Height-Related Polygenic Variants Are Associated with Metabolic Syndrome Risk and Interact with Energy Intake and a Rice-Main Diet to Influence Height in KoGES
Abstract
:1. Introduction
2. Methods and Materials
2.1. Participants
2.2. Adult Height Criteria
2.3. Survey Questionnaires and Anthropometric and Biochemical Measurements
2.4. Usual Food Intake Measurement
2.5. Dietary Patterns and Dietary Inflammatory Index (DII)
2.6. Genotyping, Its Quality Control, and Genotype-Tissue Expression (GTEx)
2.7. Selection of Genetic Variants to Influence Adult Height and Their Optimal Model with the SNP–SNP Interaction
2.8. Molecular Docking of Wild and Mutated GDF5 with Food Compounds
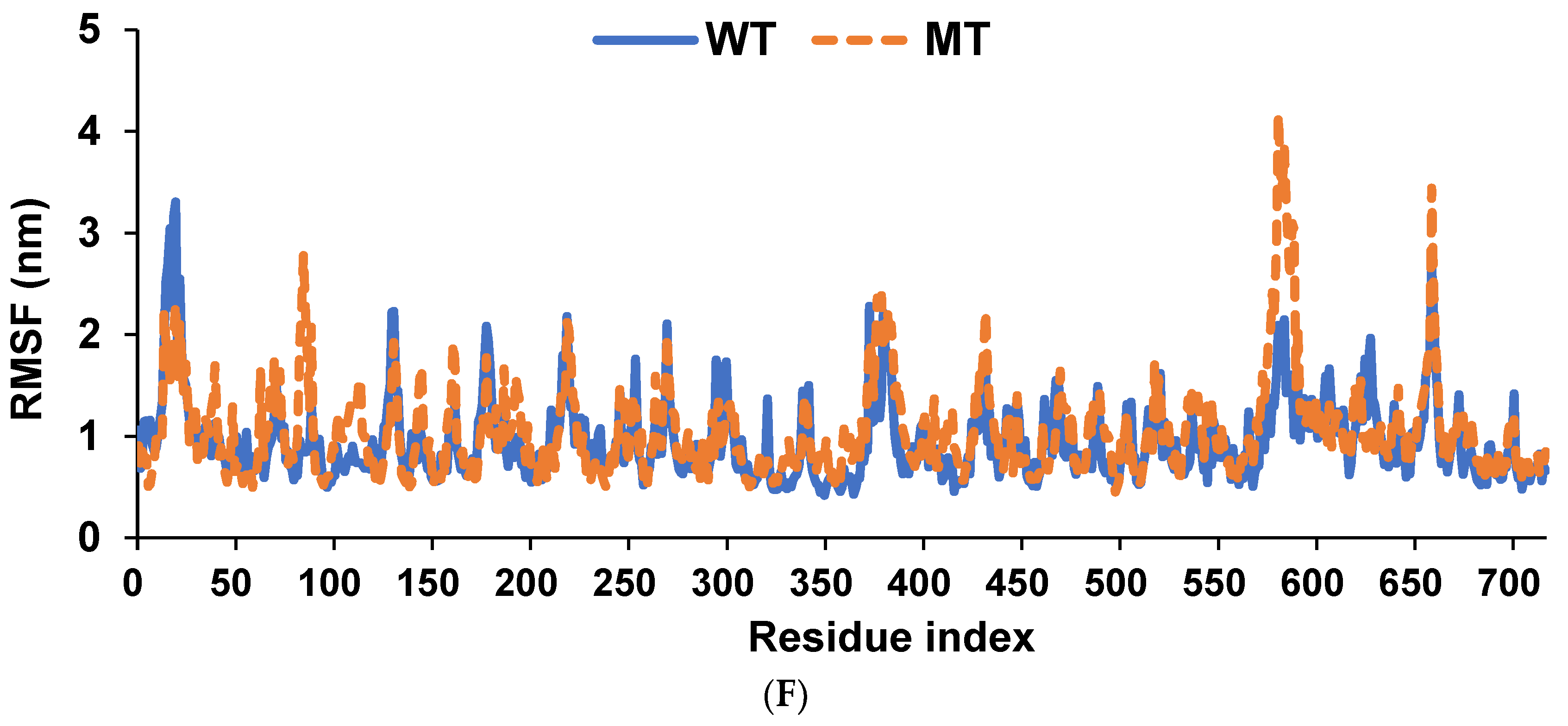
2.9. Molecular Dynamics Simulation (MDS)
2.10. Statistical Analysis
3. Results
3.1. Demographic Characteristics and Lifestyles According to Gender and Adult Height
3.2. Nutrient Intake According to Gender and Adult Height
3.3. Prevalence of MetS and Its Related Parameters According to Gender and Adult Height
3.4. Genetic Variants Linked to Adult Height
3.5. SNP–SNP Interaction by GMDR
3.6. Expression of Quantitative Trait Loci (eQTL) of the Selected Genes According to the Alleles
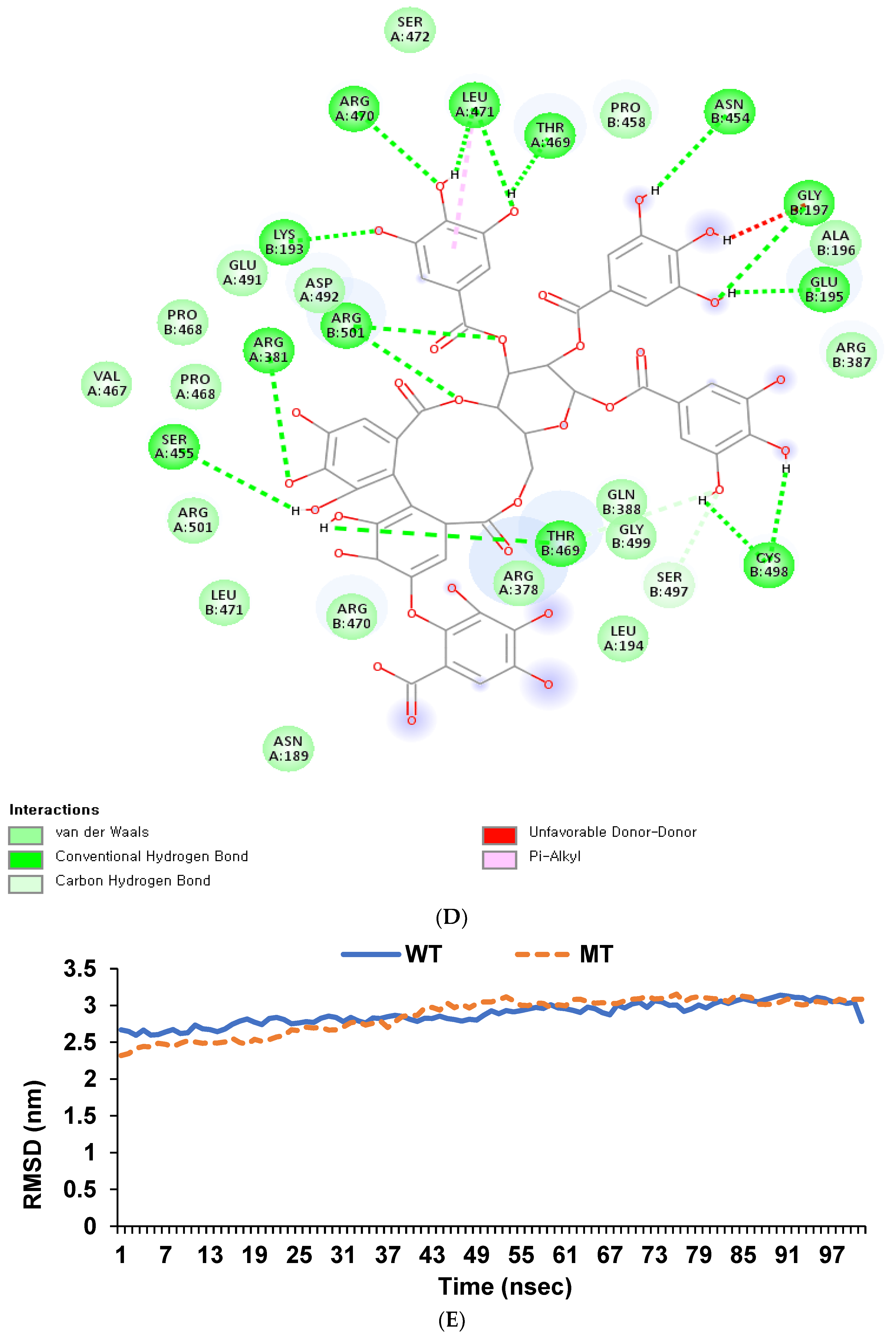
3.7. Binding Affinity of Hydrolyzable Tannins to GDF5_rs224331
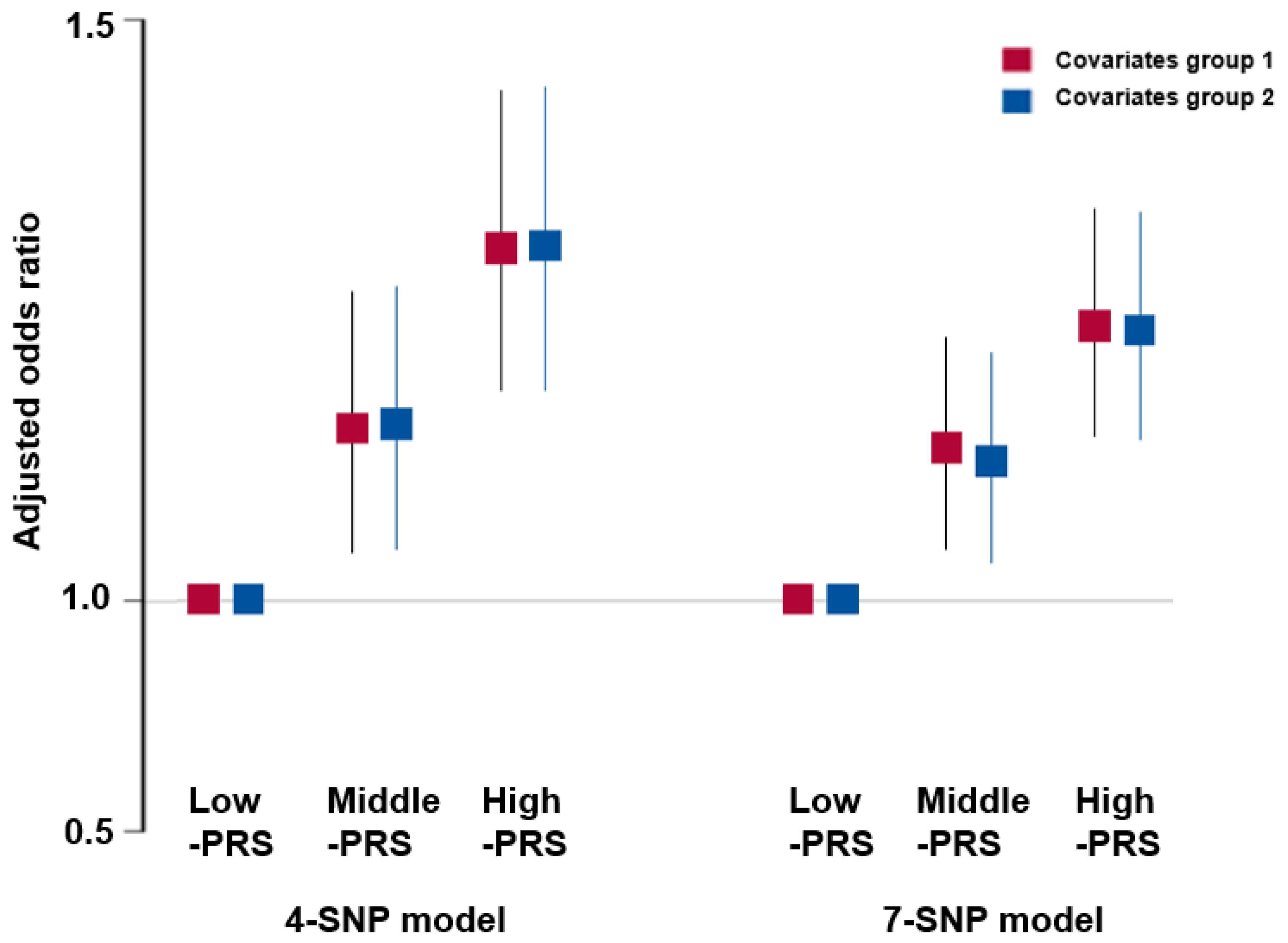
3.8. Association of PRS with the Risk of Metabolic Syndrome and Its Components
3.9. Interaction between PRS and Lifestyle Factors for Adult Height
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zoledziewska, M.; Sidore, C.; Chiang, C.W.K.; Sanna, S.; Mulas, A.; Steri, M.; Busonero, F.; Marcus, J.H.; Marongiu, M.; Maschio, A.; et al. Height-reducing variants and selection for short stature in Sardinia. Nat. Genet. 2015, 47, 1352–1356. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Lei, X.; Ridder, G.; Strauss, J.; Zhao, Y. Health, Height, Height Shrinkage, and SES at Older Ages: Evidence from China. Am. Econ. J. Appl. Econ. 2013, 5, 86–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkler, T.W.; Day, F.R.; Croteau-Chonka, D.C.; Wood, A.R.; Locke, A.E.; Mägi, R.; Ferreira, T.; Fall, T.; Graff, M.; Justice, A.E.; et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 2014, 9, 1192–1212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.Y.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiyama, M.; Ishigaki, K.; Sakaue, S.; Momozawa, Y.; Horikoshi, M.; Hirata, M.; Matsuda, K.; Ikegawa, S.; Takahashi, A.; Kanai, M.; et al. Characterizing rare and low-frequency height-associated variants in the Japanese population. Nat. Commun. 2019, 10, 4393. [Google Scholar] [CrossRef] [Green Version]
