Molecular Mechanisms for the Carnosine-Induced Activation of Muscle–Brain Interaction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Reagents
2.2. Differentiation of C2C12
2.3. Reverse Transcriptase-Quantitative Polymerase Chain Reaction (RT-qPCR)
2.4. Fluorescent Immunocytochemistry
2.5. Quantitative Evaluation of Neurite Length
2.6. Exosome Isolation and Treatment
2.7. miRNA Microarray Assay
2.8. Statistical Analysis
3. Results
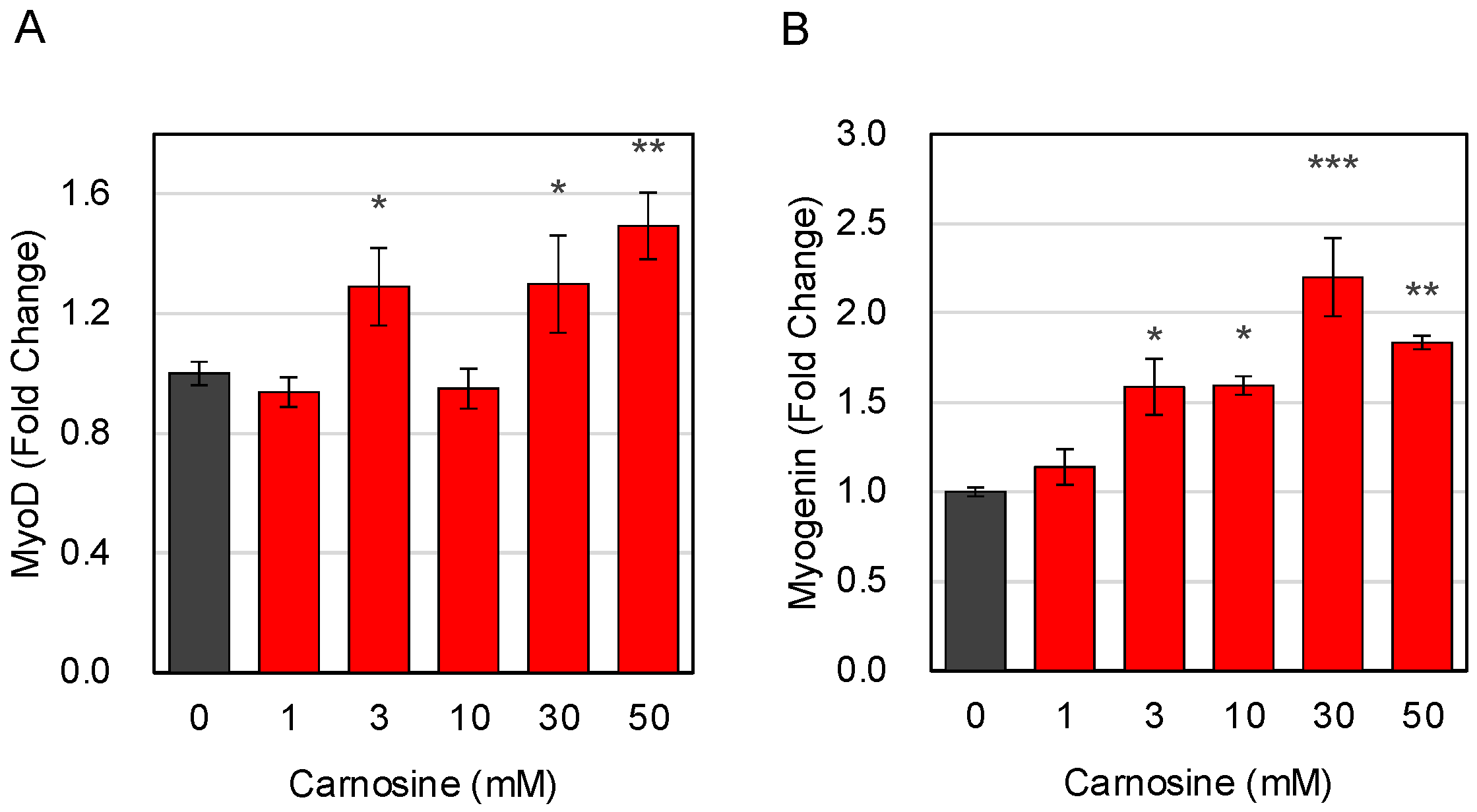
3.1. Effects of Carnosine on Muscle Cell Differentiation
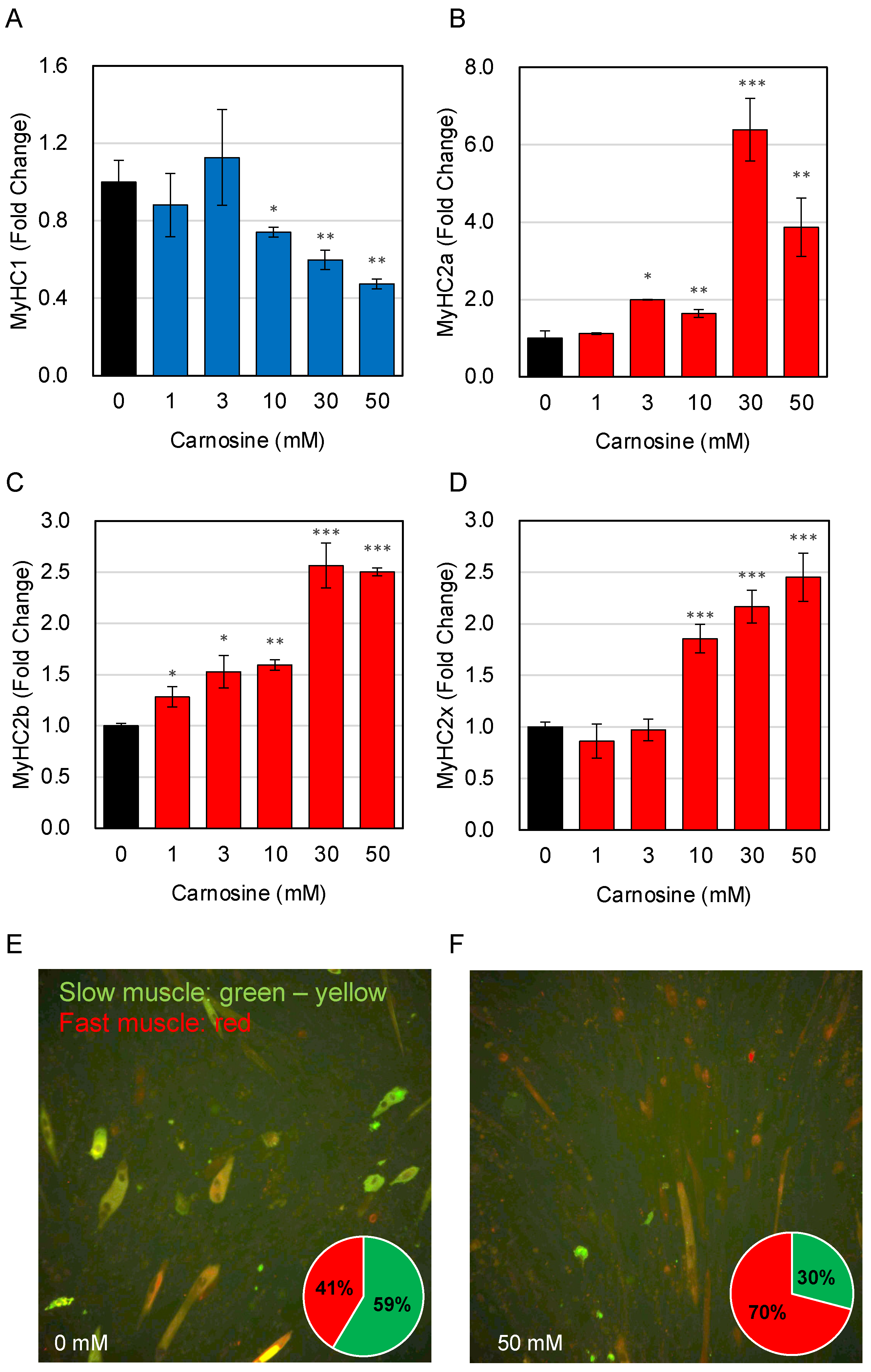
3.2. Effects of Carnosine on Muscle Fiber Type Change
3.3. Effects of Carnosine on the Expression of Factors Secreted from Myotube Cells
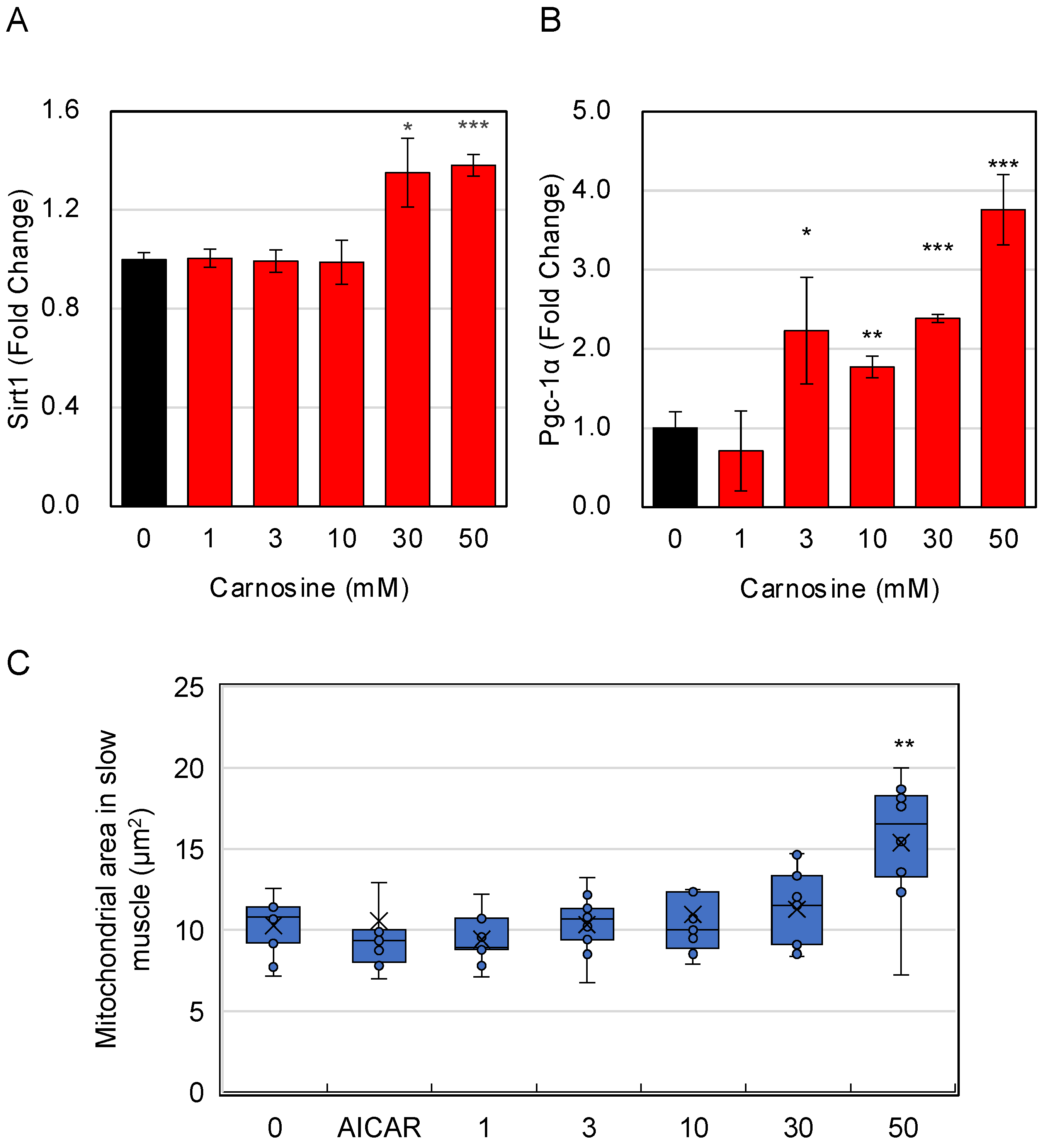
3.4. Effects of Carnosine on Mitochondria in Myotube Cells
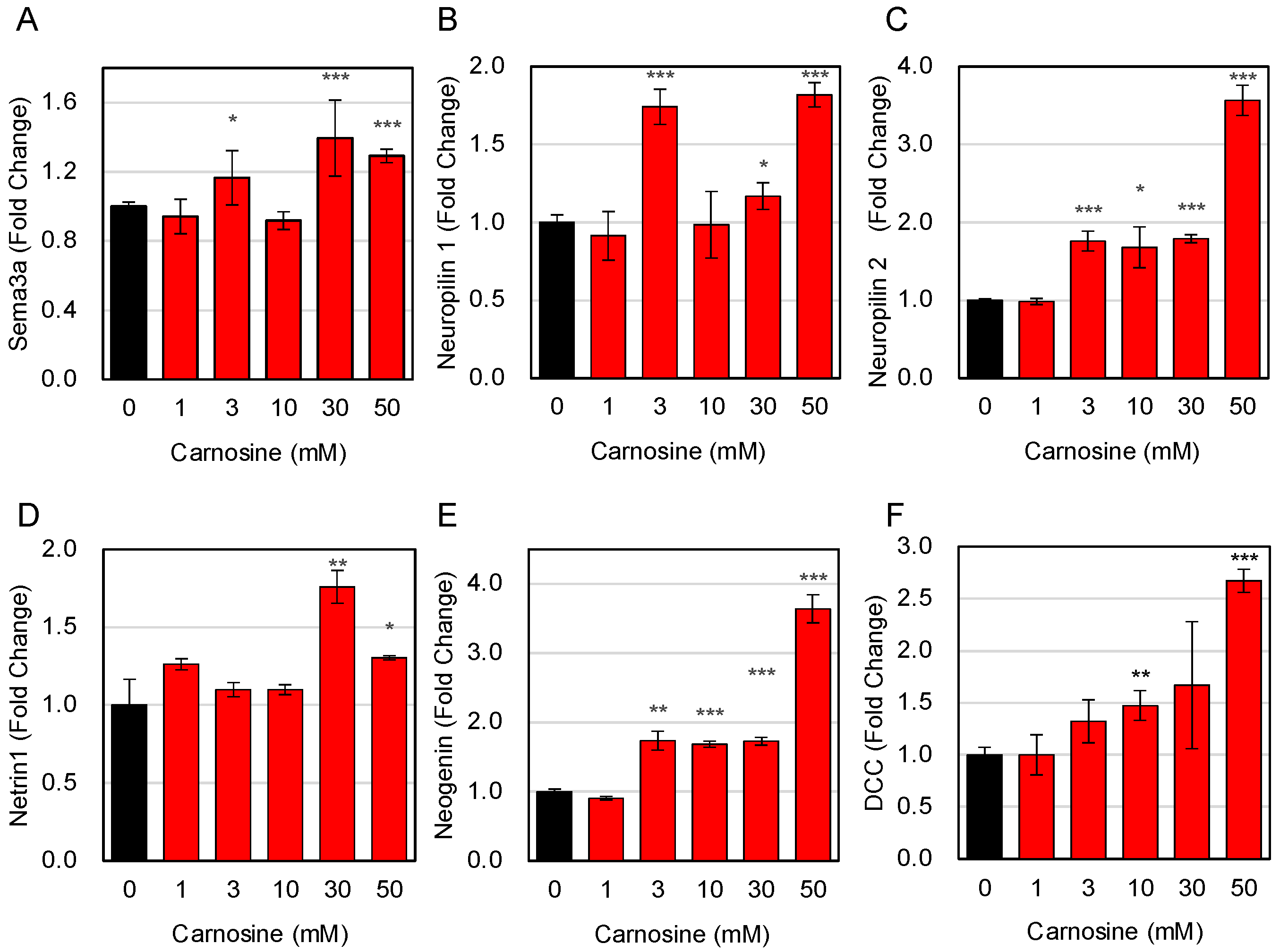
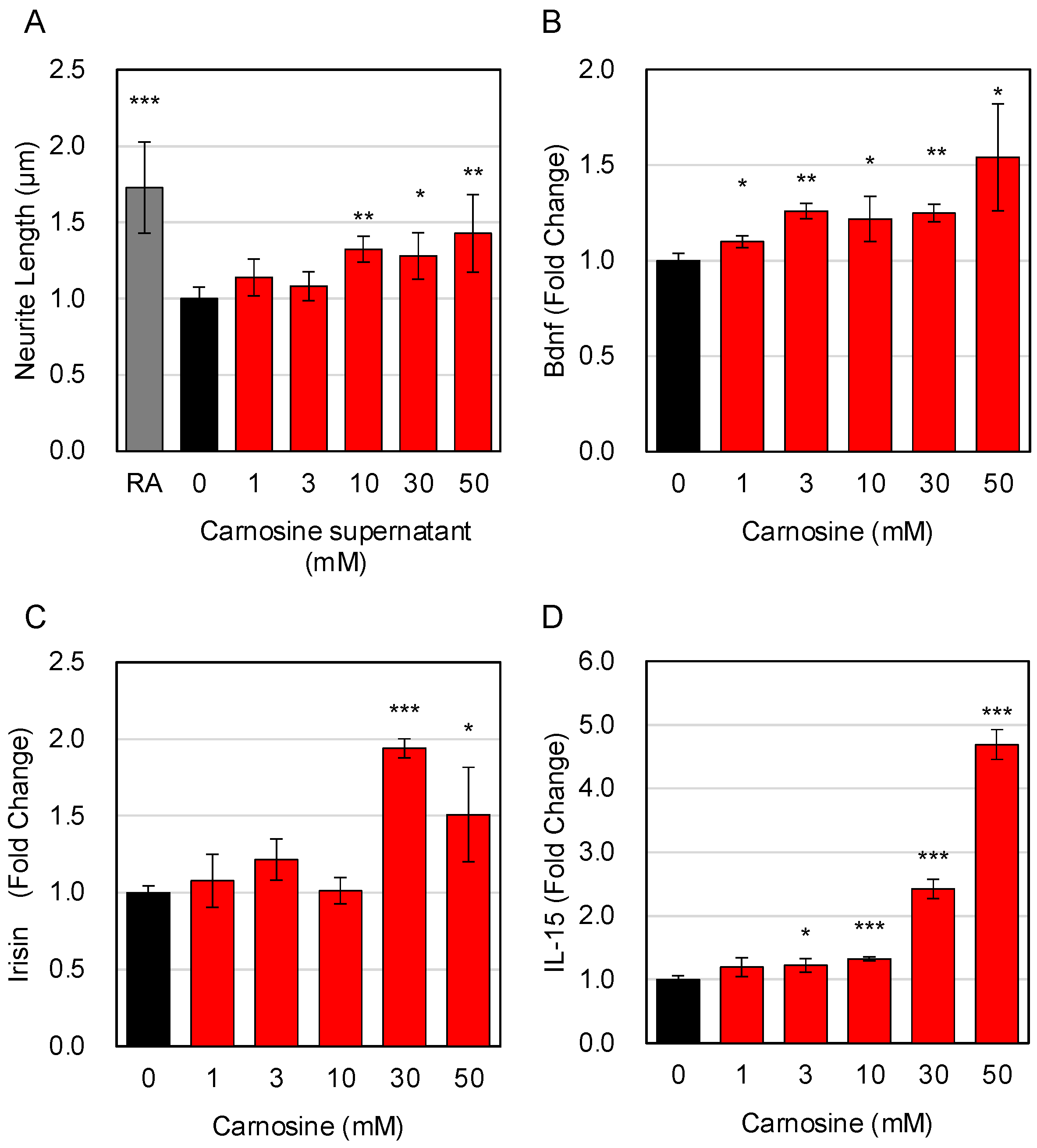
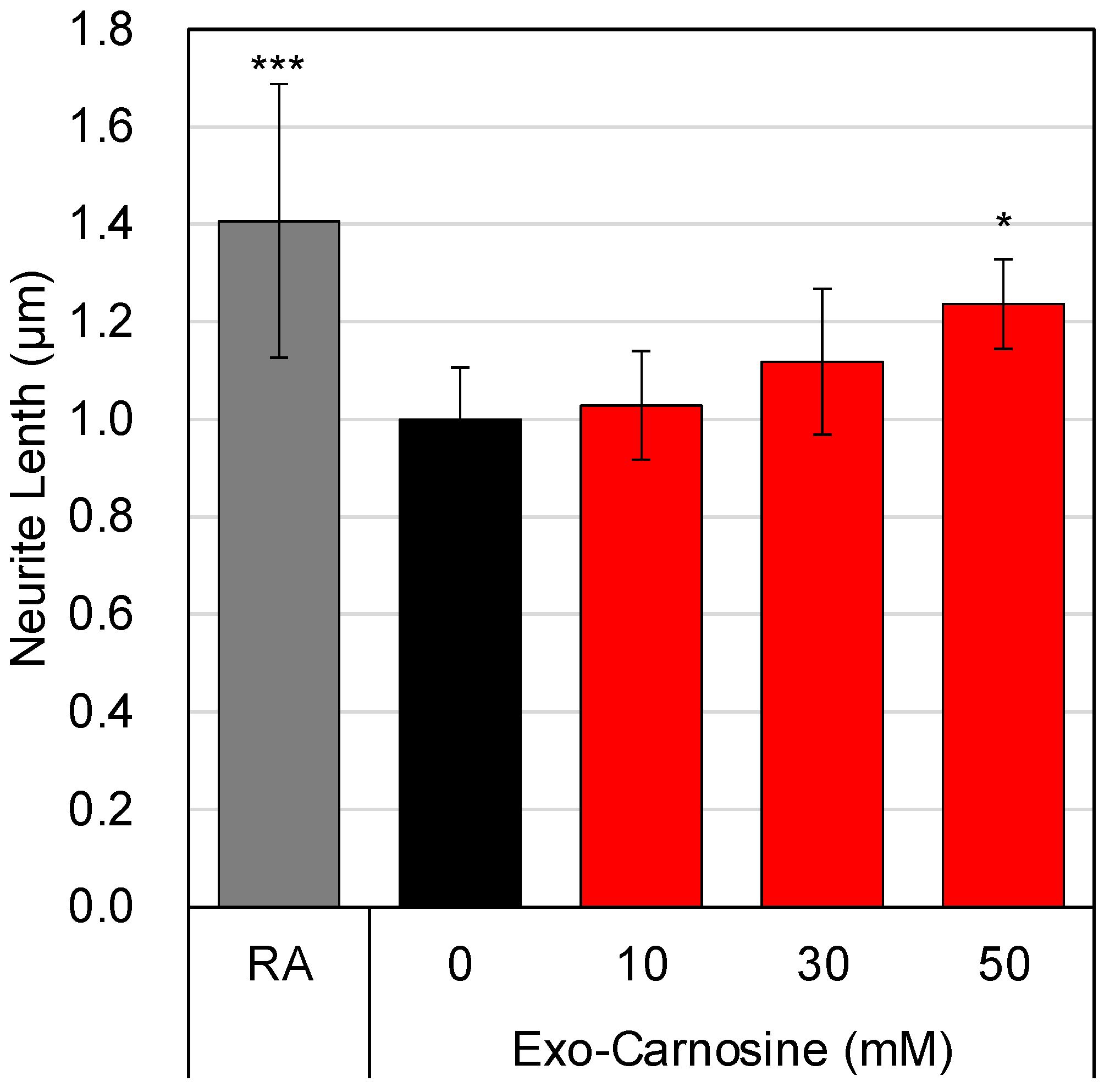
3.5. Effects of Carnosine on the Interaction between Muscle Cells and Neuronal Cells
3.6. Molecular Basis for the Carnosine-Induced Interaction between Muscle Cells and Neuronal Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boldyrev, A.A.; Aldini, G.; Derave, W. Physiology and pathophysiology of carnosine. Physiol. Rev. 2013, 93, 1803–1845. [Google Scholar] [CrossRef]
