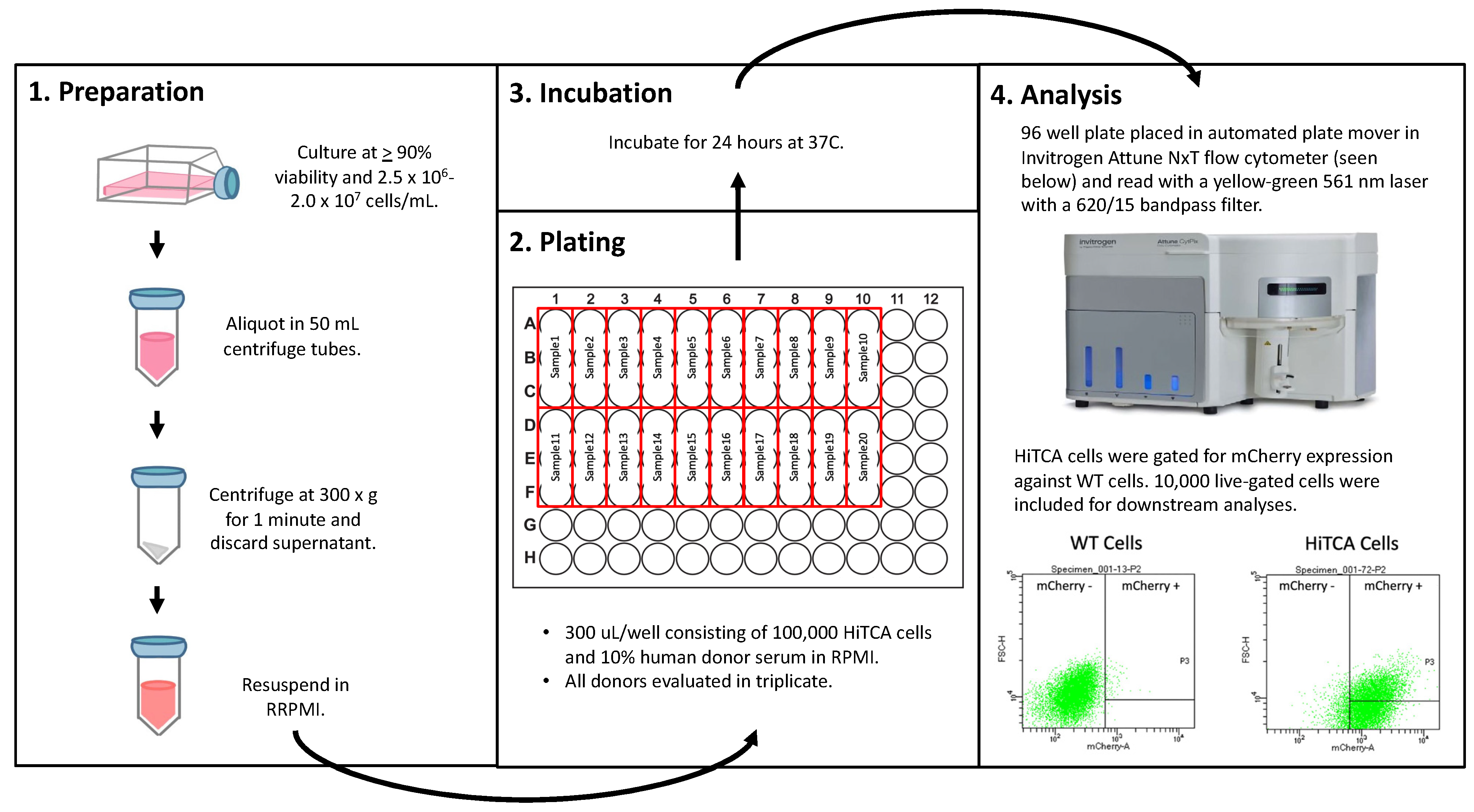
2.3. HiTCA Protocol
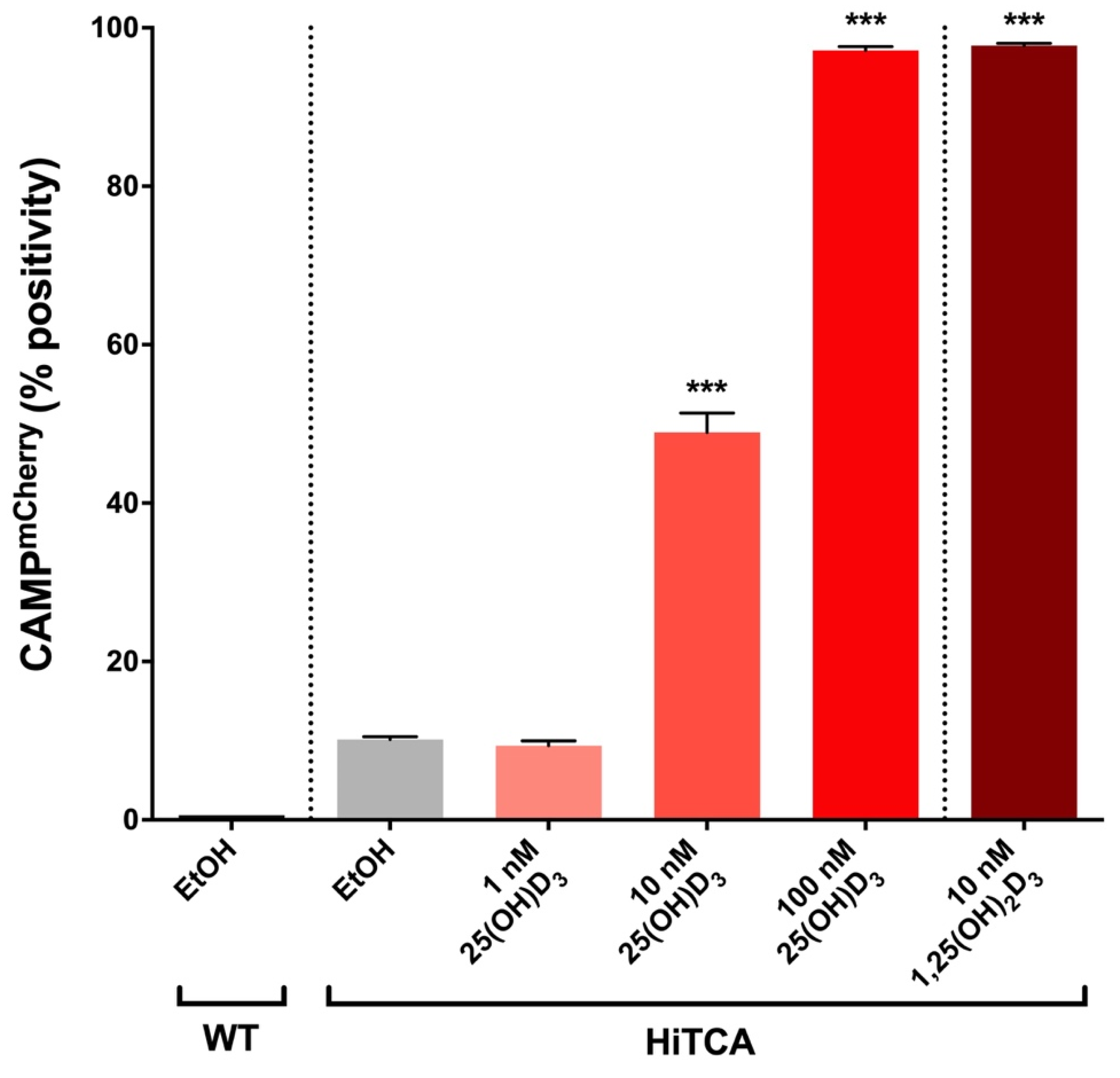
2.4. Induction of CAMPmCherry in Human SC Cell Clones by 25(OH)D3 and 1,25(OH)2D3
2.5. Application of Pooled Human Serum to HiTCA
2.6. Application of Donor-Specific Serum to HiTCA
3. Results
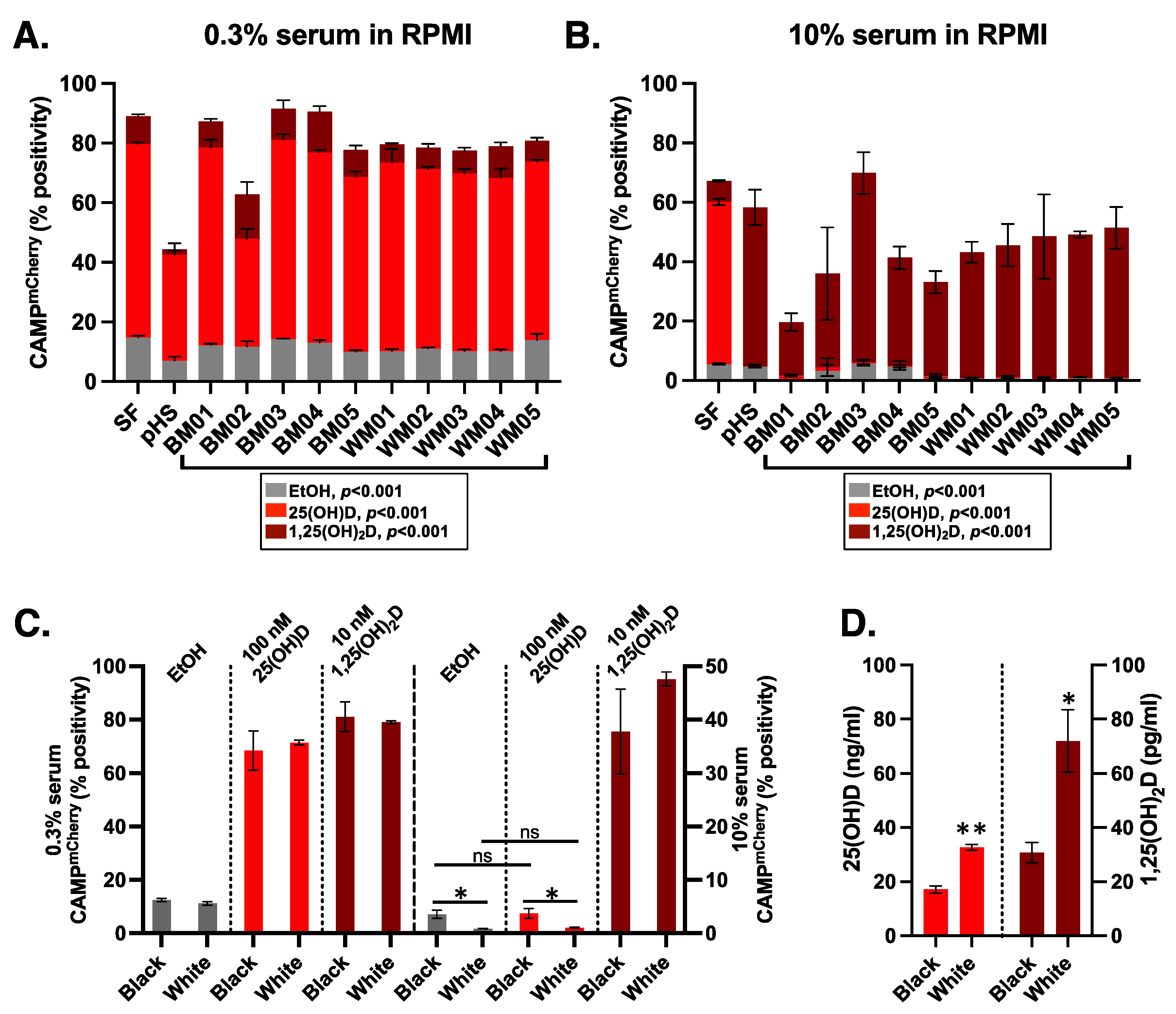
3.1. Endogenous 25(OH)D3-to-1,25(OH)2D3 Activation in HiTCA Cells
3.2. Higher Human Serum Concentration Attenuates the HiTCA Response
3.3. Donor Specific Serum Yielded Variable Responses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chieosilapatham, P.; Ikeda, S.; Ogawa, H.; Niyonsaba, F. Tissue-specific regulation of innate immune responses by human cathelicidin LL-37. Curr. Pharm. Des. 2018, 24, 1079–1091. [Google Scholar] [CrossRef] [PubMed]
- Wang, C. Human cathelicidin inhibits SARS-CoV-2 infection: Killing two birds with one stone. ACS Infect. Dis. 2021, 7, 1545–1554. [Google Scholar] [CrossRef] [PubMed]
- Hertog, A.L.D.; Van Marle, J.; Van Veen, H.A.; Hof, W.V.; Bolscher, J.G.M.; Veerman, E.C.I.; Amerongen, A.V.N. Candidacidal effects of two antimicrobial peptides: Histatin 5 causes small membrane defects, but LL-37 causes massive disruption of the cell membrane. Biochem. J. 2005, 388, 689–695. [Google Scholar] [CrossRef] [Green Version]
- Shahmiri, M.; Enciso, M.; Adda, C.G.; Smith, B.J.; Perugini, M.A.; Mechler, A. Membrane core-specific antimicrobial action of cathelicidin LL-37 peptide switches between pore and nanofibre formation. Sci. Rep. 2016, 6, 38184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howell, M.D.; Jones, J.F.; Kisich, K.O.; Streib, J.E.; Gallo, R.L.; Leung, D.Y.M. Selective killing of vaccinia virus by LL-37: Implications for eczema vaccinatum. J. Immunol. 2004, 172, 1763–1767. [Google Scholar] [CrossRef]
- Scott, M.G.; Davidson, D.J.; Gold, M.R.; Bowdish, D.; Hancock, R.E.W. The human antimicrobial peptide LL-37 is a multifunctional modulator of innate immune responses. J. Immunol. 2002, 169, 3883–3891. [Google Scholar] [CrossRef] [Green Version]