- Benonisdottir, S.; Oddsson, A.; Helgason, A.; Kristjansson, R.P.; Sveinbjornsson, G.; Oskarsdottir, A.; Thorleifsson, G.; Davidsson, O.B.; Arnadottir, G.A.; Sulem, G.; et al. Epigenetic and genetic components of height regulation. Nat. Commun. 2016, 7, 13490. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.J.; Lee, H.I.; Park, T.; Kim, K.; Lee, J.E.; Cho, N.H.; Shin, C.; Cho, Y.S.; Lee, J.Y.; Han, B.G.; et al. Identification of 15 loci influencing height in a Korean population. J. Hum. Genet. 2010, 55, 27–31. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Li, M.; Bradfield, J.P.; Zhang, H.; Mentch, F.D.; Wang, K.; Sleiman, P.M.; Kim, C.E.; Glessner, J.T.; Hou, C.; et al. The role of height-associated loci identified in genome-wide association studies in the determination of pediatric stature. BMC Med. Genet. 2010, 11, 96. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.-J.; Liao, W.-L.; Wang, C.-H.; Tsai, L.-P.; Tang, C.-H.; Chen, C.-H.; Wu, J.-Y.; Liang, W.-M.; Hsieh, A.-R.; Cheng, C.-F.; et al. Association of human height-related genetic variants with familial short stature in Han Chinese in Taiwan. Sci. Rep. 2017, 7, 6372. [Google Scholar] [CrossRef] [Green Version]
- Rehunen, S.K.J.; Kautiainen, H.; Eriksson, J.G.; Korhonen, P.E. Adult height and glucose tolerance: A re-appraisal of the importance of body mass index. Diabet. Med. 2017, 34, 1129–1135. [Google Scholar] [CrossRef]
- Rosenberg, M.A.; Kaplan, R.C.; Siscovick, D.S.; Psaty, B.M.; Heckbert, S.R.; Newton-Cheh, C.; Mukamal, K.J. Genetic variants related to height and risk of atrial fibrillation: The cardiovascular health study. Am. J. Epidemiol. 2014, 180, 215–222. [Google Scholar] [CrossRef] [Green Version]
- Lai, F.Y.; Nath, M.; Hamby, S.E.; Thompson, J.R.; Nelson, C.P.; Samani, N.J. Adult height and risk of 50 diseases: A combined epidemiological and genetic analysis. BMC Med. 2018, 16, 187. [Google Scholar] [CrossRef]
- Ryu, J.; Lee, C. Association of glycosylated hemoglobin with the gene encoding CDKAL1 in the Korean Association Resource (KARE) study. Hum. Mutat. 2012, 33, 655–659. [Google Scholar] [CrossRef]
- Wu, X.; Park, S. An Inverse Relation between Hyperglycemia and Skeletal Muscle Mass Predicted by Using a Machine Learning Approach in Middle-Aged and Older Adults in Large Cohorts. J. Clin. Med. 2021, 10, 2133. [Google Scholar] [CrossRef]
- Park, S.; Yang, H.J.; Kim, M.J.; Hur, H.J.; Kim, S.H.; Kim, M.S. Interactions between Polygenic Risk Scores, Dietary Pattern, and Menarche Age with the Obesity Risk in a Large Hospital-Based Cohort. Nutrients 2021, 13, 3772. [Google Scholar] [CrossRef]
- Stevens, L.A.; Coresh, J.; Feldman, H.I.; Greene, T.; Lash, J.P.; Nelson, R.G.; Rahman, M.; Deysher, A.E.; Zhang, Y.L.; Schmid, C.H.; et al. Evaluation of the modification of diet in renal disease study equation in a large, diverse population. J. Am. Soc. Nephrol. 2007, 18, 2749–2757. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Kim, C.; Wu, X. Development and Validation of an Insulin Resistance Predicting Model Using a Machine-Learning Approach in a Population-Based Cohort in Korea. Diagnostics 2022, 12, 212. [Google Scholar] [CrossRef]