- Hisatsune, T.; Kaneko, J.; Kurashige, H.; Cao, Y.; Satsu, H.; Totsuka, M.; Katakura, Y.; Imabayashi, E.; Matsuda, H. Effect of anserine/carnosine supplementation on verbal episodic memory in elderly people. J. Alzheimer’s Dis. 2015, 50, 149–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Q.; Tanigawa, K.; Kaneko, J.; Totsuka, M.; Katakura, Y.; Imabayashi, E.; Matsuda, H.; Hisatsune, T. Anserine/carnosine supplementation preserves blood flow in the prefrontal brain of elderly people carrying APOE e4. Aging Dis. 2018, 9, 334–345. [Google Scholar] [CrossRef] [Green Version]
- Katakura, Y.; Totsuka, M.; Imabayashi, E.; Matsuda, H.; Hisatsune, T. Anserine/carnosine supplementation suppresses the expression of the inflammatory chemokine CCL24 in peripheral blood mononuclear cells from elderly people. Nutrients 2017, 9, 1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugihara, Y.; Onoue, S.; Tashiro, K.; Sato, M.; Hasegawa, T.; Katakura, Y. Carnosine induces intestinal cells to secrete exosomes that activate neuronal cells. PLoS ONE 2019, 14, e0217394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isaac, A.R.; Lima-Filho, R.A.S.; Lourenco, M.V. How does the skeletal muscle communicate with the brain in health and disease? Neuropharmacology 2021, 197, 108744. