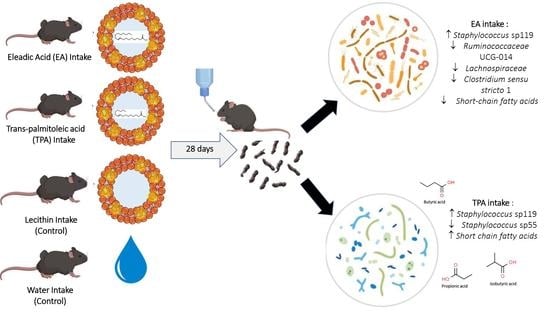
Industrial and Ruminant Trans-Fatty Acids-Enriched Diets Differentially Modulate the Microbiome and Fecal Metabolites in C57BL/6 Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. tFA Preparation
2.3. Animals and Diets
2.4. 16S rRNA Sequencing and Amplicon Sequence Variant (ASV) Processing
2.5. Metabolites Analysis
2.5.1. Analysis of SCFA
2.5.2. Analysis of Other Metabolites
2.6. Statistical Analyses
2.7. Machine Learning Analyses
3. Results
3.1. Fatty Acid Composition of Vesicles Is Stable
3.2. Animal Weight Is Similar in All Groups
3.3. tFA Intake Does Not Impact Alpha Diversity but Modifies Beta Diversity
3.4. tFA Intake Impacts Abundance of Select ASVs
3.5. tFA Intake Impacts on Fecal Metabolites
3.5.1. Lipids and Fatty Acids
3.5.2. Carbohydrates and Derivatives
3.5.3. Amino Acids and Derivatives
3.5.4. Vitamins
3.5.5. Bile Acids
3.5.6. Purine Compounds
3.5.7. Organic Compounds
3.6. Features Identified from Machine Learning Analysis as Markers of TFA Intake
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ge, Y.; Liu, W.; Tao, H.; Zhang, Y.; Liu, L.; Liu, Z.; Qiu, B.; Xu, T. Effect of industrial trans-fatty acids-enriched diet on gut microbiota of C57BL/6 mice. Eur. J. Nutr. 2019, 58, 2625–2638. [Google Scholar] [CrossRef] [PubMed]
- Kuhnt, K.; Degen, C.; Jahreis, G. Evaluation of the impact of ruminant trans fatty acids on human health: Important aspects to consider. Crit. Rev. Food Sci. Nutr. 2016, 56, 1964–1980. [Google Scholar] [CrossRef] [PubMed]
- Bendsen, N.; Christensen, R.; Bartels, E.; Astrup, A. Consumption of industrial and ruminant trans fatty acids and risk of coronary heart disease: A systematic review and meta-analysis of cohort studies. Eur. J. Clin. Nutr. 2011, 65, 773–783. [Google Scholar] [CrossRef]
- O’Connor, S.; Rudkowska, I. Dietary Fatty Acids and the Metabolic Syndrome: A Personalized Nutrition Approach. Adv. Food Nutr. Res. 2019, 87, 43–146. [Google Scholar] [PubMed]
- Micha, R.; Mozaffarian, D. Trans fatty acids: Effects on metabolic syndrome, heart disease and diabetes. Nat. Rev. Endocrinol. 2009, 5, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Mozaffarian, D.; Cao, H.; King, I.B.; Lemaitre, R.N.; Song, X.; Siscovick, D.S.; Hotamisligil, G.S. Trans-palmitoleic acid, metabolic risk factors, and new-onset diabetes in US adults: A cohort study. Ann. Intern. Med. 2010, 153, 790–799. [Google Scholar] [CrossRef] [Green Version]
- Khorraminezhad, L.; Leclercq, M.; O’Connor, S.; Julien, P.; Weisnagel, S.J.; Gagnon, C.; Droit, A.; Rudkowska, I. Dairy product intake modifies gut microbiota composition among hyperinsulinemic individuals. Eur. J. Nutr. 2021, 60, 159–167. [Google Scholar] [CrossRef]
- Chen, X.-Q.; Lv, X.-Y.; Liu, S.-J. Baitouweng decoction alleviates dextran sulfate sodium-induced ulcerative colitis by regulating intestinal microbiota and the IL-6/STAT3 signaling pathway. J. Ethnopharmacol. 2021, 265, 113357. [Google Scholar]
- Chávez-Carbajal, A.; Nirmalkar, K.; Pérez-Lizaur, A.; Hernández-Quiroz, F.; Ramírez-del-Alto, S.; García-Mena, J.; Hernández-Guerrero, C. Gut microbiota and predicted metabolic pathways in a sample of Mexican women affected by obesity and obesity plus metabolic syndrome. Int. J. Mol. Sci. 2019, 20, 438. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Wang, J.; He, T.; Becker, S.; Zhang, G.; Li, D.; Ma, X. Butyrate: A Double-Edged Sword for Health? Adv. Nutr. 2018, 9, 21–29. [Google Scholar] [CrossRef] [Green Version]