- van Woudenbergh, G.J.; Theofylaktopoulou, D.; Kuijsten, A.; Ferreira, I.; van Greevenbroek, M.M.; van der Kallen, C.J.; Schalkwijk, C.G.; Stehouwer, C.D.; Ocké, M.C.; Nijpels, G.; et al. Adapted dietary inflammatory index and its association with a summary score for low-grade inflammation and markers of glucose metabolism: The Cohort study on Diabetes and Atherosclerosis Maastricht (CODAM) and the Hoorn study. Am. J. Clin. Nutr. 2013, 98, 1533–1542. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.K.; Kim, M.K. Relationship of sodium intake with obesity among Korean children and adolescents: Korea National Health and Nutrition Examination Survey. Br. J. Nutr. 2016, 115, 834–841. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Han, B.-G.; KoGES Group. Cohort Profile: The Korean Genome and Epidemiology Study (KoGES) Consortium. Int. J. Epidemiol. 2017, 46, e20. [Google Scholar] [CrossRef]
- Rabbee, N.; Speed, T.P. A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics 2006, 22, 7–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uma Jyothi, K.; Reddy, B.M. Gene-gene and gene-environment interactions in the etiology of type 2 diabetes mellitus in the population of Hyderabad, India. Meta Gene 2015, 5, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Khera, A.V.; Chaffin, M.; Wade, K.H.; Zahid, S.; Brancale, J.; Xia, R.; Distefano, M.; Senol-Cosar, O.; Haas, M.E.; Bick, A.; et al. Polygenic Prediction of Weight and Obesity Trajectories from Birth to Adulthood. Cell 2019, 177, 587–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, C.M.; Vassos, E. Polygenic risk scores: From research tools to clinical instruments. Genome Med. 2020, 12, 44. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.-Y.; Song, M.Y.; Park, S. Carbohydrate and sodium intake and physical activity interact with genetic risk scores of four genetic variants mainly related to lipid metabolism to modulate metabolic syndrome risk in Korean middle-aged adults. Br. J. Nutr. 2019, 122, 919–927. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Liu, L.; Zhou, J.; Zhang, T.; Daily, J.W.; Park, S. Bioactive Components of Houttuynia cordata Thunb and Their Potential Mechanisms Against COVID-19 Using Network Pharmacology and Molecular Docking Approaches. J. Med. Food 2022, 25, 355–366. [Google Scholar] [CrossRef]
- Yang, Y.; Shi, C.-Y.; Xie, J.; Dai, J.-H.; He, S.-L.; Tian, Y. Identification of potential dipeptidyl peptidase (DPP)-IV inhibitors among Moringa oleifera phytochemicals by virtual screening, molecular docking analysis, ADME/T-based prediction, and in vitro analyses. Molecules 2020, 25, 189. [Google Scholar] [CrossRef] [Green Version]
- Committee of SAS. SAS/STAT® 14.2 User’s Guide; SAS Inc.: Cary, NC, USA, 2016. [Google Scholar]
- Kwon, O.; Kim, H.; Kim, J.; Hwang, J.-Y.; Lee, J.; Yoon, M.O. The development of the 2020 Dietary Reference Intakes for Korean population: Lessons and challenges. J. Nutr. Health 2021, 54, 425. [Google Scholar] [CrossRef]
- Yengo, L.; Vedantam, S.; Marouli, E.; Sidorenko, J.; Bartell, E.; Sakaue, S.; Graff, M.; Eliasen, A.U.; Jiang, Y.; Raghavan, S.; et al. A saturated map of common genetic variants associated with human height. Nature 2022, 610, 704–712. [Google Scholar] [CrossRef]