- Durr, U.H.; Sudheendra, U.S.; Ramamoorthy, A. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim. Biophys. Acta 2006, 1758, 1408–1425. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Ma, N.; Johnston, L.J.; Ma, X. Dietary nutrients mediate intestinal host defense peptide expression. Adv. Nutr. Int. Rev. J. 2020, 11, 92–102. [Google Scholar] [CrossRef]
- Gombart, A.F.; Borregaard, N.; Koeffler, H.P. Human cathelicidin antimicrobial peptide (CAMP) gene is a direct target of the vitamin D receptor and is strongly up-regulated in myeloid cells by 1,25-dihydroxyvitamin D3. FASEB J. 2005, 19, 1067–1077. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.T.; Stenger, S.; Li, H.; Wenzel, L.; Tan, B.H.; Krutzik, S.R.; Ochoa, M.T.; Schauber, J.; Wu, K.; Meinken, C.; et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science 2006, 311, 1770–1773. [Google Scholar] [CrossRef]
- Adams, J.S.; Hewison, M. Unexpected actions of vitamin D: New perspectives on the regulation of innate and adaptive immunity. Nat. Clin. Pract. Endocrinol. Metab. 2008, 4, 80–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauhan, V.S. Vitamin D and the Immune System, in Vitamin D; Özdemir, Ö., Ed.; Intech Open: London, UK, 2021. [Google Scholar]
- Hewison, M. Vitamin D and innate and adaptive immunity. Vitam. Horm. 2011, 86, 23–62. [Google Scholar] [CrossRef]
- Cannell, J.J.; Vieth, R.; Umhau, J.C.; Holick, M.F.; Grant, W.B.; Madronich, S.; Garland, C.F.; Giovannucci, E. Epidemic influenza and vitamin D. Epidemiol. Infect. 2006, 134, 1129–1140. [Google Scholar] [CrossRef] [PubMed]
- Laaksi, I.; Ruohola, J.-P.; Tuohimaa, P.; Auvinen, A.; Haataja, R.; Pihlajamäki, H.; Ylikomi, T. An association of serum vitamin D concentrations < 40 nmol/L with acute respiratory tract infection in young Finnish men. Am. J. Clin. Nutr. 2007, 86, 714–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leow, L.; Simpson, T.; Cursons, R.T.; Karalus, N.; Hancox, R.J. Vitamin D, innate immunity and outcomes in community acquired pneumonia. Respirology 2011, 16, 611–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bodnar, L.M.; Krohn, M.A.; Simhan, H.N. Maternal vitamin D deficiency is associated with bacterial vaginosis in the first trimester of pregnancy. J. Nutr. 2009, 139, 1157–1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, M.; Daniels, B.; Gunawardene, S.; Robbins, G.K. High frequency of vitamin D deficiency in ambulatory HIV-positive patients. AIDS Res. Hum. Retroviruses 2009, 25, 9–14. [Google Scholar] [CrossRef]