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT. Method 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Inotsuka, R.; Udono, M.; Yamatsu, A.; Kim, M.; Katakura, Y. Exosome-mediated activation of neuronal cells triggered by γ-aminobutyric acid (GABA). Nutrients 2021, 13, 2544. [Google Scholar] [CrossRef]
- Ogawa, M.; Udono, M.; Teruya, K.; Uehara, N.; Katakura, Y. Exosomes derived from fisetin-treated keratinocytes mediate hair growth promotion. Nutrients 2021, 13, 2087. [Google Scholar] [CrossRef]
- Bolstad, B.M.; Irizarry, R.A.; Astrand, M.; Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 19033363. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Tatsumi, R.; Suzuki, T.; Do, M.J.Q.; Ohya, Y.; Anderson, J.E.; Shibata, A.; Kawaguchi, M.; Ohya, S.; Ohtsubo, H.; Mizunoya, W.; et al. Slow-myofiber commitment by semaphorin 3A secreted from myogenic stem cells. Stem Cells 2017, 35, 1815–1834. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, T.; Mori, A.; Maeno, T.; Arimatsu, R.; Ichimura, E.; Nishi, Y.; Hisaeda, K.; Yamaya, Y.; Kobayashi, K.; Nakamura, M.; et al. Abundant synthesis of netrin-1 in satellite cell-derived myoblasts isolated from EDL rather than soleus muscle regulates fast-type myotube formation. Int. J. Mol. Sci. 2021, 22, 94499. [Google Scholar] [CrossRef] [PubMed]