- Sözen, T.; Özışık, L.; Başaran, N. An overview and management of osteoporosis. Eur. J. Rheumatol. 2017, 4, 46–56. [Google Scholar] [CrossRef]
- Curtis, E.; Litwic, A.; Cooper, C.; Dennison, E. Determinants of Muscle and Bone Aging. J. Cell. Physiol. 2015, 230, 2618–2625. [Google Scholar] [CrossRef] [Green Version]
- Wittenbecher, C.; Kuxhaus, O.; Boeing, H.; Stefan, N.; Schulze, M.B. Associations of short stature and components of height with incidence of type 2 diabetes: Mediating effects of cardiometabolic risk factors. Diabetologia 2019, 62, 2211–2221. [Google Scholar] [CrossRef] [Green Version]
- Toro-Huamanchumo, C.J.; Pérez-Zavala, M.; Urrunaga-Pastor, D.; De La Fuente-Carmelino, L.; Benites-Zapata, V.A. Relationship between the short stature and the prevalence of metabolic syndrome and insulin resistance markers in workers of a private educational institution in Peru. Diabetol. Metab. Syndr. 2020, 14, 1339–1345. [Google Scholar] [CrossRef]
- Chung, S. Growth and Puberty in Obese Children and Implications of Body Composition. J. Obes. Metab. Syndr. 2017, 26, 243–250. [Google Scholar] [CrossRef]
- Yang, T.L.; Guo, Y.; Zhang, L.S.; Tian, Q.; Yan, H.; Guo, Y.F.; Deng, H.W. HMGA2 is confirmed to be associated with human adult height. Ann. Hum. Genet. 2010, 74, 11–16. [Google Scholar] [CrossRef] [Green Version]
- Hendriks, A.E.; Brown, M.R.; Boot, A.M.; Oostra, B.A.; Drop, S.L.; Parks, J.S. Genetic variation in candidate genes like the HMGA2 gene in the extremely tall. Horm. Res. Paediatr. 2011, 76, 307–313. [Google Scholar] [CrossRef]
- Chiou, J.-S.; Cheng, C.-F.; Liang, W.-M.; Chou, C.-H.; Wang, C.-H.; Lin, W.-D.; Chiu, M.-L.; Cheng, W.-C.; Lin, C.-W.; Lin, T.-H.; et al. Your height affects your health: Genetic determinants and health-related outcomes in Taiwan. BMC Med. 2022, 20, 250. [Google Scholar] [CrossRef]
- Okada, Y.; Kamatani, Y.; Takahashi, A.; Matsuda, K.; Hosono, N.; Ohmiya, H.; Daigo, Y.; Yamamoto, K.; Kubo, M.; Nakamura, Y.; et al. A genome-wide association study in 19 633 Japanese subjects identified LHX3-QSOX2 and IGF1 as adult height loci. Hum. Mol. Genet. 2010, 19, 2303–2312. [Google Scholar] [CrossRef] [Green Version]
- Lechner, K.; Lechner, B.; Crispin, A.; Schwarz, P.E.H.; von Bibra, H. Waist-to-height ratio and metabolic phenotype compared to the Matsuda index for the prediction of insulin resistance. Sci. Rep. 2021, 11, 8224. [Google Scholar] [CrossRef]
- Cubbon, R.M.; Kearney, M.T.; Wheatcroft, S.B. Endothelial IGF-1 Receptor Signalling in Diabetes and Insulin Resistance. Trends Endocrinol. Metab. 2016, 27, 96–104. [Google Scholar] [CrossRef]
- Zhang, W.; Wu, X.; Pei, Z.; Kiess, W.; Yang, Y.; Xu, Y.; Chang, Z.; Wu, J.; Sun, C.; Luo, F. GDF5 Promotes White Adipose Tissue Thermogenesis via p38 MAPK Signaling Pathway. DNA Cell Biol. 2019, 38, 1303–1312. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Gong, J.; Jie, Y.; Cao, J.; Chen, Z.; Li, R.; Chong, Y.; Hu, B.; Zhang, Q. NCAPG Is a Promising Therapeutic Target Across Different Tumor Types. Front. Pharmacol. 2020, 11, 387. [Google Scholar] [CrossRef] [PubMed]