- Sanada, S.; Suzuki, T.; Nagata, A.; Hashidume, T.; Yoshikawa, Y.; Miyoshi, N. Intestinal microbial metabolite stercobilin involvement in the chronic inflammation of ob/ob mice. Sci. Rep. 2020, 10, 6479. [Google Scholar] [CrossRef] [Green Version]
- Mosele, J.I.; Gosalbes, M.J.; Macià, A.; Rubió, L.; Vázquez-Castellanos, J.F.; Jimenez Hernandez, N.; Moya, A.; Latorre, A.; Motilva, M.J. Effect of daily intake of pomegranate juice on fecal microbiota and feces metabolites from healthy volunteers. Mol. Nutr. Food Res. 2015, 59, 1942–1953. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhao, K.; Yang, X.; Zhao, Y. Gut microbiota and metabolome response of Decaisnea insignis seed oil on metabolism disorder induced by excess alcohol consumption. J. Agric. Food Chem. 2019, 67, 10667–10677. [Google Scholar] [CrossRef] [PubMed]
- Kasper, J.M.; Johnson, S.B.; Hommel, J.D. Fat Preference: A novel model of eating behavior in rats. JoVE (J. Vis. Exp.) 2014, 88, e51575. [Google Scholar]
- Vasconcelos, T.; Sarmento, B.; Costa, P. Solid dispersions as strategy to improve oral bioavailability of poor water soluble drugs. Drug Discov. Today 2007, 12, 1068–1075. [Google Scholar] [CrossRef]
- Bertrand, N.; Simard, P.; Leroux, J.-C. Serum-stable, long-circulating, pH-sensitive PEGylated liposomes. In Liposomes; Springer: Berlin/Heidelberg, Germany, 2010; pp. 545–558. [Google Scholar]
- Chotard, É.; Mohammadi, F.; Julien, P.; Berthiaume, L.; Rudkowska, I.; Bertrand, N. Drinkable lecithin nanovesicles to study the biological effects of individual hydrophobic macronutrients and food preferences. Food Chem. 2020, 322, 126736. [Google Scholar] [CrossRef]
- Tang, W.; Pan, L.; Cheng, J.; Wang, X.; Zheng, L.; Wang, S.; Zhou, Y.; Wang, H. High-fat-diet-induced gut microbiome changes in mice. Stress Brain 2022, 2, 17–30. [Google Scholar] [CrossRef]
- Hassan, A.M.; Mancano, G.; Kashofer, K.; Liebisch, G.; Farzi, A.; Zenz, G.; Claus, S.P.; Holzer, P. Anhedonia induced by high-fat diet in mice depends on gut microbiota and leptin. Nutr. Neurosci. 2022, 25, 299–312. [Google Scholar] [CrossRef] [Green Version]
- Cheng, M.; Zhang, X.; Zhu, J.; Cheng, L.; Cao, J.; Wu, Z.; Weng, P.; Zheng, X. A metagenomics approach to the intestinal microbiome structure and function in high fat diet-induced obesity mice fed with oolong tea polyphenols. Food Funct. 2018, 9, 1079–1087. [Google Scholar] [CrossRef]
- Stearns, J.C.; Davidson, C.J.; McKeon, S.; Whelan, F.J.; Fontes, M.E.; Schryvers, A.B.; Bowdish, D.M.; Kellner, J.D.; Surette, M.G. Culture and molecular-based profiles show shifts in bacterial communities of the upper respiratory tract that occur with age. ISME J. 2015, 9, 1246–1259. [Google Scholar] [CrossRef] [Green Version]
- Whelan, F.J.; Surette, M.G. A comprehensive evaluation of the sl1p pipeline for 16S rRNA gene sequencing analysis. Microbiome 2017, 5, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whelan, F.J.; Verschoor, C.P.; Stearns, J.C.; Rossi, L.; Luinstra, K.; Loeb, M.; Smieja, M.; Johnstone, J.; Surette, M.G.; Bowdish, D.M. The loss of topography in the microbial communities of the upper respiratory tract in the elderly. Ann. Am. Thorac. Soc. 2014, 11, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, C.; Beall, C.J.; Griffen, A.L.; Leys, E.J. High-resolution ISR amplicon sequencing reveals personalized oral microbiome. Microbiome 2018, 6, 153. [Google Scholar] [CrossRef] [Green Version]