- Villamor, E. A potential role for vitamin D on HIV infection? Nutr. Rev. 2006, 64, 226–233. [Google Scholar] [CrossRef]
- Talat, N.; Perry, S.; Parsonnet, J.; Dawood, G.; Hussain, R. Vitamin D deficiency and tuberculosis progression. Emerg. Infect. Dis. 2010, 16, 853–855. [Google Scholar] [CrossRef]
- Gunville, C.F.; Mourani, P.M.; Ginde, A.A. The role of vitamin D in prevention and treatment of infection. Inflamm. Allergy-Drug Targets 2013, 12, 239–245. [Google Scholar] [CrossRef]
- Grant, W.B. Evidence that vitamin D supplementation could reduce risk of influenza and COVID-19 infections and deaths. Nutrients 2020, 12, 988. [Google Scholar] [CrossRef] [Green Version]
- Kaufman, H.W. SARS-CoV-2 positivity rates associated with circulating 25-hydroxyvitamin D levels. PLoS ONE 2020, 15, e0239252. [Google Scholar] [CrossRef]
- Jude, E.B.; Ling, S.F.; Allcock, R.; Yeap, B.X.Y.; Pappachan, J.M. Vitamin D deficiency is associated with higher hospitalization risk from COVID-19: A retrospective case-control study. J. Clin. Endocrinol. Metab. 2021, 106, e4708–e4715. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Joshi, A.; Leopold, K.; Jackson, S.; Christensen, S.; Nayfeh, T.; Mohammed, K.; Creo, A.; Tebben, P.; Kumar, S. Association of vitamin D deficiency with COVID-19 infection severity: Systematic review and meta-analysis. Clin. Endocrinol. 2022, 96, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Hastie, C.E.; Mackay, D.F.; Ho, F.; Celis-Morales, C.A.; Katikireddi, S.V.; Niedzwiedz, C.L.; Jani, B.D.; Welsh, P.; Mair, F.S.; Gray, S.R.; et al. Vitamin D concentrations and COVID-19 infection in UK biobank. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Raisi-Estabragh, Z.; McCracken, C.; Bethell, M.S.; Cooper, J.; Cooper, C.; Caulfield, M.J.; Munroe, P.B.; Harvey, N.C.; Petersen, S.E. Greater risk of severe COVID-19 in black, Asian and minority ethnic populations is not explained by cardiometabolic, socioeconomic or behavioural factors, or by 25(OH)-vitamin D status: Study of 1326 cases from the UK biobank. J. Public Health 2020, 42, 451–460. [Google Scholar] [CrossRef]