- Soranzo, N.; Rivadeneira, F.; Chinappen-Horsley, U.; Malkina, I.; Richards, J.B.; Hammond, N.; Stolk, L.; Nica, A.; Inouye, M.; Hofman, A.; et al. Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet. 2009, 5, e1000445. [Google Scholar] [CrossRef] [PubMed]
- Metzger, J.; Schrimpf, R.; Philipp, U.; Distl, O. Expression Levels of LCORL Are Associated with Body Size in Horses. PLoS ONE 2013, 8, e56497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindholm-Perry, A.K.; Sexten, A.K.; Kuehn, L.A.; Smith, T.P.; King, D.A.; Shackelford, S.D.; Wheeler, T.L.; Ferrell, C.L.; Jenkins, T.G.; Snelling, W.M.; et al. Association, effects and validation of polymorphisms within the NCAPG—LCORL locus located on BTA6 with feed intake, gain, meat and carcass traits in beef cattle. BMC Genet. 2011, 12, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sergi, C.; Shen, F.; Liu, S.M. Insulin/IGF-1R, SIRT1, and FOXOs Pathways-An Intriguing Interaction Platform for Bone and Osteosarcoma. Front. Endocrinol. 2019, 10, 93. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Huang, H.; Chen, K.; Yang, L.; Xie, L.L.; Xiong, T.; Wu, X. Novel mutation of type-1 insulin-like growth factor receptor (IGF-1R) gene in a severe short stature pedigree identified by targeted next-generation sequencing. J. Genet. 2019, 98, 16. [Google Scholar] [CrossRef]
- Campbell, W.W., Jr.; Ward, L.C.; Swift, T.R. Nerve conduction velocity varies inversely with height. Muscle Nerve 1981, 4, 520–523. [Google Scholar] [CrossRef]
- Lettre, G. The osteoarthritis and height GDF5 locus yields its secrets. Nat. Genet. 2017, 49, 1165–1166. [Google Scholar] [CrossRef]
- Sanna, S.; Jackson, A.U.; Nagaraja, R.; Willer, C.J.; Chen, W.M.; Bonnycastle, L.L.; Shen, H.; Timpson, N.; Lettre, G.; Usala, G.; et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat. Genet. 2008, 40, 198–203. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.D.; Li, G.M.; Jin, W.; Li, Y.; Zhang, Y.P. Positive selection on the osteoarthritis-risk and decreased-height associated variants at the GDF5 gene in East Asians. PLoS ONE 2012, 7, e42553. [Google Scholar] [CrossRef]
- Ekambaram, S.P.; Perumal, S.S.; Erusappan, T.; Srinivasan, A. Hydrolysable tannin-rich fraction from Terminalia chebula Retz. fruits ameliorates collagen-induced arthritis in BALB/c mice. Inflammopharmacol 2020, 28, 275–287. [Google Scholar] [CrossRef]
- Li, K.; Zhang, X.; He, B.; Yang, R.; Zhang, Y.; Shen, Z.; Chen, P.; Du, W. Geraniin promotes osteoblast proliferation and differentiation via the activation of Wnt/β-catenin pathway. Biomed. Pharmacother. 2018, 99, 319–324. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Park, J.-Y.; Lee, S.H.; Woo, J.T.; Kima, T.H.; Park, E.K. Furosin, an ellagitannin, suppresses RANKL-induced osteoclast differentiation and function through inhibition of MAP kinase activation and actin ring formation. Biochem. Biophys. Res. Commun. 2004, 325, 1471–1480. [Google Scholar]