- Cantó, C.; Gerhart-Hines, Z.; Feige, J.N.; Lagouge, M.; Noriega, L.; Milne, J.C.; Elliott, P.J.; Puigserver, P.; Auwerx, J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 2009, 458, 1056–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zierath, J.R.; Hawley, J.A. Skeletal muscle fiber type: Influence on contractile and metabolic properties. PLoS Biol. 2004, 2, e348. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Chen, X.; Huang, Z.; Chen, D.; Chen, H.; Luo, Y.; Zhen, P.; He, J.; Yu, J.; Yu, B. Procyanidin B2 promotes skeletal slow-twitch myofiber gene expression through the AMPK signaling pathway in C2C12 myotubes. J. Agric. Food Chem. 2020, 68, 1306–1314. [Google Scholar] [CrossRef]
- Cripps, M.J.; Hanna, K.; Lavilla, C.; Sayers, S.R.; Caton, P.W.; Sims, C.; De Girolamo, L.; Sale, C.; Turner, M.D. Carnosine scavenging of glucolipotoxic free radicals enhances insulin secretion and glucose uptake. Sci. Rep. 2017, 7, 13313–13317. [Google Scholar] [CrossRef] [Green Version]
- Matthews, J.J.; Artioli, G.G.; Turner, M.D.; Sale, C. The physiological roles of carnosine and β-alanine in exercising human skeletal muscle. Med. Sci. Sport Exerc. 2019, 51, 2098–2108. [Google Scholar] [CrossRef]