- Francella, C.; Green, M.; Caspani, G.; Lai, J.K.Y.; Rilett, K.C.; Foster, J.A. Microbe–Immune–Stress Interactions Impact Behaviour during Postnatal Development. Int. J. Mol. Sci. 2022, 23, 15064. [Google Scholar] [CrossRef]
- Zhou, H.; He, K.; Chen, J.; Zhang, X. LinDA: Linear models for differential abundance analysis of microbiome compositional data. Genome Biol. 2022, 23, 95. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [Green Version]
- Sannasiddappa, T.H.; Costabile, A.; Gibson, G.R.; Clarke, S.R. The influence of Staphylococcus aureus on gut microbial ecology in an in vitro continuous culture human colonic model system. PLoS ONE 2011, 6, e23227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-García, C.; Sánchez-Quesada, C.; Algarra, I.; Gaforio, J.J. The high-fat diet based on extra-virgin olive oil causes dysbiosis linked to colorectal cancer prevention. Nutrients 2020, 12, 1705. [Google Scholar] [CrossRef]
- Hsu, C.N.; Hou, C.Y.; Chang-Chien, G.P.; Lin, S.; Tain, Y.L. Maternal Garlic Oil Supplementation Prevents High-Fat Diet-Induced Hypertension in Adult Rat Offspring: Implications of H2S-Generating Pathway in the Gut and Kidneys. Mol. Nutr. Food Res. 2021, 65, 2001116. [Google Scholar] [CrossRef]
- Sepp, E.; Kolk, H.; Lõivukene, K.; Mikelsaar, M. Higher blood glucose level associated with body mass index and gut microbiota in elderly people. Microb. Ecol. Health Dis. 2014, 25, 22857. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Guo, J.; Zhao, C.; Jiang, P.; Maimai, T.; Yanyi, L.; Cao, Y.; Fu, Y.; Zhang, N. The gut microbiota contributes to the development of Staphylococcus aureus-induced mastitis in mice. ISME J. 2020, 14, 1897–1910. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Sim, J.X.; Lee, W.L.; Cui, L.; Chan, Y.F.; Chang, E.D.; Teh, Y.E.; Zhang, A.-N.; Armas, F.; Chandra, F. Gut Ruminococcaceae levels at baseline correlate with risk of antibiotic-associated diarrhea. Iscience 2022, 25, 103644. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar]
- Hu, B.; Ye, C.; Leung, E.L.-H.; Zhu, L.; Hu, H.; Zhang, Z.; Zheng, J.; Liu, H. Bletilla striata oligosaccharides improve metabolic syndrome through modulation of gut microbiota and intestinal metabolites in high fat diet-fed mice. Pharmacol. Res. 2020, 159, 104942. [Google Scholar] [CrossRef]
- Horne, R.G.; Yu, Y.; Zhang, R.; Abdalqadir, N.; Rossi, L.; Surette, M.; Sherman, P.M.; Adeli, K. High fat-high fructose diet-induced changes in the gut microbiota associated with dyslipidemia in Syrian hamsters. Nutrients 2020, 12, 3557. [Google Scholar] [CrossRef] [PubMed]
- Rossell, J.; Brindefalk, B.; Baena-Fustegueras, J.A.; Peinado-Onsurbe, J.; Udekwu, K.I. Diet change affects intestinal microbiota restoration and improves vertical sleeve gastrectomy outcome in diet-induced obese rats. Eur. J. Nutr. 2020, 59, 3555–3564. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Wu, M.; Xiong, W.; Li, J.; An, Y.; Ren, J.; Xie, Y.; Xue, H.; Yan, D.; Li, M. Saikosaponin-d ameliorates dextran sulfate sodium-induced colitis by suppressing NF-κB activation and modulating the gut microbiota in mice. Int. Immunopharmacol. 2020, 81, 106288. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, Q.; Ma, W.; Tian, F.; Shen, H.; Zhou, M. A combination of quercetin and resveratrol reduces obesity in high-fat diet-fed rats by modulation of gut microbiota. Food Funct. 2017, 8, 4644–4656. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.K.; Vasquez, R.; Kim, S.H.; Lee, J.H.; Kim, E.J.; Hong, S.-K.; Kang, D.-K. Neoagarooligosaccharides modulate gut microbiota and alleviate body weight gain and metabolic syndrome in high-fat diet-induced obese rats. J. Funct. Foods 2022, 88, 104869. [Google Scholar] [CrossRef]