- Tong, Z.; He, W.; Fan, X.; Guo, A. Biological Function of Plant Tannin and Its Application in Animal Health. Front. Vet. Sci. 2022, 8, 803657. [Google Scholar] [CrossRef]




| Men | Women | |||
|---|---|---|---|---|
| Short-Stature (n = 17,305) | Tall-Stature (n = 2988) | Short-Stature (n = 33,860) | Tall-Stature (n = 4548) | |
| Age (years) | 56.4 ± 0.06 b | 53.2 ± 0.18 a | 52.8 ± 0.04 c | 50.2 ± 0.11 c***+++ |
| Gender (%) | 33.82 | 39.65 ‡‡‡ | 66.18 | 60.35 ‡‡‡ |
| Education ≤Middle school High school ≥College | 1654 (15.1) 8261 (75.4) 1045 (9.53) | 99 (6.52) ‡‡‡ 1173 (77.3) 246 (16.2) | 6409 (23.5) 19,373 (71.1) 1486 (5.45) | 329 (10.5) ‡‡‡ 2498 (80.0) 296 (9.48) |
| Income ≤$2000 $2000–4000 >$4000 | 1483 (9.03) 7184 (43.7) 7764 (47.3) | 268 (10.7) ‡‡‡ 1125 (44.9) 1111 (44.4) | 3911 (12.3) 14,278 (44.9) 13,599 (42.8) | 261 (6.02) ‡‡‡ 1707 (39.3) 2371(54.6) |
| Former smoking (%) Smoking (%) | 2237 (12.9) 1447 (8.36) | 349 (11.7) 275 (9.21) | 100 (0.30) 172 (0.51) | 17 (0.37) 19 (0.42) |
| Physical exercise (%) | 10,168 (59.0) | 1784 (59.9) | 17,579 (52.1) | 2445 (53.8) ‡ |
| Alcohol (g) | 35.2 ± 0.38 b | 39.8 ± 0.88 a | 5.24 ± 0.27 b | 5.89 ± 0.71 b***+++## |
| Energy intake (EER %) | 89.4 ± 0.26 d | 93.3 ± 0.58 c | 99.1 ± 0.18 b | 101.6 ± 0.47 a***### |
| Carbohydrates (En%) | 71.3 ± 0.06 b | 71.2 ± 0.13 b | 72.0 ± 0.04 a | 71.6 ± 0.11 b***+ |
| Fat (En%) | 14.2 ± 0.04 a | 14.3 ± 0.10 a | 13.7 ± 0.03 b | 14.1 ± 0.08 a***++# |
| Protein (En%) | 13.3 ± 0.02 b | 13.3 ± 0.05 b | 13.5 ± 0.02 a | 13.5 ± 0.04 a*** |
| Fiber (g) | 15.2 ± 0.08 b | 15.8 ± 0.18 a | 14.3 ± 0.06 c | 14.6 ± 0.15 c***+++ |
| Calcium (mg) | 411 ± 2.15 d | 427 ± 4.89 c | 456 ± 1.50 b | 471 ± 3.95 a***+++ |
| Vitamin C (mg) | 95.1 ± 0.56 d | 98.9 ± 1.28 c | 110 ± 0.39 b | 113 ± 1.04 a***++ |
| Vitamin D (ug) | 5.59 ± 0.05 d | 5.89 ± 0.11 c | 6.81 ± 0.03 b | 7.09 ± 0.09 a***+++ |
| DII (scores) | −13.8 ± 0.39 | −14.2 ± 0.89 | −13.4 ± 0.27 | −15.5 ± 0.72 |
| Flavonoids (mg) | 32.1 ± 0.27 b | 32.8 ± 0.61 b | 41.5 ± 0.19 a | 42.6 ± 0.49 a***+ |
| KBD (%) | 6832 (39.5) | 1269 (42.5) ‡‡ | 10,059 (29.7) | 1409 (31.0) |
| PBD (%) | 3551 (20.5) | 647 (21.7) | 13,316 (39.3) | 2062 (45.3) ‡‡‡ |
| WSD (%) | 8619 (49.8) | 1812 (60.6) ‡‡‡ | 11,155 (32.9) | 1966 (43.2) ‡‡‡ |
| RMD (%) | 5482 (31.7) | 983 (32.9) | 11,480 (33.9) | 1626 (35.8) ‡ |
| Coffee intake (g/day) | 4.23 ± 0.03 a | 4.25 ± 0.06 a | 3.34 ± 0.02 c | 3.48 ± 0.05 b***+ |
| Tea (g/day) | 43.5 ± 0.71 ab | 47.7 ± 1.63 a | 43.2 ± 0.50 b | 42.2 ± 1.32 b*** |
| Men | Women | ||||
|---|---|---|---|---|---|
| Short- Stature (n = 17,305) | Tall- Stature (n = 2988) | Short- Stature (n = 33,860) | Tall-Stature (n = 4548) | Adjusted OR and 95% CI | |
| Height (cm) 1 | 167.3 ± 0.03 b | 177.2 ± 0.08 a | 155.5 ± 0.02 d | 164.7 ± 0.06 c***+++### | |
| BMI (kg/m2) 2 | 24.5 ± 0.04 a | 24.6 ± 0.07 a | 23.6 ± 0.03 b | 23.0 ± 0.06 c***+++### | 0.908 (0.857–0.962) |
| Waist (cm) 3 | 80.1 ± 0.07 c | 76.9 ± 0.13 a | 81.5 ± 0.05 b | 78.7 ± 0.10 d***+++# | 0.327 (0.299–0.357) |
| Weight at age 18 (kg) 4 | 55.4 ± 0.14 | 56.1 ± 0.31 | 55.0 ± 0.11 | 55.1 ± 0.20 *** | 1.025 (0.956–1.099) |