- Herculano, B.; Tamura, M.; Ohba, A.; Shimatani, M.; Kutsuna, N.; Hisatsune, T. β-alanyl-L-histidine rescues cognitive deficits caused by feeding a high fat diet in a transgenic mouse model of Alzheimer’s disease. J. Alzheimer's Dis. 2013, 33, 983–997. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Jiao, J.; Zhang, Y. Therapeutic approaches for improving cognitive function in the aging brain. Front. Neurosci. 2022, 16, 1060556. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Ito, O.; Takahashi, H.; Takamatsu, K. The effect of sprint training on skeletal muscle carnosine in humans. Int. J. Sport Health Sci. 2004, 2, 105–110. [Google Scholar] [CrossRef] [Green Version]
- Yue, B.; Wang, H.; Cai, X.; Wang, J.; Chai, Z.; Peng, W.; Shu, S.; Fu, C.; Zhong, J. Adipose-secreted exosomes and their pathophysiologic effects on skeletal muscle. Int. J. Mol. Sci. 2022, 23, 12411. [Google Scholar] [CrossRef]
- Zhou, S.; Cheng, G.L.Y.; Cheung, A.K.K. Role of exosomes and exosomal microRNA in muscle-Kidney crosstalk in chronic kidney disease. Front. Cell Dev. Biol. 2022, 10, 951837. [Google Scholar] [CrossRef] [PubMed]
- Pavlic, A.; Sangani, N.B.; Kerins, J.; Nicolaes, G.; Schurgers, L.; Reutelingsperger, C. Vascular Smooth Muscle Cell Neutral Sphingomyelinase 2 in the Release of Exosomes and Vascular Calcification. Int. J. Mol. Sci. 2022, 23, 69178. [Google Scholar] [CrossRef]
| miRNA | Ratio | Functional Annotation | Enrichment Score |
|---|---|---|---|