- Liu, Y.; Li, S.; Wang, X.; Xing, T.; Li, J.; Zhu, X.; Zhang, L.; Gao, F. Microbiota populations and short-chain fatty acids production in cecum of immunosuppressed broilers consuming diets containing γ-irradiated Astragalus polysaccharides. Poult. Sci. 2021, 100, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Chuang, S.-T.; Li, K.-Y.; Tu, P.-W.; Ho, S.-T.; Hsu, C.-C.; Hsieh, J.-C.; Chen, M.-J. Investigating the reciprocal interrelationships among the ruminal microbiota, metabolome, and mastitis in early lactating holstein dairy cows. Animals 2021, 11, 3108. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Zhang, K.; Ma, X.; He, P. Clostridium species as probiotics: Potentials and challenges. J. Anim. Sci. Biotechnol. 2020, 11, 24. [Google Scholar] [CrossRef]
- O’keefe, S.J. Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691–706. [Google Scholar] [CrossRef]
- Luo, H.; Yang, R.; Zhao, Y.; Wang, Z.; Liu, Z.; Huang, M.; Zeng, Q. Recent advances and strategies in process and strain engineering for the production of butyric acid by microbial fermentation. Bioresour. Technol. 2018, 253, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Vital, M.; Howe, A.C.; Tiedje, J.M. Revealing the bacterial butyrate synthesis pathways by analyzing (meta) genomic data. MBio 2014, 5, e00889-14. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.Y.; Lee, Y.; Lu, H.; Chou, C.H.; Wang, C. Analysis of gut microbiota and the effect of lauric acid against necrotic enteritis in Clostridium perfringens and Eimeria side-by-side challenge model. PLoS ONE 2019, 14, e0205784. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; Niu, X.; Chen, S.; Wen, J.; Bao, M.; Mohyuddin, S.G.; Yong, Y.; Liu, X.; Wu, L.; Yu, Z.; et al. A Comprehensive Analysis of the Colonic Flora Diversity, Short Chain Fatty Acid Metabolism, Transcripts, and Biochemical Indexes in Heat-Stressed Pigs. Front. Immunol. 2021, 12, 717723. [Google Scholar] [CrossRef]
- Milton-Laskibar, I.; Marcos-Zambrano, L.J.; Gómez-Zorita, S.; de Santa Pau, E.C.; Fernández-Quintela, A.; Martínez, J.A.; Portillo, M.P. Involvement of microbiota and short-chain fatty acids on non-alcoholic steatohepatitis when induced by feeding a hypercaloric diet rich in saturated fat and fructose. Gut Microbiome 2022, 3, e5. [Google Scholar] [CrossRef]
- Martin-Gallausiaux, C.; Marinelli, L.; Blottière, H.M.; Larraufie, P.; Lapaque, N. SCFA: Mechanisms and functional importance in the gut. Proc. Nutr. Soc. 2021, 80, 37–49. [Google Scholar] [CrossRef]
- Den Besten, G.; Van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Z.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009, 58, 1509–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.V.; Frassetto, A.; Kowalik, E.J., Jr.; Nawrocki, A.R.; Lu, M.M.; Kosinski, J.R.; Hubert, J.A.; Szeto, D.; Yao, X.; Forrest, G. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS ONE 2012, 7, e35240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heimann, E.; Nyman, M.; Pålbrink, A.-K.; Lindkvist-Petersson, K.; Degerman, E. Branched short-chain fatty acids modulate glucose and lipid metabolism in primary adipocytes. Adipocyte 2016, 5, 359–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef] [Green Version]