| SMI (kg/m) 5 | 7.34 ± 0.004 a | 7.04 ± 0.010 b | 6.90 ± 0.003 c | 6.46 ± 0.006 d***+++# | 0.402 (0.376–0.430) |
| Fat mass (%) 6 | 20.4 ± 0.02 c | 17.3 ± 0.05 d | 32.8 ± 0.02 a | 30.1 ± 0.04 b***+++### | 0.479 (0.445–0.515) |
| WBC (109/L) 7 | 5.79 ± 0.02 a | 5.61 ± 0.04 b | 5.67 ± 0.01 b | 5.60 ± 0.03 c*+++# | 0.813 (0.767–0.861) |
| hs-CRP (mg/dL) 8 | 0.14 ± 0.004 ab | 0.16 ± 0.009 a | 0.14 ± 0.003 ab | 0.12 ± 0.007 b*** | 0.784 (0.612–1.005) |
| MetS 9 | 3005 (17.4) | 593 (19.9) ‡‡ | 4275 (12.6) | 427 (9.39) ‡‡‡ | 0.494 (0.452–0.540) |
| CVD 9 | 1107 (6.41) ‡‡ | 109 (3.65) | 1030 (3.05) | 68 (1.50) ‡‡‡ | 0.669 (0.563–0.794) |
| Glucose (mg/dL) 10 | 96.38 ± 0.27 a | 93.4 ± 0.55 b | 95.4 ± 0.19 a | 93.4 ± 0.42 b+++ | 0.718 (0.657–0.785) |
| HbA1c (%) 11 | 5.63 ± 0.01 b | 5.49 ± 0.03 c | 5.79 ± 0.01 a | 5.68 ± 0.02 b***+++ | 0.659 (0.569–0.763) |
| Insulin resistance (%) 9 | 1955 (11.3) | 347 (12.0) | 20,66 (6.1) | 226 (4.97) ‡‡ | 0.542 (0.487–0.603) |
| Total cholesterol (mg/dL) 12 | 189 ± 0.35 c | 190 ± 0.71 c | 202 ± 0.23 a | 198 ± 0.56 b***+++### | 0.707 (0.658–0.758) |
| HDL (mg/dL) 13 | 52.2 ± 0.18 b | 53.9 ± 0.36 c | 55.1 ± 0.13 a | 57.3 ± 0.24 a***++### | 1.330 (1.249–1.415) |
| LDL (mg/dL) 14 | 112 ± 0.45 c | 112 ± 0.94 c | 122 ± 0.33 a | 117 ± 0.71 b***+++### | 0.702 (0.646–0.763) |
| TG (mg/dL) 15 | 120 ± 1.11 b | 105 ± 2.29 c | 127 ± 0.81 a | 109 ± 1.74 c**+++ | 0.633 (0.594–0.675) |
| SBP (mmHg) 16 | 123 ± 0.20 a | 120 ± 0.40 b | 123 ± 0.14 a | 120 ± 0.31 b+++ | 0.749 (0.704–0.796) |
| DBP (mmHg) 17 | 76.9 ± 0.13 a | 75.1 ± 0.27 b | 75.5 ± 0.09 b | 73.9 ± 0.20 c***++# | 0.698 (0.633–0.770) |
| AST (IU/L) 18 | 25.1 ± 0.20 a | 25.1 ± 0.45 a | 23.1 ± 0.14 b | 22.5 ± 0.36 b*** | 0.633 (0.550–0.729) |
| ALT (IU/L) 19 | 24.5 ± 0.16 a | 23.6 ± 0.34 b | 23.5 ± 0.12 b | 22.4 ± 0.25 c**+++ | 0.531 (0.485–0.582) |
| Egfr 20 | 84.4 ± 0.21 b | 84.1 ± 0.43 b | 86.6 ± 0.15 a | 85.3 ± 0.33 b***++ | 0.940 (0.867–1.020) |
| Arthritis (N, %) 9 | 698 (4.04) | 111 (3.72) | 3995 (11.8) | 326 (7.17) ‡‡‡ | 0.868 (0.774–0.973) |
| Osteoporosis (N, Yes%) 9 | 117 (0.68) | 16 (0.54) | 2779 (8.22) | 162(3.56) ‡‡‡ | 0.882 (0.740–1.051) |
| CHR 1 | SNP 2 | Base Pair | A1 3 | A2 4 | OR 5 | SE 6 | p for City 7 | p for Asan + Nong 8 | MAF 9 | p for HWE 10 | Gene Names | Location |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | rs4630744 | 33461375 | G | A | 1.105 | 0.01906 | 1.53 × 10−7 | 0.02458 | 0.3792 | 0.4099 | LTBP1 | Intron |
| 2 | rs13034890 | 71430542 | T | C | 0.9086 | 0.01886 | 3.72 × 10−7 | 0.0014 | 0.4567 | 0.6295 | PAIP2B | Intron |
| 2 | rs1249260 | 233046182 | C | T | 1.172 | 0.01877 | 2.99 × 10−17 | 0.00016 | 0.4515 | 0.7705 | DIS3L2 | Downstream |
| 3 | rs6762722 | 141145216 | G | A | 1.177 | 0.02095 | 6.65 × 10−15 | 0.000749 | 0.2546 | 0.5644 | ZBTB38 | Intron |
| 4 | rs3756173 | 8598698 | T | C | 0.9058 | 0.01917 | 2.48 × 10−7 | 0.000515 | 0.4135 | 0.5123 | CPZ | Intron |
| 4 | rs2074974 | 17812615 | C | A | 0.8989 | 0.01884 | 1.57 × 10−8 | 0.00103 | 0.461 | 0.8746 | NCAPG | 5′ UTR |