| miR-6240 | 4.00 | brain development | 9.305 |
| nervous system development | 7.446 | ||
| central nervous system neuron differentiation | 6.410 | ||
| dendrite | 4.859 | ||
| synapse | 4.590 | ||
| central nervous system neuron development | 3.683 | ||
| neuron spine | 3.537 | ||
| synaptic signaling | 3.423 | ||
| miR-1983 | 2.57 | central nervous system development | 11.885 |
| nervous system development | 10.593 | ||
| dendrite | 9.141 | ||
| synaptic signaling | 8.154 | ||
| synapse | 5.675 | ||
| axon part | 5.199 | ||
| learning or memory | 4.093 | ||
| miR-3963 | 2.37 | nervous system development | 6.118 |
| synapse | 4.202 | ||
| synapse part | 3.550 | ||
| synaptic signaling | 3.193 | ||
| miR-3968 | 2.25 | nervous system development | 10.328 |
| synapse | 7.524 | ||
| brain development | 6.161 | ||
| dendrite | 5.653 | ||
| axon | 3.739 | ||
| axon extension | 3.459 | ||
| miR-125a-3p | 2.23 | synapse | 16.259 |
| nervous system development | 11.463 | ||
| axon | 8.528 | ||
| synaptic signaling | 8.131 | ||
| brain development | 5.358 | ||
| neuron spine | 5.216 | ||
| learning or memory | 3.978 | ||
| dendrite development | 3.651 | ||
| miR-6366 | 2.10 | nervous system development | 10.013 |
| synapse | 8.803 | ||
| synaptic signaling | 8.568 | ||
| brain development | 6.534 | ||
| axon | 6.077 | ||