- Canfora, E.E.; Meex, R.C.; Venema, K.; Blaak, E.E. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat. Rev. Endocrinol. 2019, 15, 261–273. [Google Scholar] [CrossRef]
- Tiffany, C.R.; Lee, J.-Y.; Rogers, A.W.; Olsan, E.E.; Morales, P.; Faber, F.; Bäumler, A.J. The metabolic footprint of Clostridia and Erysipelotrichia reveals their role in depleting sugar alcohols in the cecum. Microbiome 2021, 9, 174. [Google Scholar] [CrossRef]
- Belinda, T.J. Significance of riboflavin (vitamin-B2) for health. J. Pharm. Sci. Res. 2014, 6, 285. [Google Scholar]
- Croze, M.L.; Géloën, A.; Soulage, C.O. Abnormalities in myo-inositol metabolism associated with type 2 diabetes in mice fed a high-fat diet: Benefits of a dietary myo-inositol supplementation. Br. J. Nutr. 2015, 113, 1862–1875. [Google Scholar] [CrossRef] [Green Version]
- Van Der Drift, C.; Vogels, G. Degradation of purine and pyrimidine by microorganisms. Bacteriol. Rev. 1976, 40, 403–468. [Google Scholar]
- Lan, J.C.; Li, Y.; Mo, Y.; Lu, Z.Y.; Lu, F.; Lin, Q.; Liu, X.S.; Chuan, Z.; Wu, Y. Rhubarb enema decreases circulating Trimethylamine N-oxide level and improves renal fibrosis accompanied with gut microbiota change in chronic kidney disease rats. Front. Pharmacol. 2021, 12, 3509. [Google Scholar]
- Xiao, L.; Liu, Q.; Luo, M.; Xiong, L. Gut Microbiota-Derived Metabolites in Irritable Bowel Syndrome. Front. Cell. Infect. Microbiol. 2021, 880, 729346. [Google Scholar] [CrossRef] [PubMed]
- Badoud, F.; Lam, K.P.; DiBattista, A.; Perreault, M.; Zulyniak, M.A.; Cattrysse, B.; Stephenson, S.; Britz-McKibbin, P.; Mutch, D.M. Serum and adipose tissue amino acid homeostasis in the metabolically healthy obese. J. Proteome Res. 2014, 13, 3455–3466. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Li, Y.; Yun, H.; Zhang, T.; Huang, Y.; Zhou, J.; Yan, H.; Wei, J.; Liu, Y.; Zhang, Z. Lactobacillus maintains healthy gut mucosa by producing l-ornithine. Commun. Biol. 2019, 2, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontana, M.; Pecci, L.; Duprè, S.; Cavallini, D. Antioxidant properties of sulfinates: Protective effect of hypotaurine on peroxynitrite-dependent damage. Neurochem. Res. 2004, 29, 111–116. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [Green Version]
- Berger, K.; Burleigh, S.; Lindahl, M.; Bhattacharya, A.; Patil, P.; Stålbrand, H.; Nordberg Karlsson, E.; Hållenius, F.; Nyman, M.; Adlercreutz, P. Xylooligosaccharides increase bifidobacteria and lachnospiraceae in mice on a high-fat diet, with a concomitant increase in short-chain fatty acids, especially butyric acid. J. Agric. Food Chem. 2021, 69, 3617–3625. [Google Scholar] [CrossRef]
- Li, H.; Liu, F.; Lu, J.; Shi, J.; Guan, J.; Yan, F.; Li, B.; Huo, G. Probiotic mixture of Lactobacillus plantarum strains improves lipid metabolism and gut microbiota structure in high fat diet-fed mice. Front. Microbiol. 2020, 11, 512. [Google Scholar] [CrossRef] [Green Version]
- Telle-Hansen, V.H.; Gaundal, L.; Bastani, N.; Rud, I.; Byfuglien, M.G.; Gjøvaag, T.; Retterstøl, K.; Holven, K.B.; Ulven, S.M.; Myhrstad, M.C. Replacing saturated fatty acids with polyunsaturated fatty acids increases the abundance of Lachnospiraceae and is associated with reduced total cholesterol levels—A randomized controlled trial in healthy individuals. Lipids Health Dis. 2022, 21, 92. [Google Scholar] [CrossRef]