- Carter, S. The medicalization of sunlight in the early twentieth century. J. Hist. Sociol. 2012, 25, 83–105. [Google Scholar] [CrossRef] [PubMed]
- Bergman, P. Vitamin D and respiratory tract infections: A systematic review and meta-analysis of randomized controlled trials. PLoS ONE 2013, 8, e65835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urashima, M.; Segawa, T.; Okazaki, M.; Kurihara, M.; Wada, Y.; Ida, H. Randomized trial of vitamin D supplementation to prevent seasonal influenza A in schoolchildren. Am. J. Clin. Nutr. 2010, 91, 1255–1260. [Google Scholar] [CrossRef] [Green Version]
- Laaksi, I.; Ruohola, J.; Mattila, V.; Auvinen, A.; Ylikomi, T.; Pihlajamäki, H. Vitamin D supplementation for the prevention of acute respiratory tract infection: A randomized, double-blinded trial among young Finnish men. J. Infect. Dis. 2010, 202, 809–814. [Google Scholar] [CrossRef] [Green Version]
- Martineau, A.R. Vitamin D supplementation to prevent acute respiratory tract infections: Systematic review and meta-analysis of individual participant data. BMJ 2017, 356, i6583. [Google Scholar] [CrossRef] [Green Version]
- Castillo, M.E. Effect of calcifediol treatment and best available therapy versus best available therapy on intensive care unit admission and mortality among patients hospitalized for COVID-19: A pilot randomized clinical study. J. Steroid Biochem. Mol. Biol. 2020, 203, 105751. [Google Scholar] [CrossRef]
- Bishop, C.W.; Ashfaq, A.; Melnick, J.Z.; Vazquez-Escarpanter, E.; Fialkow, J.A.; Strugnell, S.A.; Choe, J.; Kalantar-Zadeh, K.; Federman, N.C.; Ng, D.; et al. REsCue trial: Randomized controlled clinical trial with extended-release calcifediol in symptomatic COVID-19 outpatients. Nutrition 2022, 107, 111899. [Google Scholar] [CrossRef]
- Rastogi, A.; Bhansali, A.; Khare, N.; Suri, V.; Yaddanapudi, N.; Sachdeva, N.; Puri, G.D.; Malhotra, P. Short term, high-dose vitamin D supplementation for COVID-19 disease: A randomised, placebo-controlled, study (SHADE study). Postgrad. Med. J. 2020, 98, 87–90. [Google Scholar] [CrossRef]