| dendrite | 5.448 | ||
| synapse part | 3.737 | ||
| central nervous system neuron development | 3.580 | ||
| learning or memory | 3.368 | ||
| miR-3072-3p | 2.01 | synapse | 11.804 |
| nervous system development | 10.568 | ||
| synaptic signaling | 10.263 | ||
| central nervous system development | 6.600 | ||
| axon | 6.092 | ||
| neuron spine | 5.688 | ||
| learning or memory | 4.474 |
| miRNA | KEGG Pathway | p-Value |
|---|---|---|
| miR-6240 | Long-term potentiation | 4.957 × 10−4 |
| Axon guidance | 1.005 × 10−3 | |
| cAMP signaling pathway | 1.318 × 10−3 | |
| miR-1983 | FoxO signaling pathway | 6.262 × 10−9 |
| cAMP signaling pathway | 1.010 × 10−3 | |
| Calcium signaling pathway | 1.287 × 10−3 | |
| Long-term depression | 1.767 × 10−3 | |
| Axon guidance | 2.549 × 10−3 | |
| AMPK signaling pathway | 9.073 × 10−3 | |
| GABAergic synapse | 2.809 × 10−2 | |
| miR-3963 | Axon guidance | 3.766 × 10−3 |
| cAMP signaling pathway | 6.143 × 10−3 | |
| MAPK signaling pathway | 1.322 × 10−2 | |
| Long-term potentiation | 3.981 × 10−2 | |
| miR-3968 | Axon guidance | 1.285 × 10−4 |
| MAPK signaling pathway | 5.188 × 10−3 | |
| FoxO signaling pathway | 1.429 × 10−2 | |
| Circadian entrainment | 3.075 × 10−2 | |
| miR-125a-3p | MAPK signaling pathway | 1.032 × 10−4 |
| Dopaminergic synapse | 1.213 × 10−4 | |