- Chiang, S.-S.; Pan, T.-M. Beneficial effects of Lactobacillus paracasei subsp. paracasei NTU 101 and its fermented products. Appl. Microbiol. Biotechnol. 2012, 93, 903–916. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.-N.; Zhu, J.; Pan, W.-s.; Shen, S.-R.; Shan, W.-G.; Das, U.N. Effects of fish oil with a high content of n-3 polyunsaturated fatty acids on mouse gut microbiota. Arch. Med. Res. 2014, 45, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Shah, N.P. Effects of elaidic acid, a predominant industrial trans fatty acid, on bacterial growth and cell surface hydrophobicity of lactobacilli. J. Food Sci. 2014, 79, M2485–M2490. [Google Scholar] [CrossRef] [PubMed]
- Endo, Y.; Kamisada, S.; Fujimoto, K.; Saito, T. Trans fatty acids promote the growth of some Lactobacillus strains. J. Gen. Appl. Microbiol. 2006, 52, 29–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, U.N. Arachidonic acid and lipoxin A4 as possible endogenous anti-diabetic molecules. Prostaglandins Leukot. Essent. Fat. Acids 2013, 88, 201–210. [Google Scholar] [CrossRef]
- Das, U.N. Is there a role for bioactive lipids in the pathobiology of diabetes mellitus? Front. Endocrinol. 2017, 8, 182. [Google Scholar] [CrossRef] [Green Version]
- McCoy, K.D.; Geuking, M.B.; Ronchi, F. Gut microbiome standardization in control and experimental mice. Curr. Protoc. Immunol. 2017, 117, 23.1.1–23.1.13. [Google Scholar] [CrossRef] [PubMed]
- Dikpati, A.; Mohammadi, F.; Greffard, K.; Quéant, C.; Arnaud, P.; Bastiat, G.; Rudkowska, I.; Bertrand, N. Residual solvents in nanomedicine and lipid-based drug delivery systems: A case study to better understand processes. Pharm. Res. 2020, 37, 149. [Google Scholar] [CrossRef]
- Hoyles, L.; Wallace, R.J. Gastrointestinal Tract: Fat Metabolism in the Colon; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammadi, F.; Green, M.; Tolsdorf, E.; Greffard, K.; Leclercq, M.; Bilodeau, J.-F.; Droit, A.; Foster, J.; Bertrand, N.; Rudkowska, I. Industrial and Ruminant Trans-Fatty Acids-Enriched Diets Differentially Modulate the Microbiome and Fecal Metabolites in C57BL/6 Mice. Nutrients 2023, 15, 1433. https://doi.org/10.3390/nu15061433
Mohammadi F, Green M, Tolsdorf E, Greffard K, Leclercq M, Bilodeau J-F, Droit A, Foster J, Bertrand N, Rudkowska I. Industrial and Ruminant Trans-Fatty Acids-Enriched Diets Differentially Modulate the Microbiome and Fecal Metabolites in C57BL/6 Mice. Nutrients. 2023; 15(6):1433. https://doi.org/10.3390/nu15061433
Chicago/Turabian StyleMohammadi, Farzad, Miranda Green, Emma Tolsdorf, Karine Greffard, Mickael Leclercq, Jean-François Bilodeau, Arnaud Droit, Jane Foster, Nicolas Bertrand, and Iwona Rudkowska. 2023. "Industrial and Ruminant Trans-Fatty Acids-Enriched Diets Differentially Modulate the Microbiome and Fecal Metabolites in C57BL/6 Mice" Nutrients 15, no. 6: 1433. https://doi.org/10.3390/nu15061433
APA StyleMohammadi, F., Green, M., Tolsdorf, E., Greffard, K., Leclercq, M., Bilodeau, J.-F., Droit, A., Foster, J., Bertrand, N., & Rudkowska, I. (2023). Industrial and Ruminant Trans-Fatty Acids-Enriched Diets Differentially Modulate the Microbiome and Fecal Metabolites in C57BL/6 Mice. Nutrients, 15(6), 1433. https://doi.org/10.3390/nu15061433