- Adams, J.S.; Ren, S.; Liu, P.T.; Chun, R.F.; Lagishetty, V.; Gombart, A.F.; Borregaard, N.; Modlin, R.L.; Hewison, M. Vitamin D-directed rheostatic regulation of monocyte antibacterial responses. J. Immunol. 2009, 182, 4289–4295. [Google Scholar] [CrossRef] [Green Version]
- Chun, R.F.; Lauridsen, A.L.; Suon, L.; Zella, L.A.; Pike, J.W.; Modlin, R.L.; Martineau, A.R.; Wilkinson, R.; Adams, J.; Hewison, M. Vitamin D-binding protein directs monocyte responses to 25-hydroxy- and 1,25-dihydroxyvitamin D. J. Clin. Endocrinol. Metab. 2010, 95, 3368–3376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zavala, K.; Gottlieb, C.A.; Teles, R.; Adams, J.S.; Hewison, M.; Modlin, R.L.; Liu, P.T. Intrinsic activation of the vitamin D antimicrobial pathway by M. leprae infection is inhibited by type I IFN. PLoS Negl. Trop. Dis. 2018, 12, e0006815. [Google Scholar] [CrossRef]
- Chun, R.F.; Liu, N.Q.; Lee, T.; Schall, J.I.; Denburg, M.R.; Rutstein, R.M.; Adams, J.S.; Zemel, B.S.; Stallings, V.A.; Hewison, M. Vitamin D supplementation and antibacterial immune responses in adolescents and young adults with HIV/AIDS. J. Steroid Biochem. Mol. Biol. 2015, 148, 290–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agerberth, B. The human antimicrobial and chemotactic peptides LL-37 and alpha-defensins are expressed by specific lymphocyte and monocyte populations. Blood 2000, 96, 3086–3093. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, M.C. A simple method for human peripheral blood monocyte isolation. Mem. Inst. Oswaldo. Cruz. 2000, 95, 221–223. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, C.; Taylor, A.E.; Hassan-Smith, Z.K.; Adams, J.S.; Stewart, P.M.; Hewison, M.; Keevil, B.G. High throughput LC-MS/MS method for the simultaneous analysis of multiple vitamin D analytes in serum. J. Chromatogr. B 2016, 1014, 56–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bower, J. 8th GCC: Consolidated feedback to US FDA on the 2013 draft FDA guidance on bioanalytical method validation. Bioanalysis 2014, 6, 2957–2963. [Google Scholar] [CrossRef]