| Axon guidance | 2.967 × 10−4 | |
| AMPK signaling pathway | 1.233 × 10−3 | |
| FoxO signaling pathway | 2.631 × 10−3 | |
| Long-term depression | 2.756 × 10−2 | |
| GABAergic synapse | 3.667 × 10−2 | |
| miR-6366 | FoxO signaling pathway | 1.938 × 10−7 |
| Axon guidance | 2.215 × 10−6 | |
| MAPK signaling pathway | 4.478 × 10−6 | |
| cAMP signaling pathway | 5.824 × 10−5 | |
| Synaptic vesicle cycle | 4.102 × 10−3 | |
| miR-3072-3p | Axon guidance | 8.508 × 10−5 |
| cAMP signaling pathway | 1.811 × 10−2 | |
| FoxO signaling pathway | 2.037 × 10−2 | |
| MAPK signaling pathway | 2.510 × 10−2 | |
| AMPK signaling pathway | 2.539 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishibashi, A.; Udono, M.; Sato, M.; Katakura, Y. Molecular Mechanisms for the Carnosine-Induced Activation of Muscle–Brain Interaction. Nutrients 2023, 15, 1479. https://doi.org/10.3390/nu15061479
Ishibashi A, Udono M, Sato M, Katakura Y. Molecular Mechanisms for the Carnosine-Induced Activation of Muscle–Brain Interaction. Nutrients. 2023; 15(6):1479. https://doi.org/10.3390/nu15061479
Chicago/Turabian StyleIshibashi, Asuka, Miyako Udono, Mikako Sato, and Yoshinori Katakura. 2023. "Molecular Mechanisms for the Carnosine-Induced Activation of Muscle–Brain Interaction" Nutrients 15, no. 6: 1479. https://doi.org/10.3390/nu15061479