- Harris, S.S. Vitamin D and African Americans. J. Nutr. 2006, 136, 1126–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawakami, M.; Imawari, M.; Goodman, D.S. Quantitative studies of the interaction of cholecalciferol (vitamin D3) and its metabolites with different genetic variants of the serum binding protein for these sterols. Biochem. J. 1979, 179, 413–423. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.J.; Zhang, F.; Richards, J.B.; Kestenbaum, B.; van Meurs, J.B.; Berry, D.; Kiel, D.P.; Streeten, E.A.; Ohlsson, C.; Koller, D.L.; et al. Common genetic determinants of vitamin D insufficiency: A genome-wide association study. Lancet 2010, 376, 180–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bikle, D. Vitamin D metabolites in captivity? Should we measure free or total 25(OH)D to assess vitamin D status? J. Steroid Biochem. Mol. Biol. 2017, 173, 105–116. [Google Scholar] [CrossRef] [Green Version]
- Boutin, B.; Galbraith, R.M.; Arnaud, P. Comparative affinity of the major genetic variants of human group-specific component (vitamin D-binding protein) for 25-(OH) vitamin D. J. Steroid Biochem. 1989, 32, 59–63. [Google Scholar] [CrossRef]
- Nielson, C.M. Free 25-hydroxyvitamin D: Impact of vitamin D binding protein assays on racial-genotypic associations. J. Clin. Endocrinol. Metab. 2016, 101, 2226–2234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chun, R.; Liu, P.T.; Modlin, R.L.; Adams, J.S.; Hewison, M. Impact of vitamin D on immune function: Lessons learned from genome-wide analysis. Front. Physiol. 2014, 5, 151. [Google Scholar] [CrossRef] [Green Version]
- Edfeldt, K.; Liu, P.T.; Chun, R.; Fabri, M.; Schenk, M.; Wheelwright, M.; Keegan, C.; Krutzik, S.R.; Adams, J.S.; Hewison, M.; et al. T-cell cytokines differentially control human monocyte antimicrobial responses by regulating vitamin D metabolism. Proc. Natl. Acad. Sci. USA 2010, 107, 22593–22598. [Google Scholar] [CrossRef] [Green Version]
- Krutzik, S.R. IL-15 links TLR2/1-induced macrophage differentiation to the vitamin D-dependent antimicrobial pathway. J. Immunol. 2008, 181, 7115–7120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.T.; Wheelwright, M.; Teles, R.; Komisopoulou, E.; Edfeldt, K.; Ferguson, B.; Mehta, M.D.; Vazirnia, A.; Rea, T.H.; Sarno, E.N.; et al. MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat. Med. 2012, 18, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Chen, J.; Ren, Y.; Fan, L.; Xiang, W.; He, X. Exosomal miR-122 promotes adipogenesis and aggravates obesity through the VDR/SREBF1 axis. Obesity 2022, 30, 666–679. [Google Scholar] [CrossRef] [PubMed]
- Carlberg, C.; Haq, A. The concept of the personal vitamin D response index. J. Steroid Biochem. Mol. Biol. 2018, 175, 12–17. [Google Scholar] [CrossRef]

| Sample | Race | Sex | Age | 25(OH)D (ng/mL) | 1,25D (pg/mL) |
|---|---|---|---|---|---|
| BM01 | Black | Male | 26 | 18.45 | 35.66 |
| BM02 | Black | Male | 38 | 21.05 | 35.66 |
| BM03 | Black | Male | 38 | 18.68 | 40.09 |
| BM04 | Black | Male | 37 | 14.38 | 24.96 |
| BM05 | Black | Male | 26 | 13.13 | 17.39 |
| WM01 | White | Male | 29 | 31.92 | 76.75 |
| WM02 | White | Male | 24 | 29.15 | 67.58 |
| WM03 | White | Male | 38 | 34.29 | 107.15 |
| WM04 | White | Male | 39 | 36.24 | 80.32 |
| WM05 | White | Male | 27 | 32.02 | 28.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gottlieb, C.; Henrich, M.; Liu, P.T.; Yacoubian, V.; Wang, J.; Chun, R.; Adams, J.S. High-Throughput CAMP Assay (HiTCA): A Novel Tool for Evaluating the Vitamin D-Dependent Antimicrobial Response. Nutrients 2023, 15, 1380. https://doi.org/10.3390/nu15061380
Gottlieb C, Henrich M, Liu PT, Yacoubian V, Wang J, Chun R, Adams JS. High-Throughput CAMP Assay (HiTCA): A Novel Tool for Evaluating the Vitamin D-Dependent Antimicrobial Response. Nutrients. 2023; 15(6):1380. https://doi.org/10.3390/nu15061380
Chicago/Turabian StyleGottlieb, Carter, Mason Henrich, Philip T. Liu, Vahe Yacoubian, Jeffery Wang, Rene Chun, and John S. Adams. 2023. "High-Throughput CAMP Assay (HiTCA): A Novel Tool for Evaluating the Vitamin D-Dependent Antimicrobial Response" Nutrients 15, no. 6: 1380. https://doi.org/10.3390/nu15061380
APA StyleGottlieb, C., Henrich, M., Liu, P. T., Yacoubian, V., Wang, J., Chun, R., & Adams, J. S. (2023). High-Throughput CAMP Assay (HiTCA): A Novel Tool for Evaluating the Vitamin D-Dependent Antimicrobial Response. Nutrients, 15(6), 1380. https://doi.org/10.3390/nu15061380