| 4 | rs7700107 | 17880416 | C | A | 0.8112 | 0.02421 | 5.34 × 10−18 | 6.78 × 10−5 | 0.2072 | 0.1078 | LCORL | Downstream |
| 15 | rs1600640 | 84603034 | T | G | 0.8711 | 0.02363 | 5.18 × 10−9 | 0.000252 | 0.2115 | 0.5692 | ADAMTSL3 | Intron |
| 15 | rs2871865 | 99194896 | G | C | 0.8078 | 0.03676 | 6.34 × 10−9 | 0.00673 | 0.0800 | 0.0640 | IGF1R | Intron |
| 20 | rs224331 | 34022387 | A | C | 1.191 | 0.02073 | 3.13 × 10−16 | 0.000956 | 0.2683 | 0.8747 | GDF5 | Missense (Ala276Ser) |
| Compounds | Wide Type | Mutated Type |
|---|---|---|
| Stachyurin | −13.7 | −13.8 |
| Lambertianin B | −13.3 | −13.3 |
| Sanguiin H6 | −13.2 | −13.3 |
| Lambertianin A | −13.2 | −13.3 |
| Mongolicain A | −12.9 | −12.3 |
| Casuariin | −12.7 | −12.7 |
| Punicacortein D | −12.5 | −11.9 |
| Rugosin E | −12.2 | −5 |
| Valolaginic acid | −12.1 | −9.7 |
| Rugosin D | −12 | −5 |
| Cinnamtannin II | −11.8 | −11.8 |
| Eugenigrandin A | −11.7 | −11.8 |
| Rugosin A | −11 | −5.1 |
| Chinese tannin | −10.9 | −11 |
| Gemin D | −10.7 | −10.6 |
| Low-PRS (n = 6107) | Middle-PRS (n = 29,668) | High-PRS (n = 22,926) | Adjusted ORs and 95 CI | |
|---|---|---|---|---|
| Height (cm) 1 | 160.4 ± 0.07 c | 160.7 ± 0.03 b | 160.9 ± 0.04 a*** | 1.293 (1.127–1.381) |
| BMI (kg/m2) 2 | 23.8 ± 0.04 | 23.8 ± 0.02 | 23.9 ± 0.02 | 1.054 (0.987–1.124) |
| Waist (cm) 3 | 80.6 ± 0.10 | 80.7 ± 0.05 | 80.7 ± 0.05 | 0.975 (0.883–1.077) |
| Weight at 18 4 | 54.8 ± 0.11 b | 55.2 ± 0.05 a | 55.1 ± 0.06 ab | 1.059 (0.994–1.129) |
| SMI 5 | 7.05 ± 0.008 a | 7.01 ± 0.004 b | 6.98 ± 0.004 c*** | 0.960 (0.896–1.029) |
| Fat mass 6 | 28.3 ± 0.04 | 28.3 ± 0.02 | 28.4 ± 0.02 | 0.959 (0.896–1.027) |
| WBC (109/L) 7 | 5.78 ± 0.02 a | 5.70 ± 0.01 b | 5.68 ± 0.01 b** | 0.894 (0.837–0.954) |
| Serum hs-CRP (mg/dL) 8 | 0.152 ± 0.006 a | 0.136 ± 0.003 b | 0.141 ± 0.003 ab* | 0.862 (0.675–1.100) |
| MetS 9 | 883 (14.5) | 4162 (14.0) | 3255 (14.2) | 0.894 (0.815–0.982) |
| Serum glucose (mg/dL) 10 | 95.5 ± 0.26 | 95.1 ± 0.12 | 95.0 ± 0.14 | 0.905 (0.828–0.990) |
| Blood HbA1c (%) 11 | 5.73 ± 0.01 a | 5.71 ± 0.01 b | 5.71 ± 0.01 b* | 0.851 (0.740–0.980) |
| Insulin resistance (%) | 1955 (11.3) | 347 (12.0) | 2066 (6.1) | 0.953 (0.854–1.064) |
| Serum total cholesterol (mg/dL) 12 | 197 ± 0.48 | 197 ± 0.22 | 197 ± 0.25 | 0.940 (0.875–1.010) |
| Serum HDL (mg/dL) 13 | 53.6 ± 0.17 | 53.8 ± 0.08 | 53.8 ± 0.09 | 1.047 (0.978–1.120) |
| Serum LDL (mg/dL) 14 | 119 ± 0.44 | 119 ± 0.20 | 119 ± 0.23 | 0.960 (0.883–1.043) |
| Serum TG (mg/dL) 15 | 126 ± 1.11 | 125 ± 0.50 | 124 ± 0.57 | 0.981 (0.917–1.048) |
| SBP (mmHg) 16 | 122 ± 0.19 | 122 ± 0.08 | 123 ± 0.10 | 1.050 (0.984–1.121) |
| DBP (mmHg) 17 | 75.6 ± 0.12 | 75.6 ± 0.06 | 75.8 ± 0.06 | 1.020 (0.919–1.132) |
| Serum AST (IU/L) 18 | 24.7 ± 0.31 a | 23.7 ± 0.14 b | 23.5 ± 0.16 b | 0.875 (0.755–1.014) |
| Serum ALT (IU/L) 19 | 23.4 ± 0.30 a | 22.4 ± 0.14 b | 22.1 ± 0.16 b | 0.893 (0.812–0.981) |
| Egfr 20 | 86.5 ± 0.20 | 86.0 ± 0.09 | 86.1 ± 0.11 | 1.059 (0.893–1.257) |
| Arthritis (N, %) | 549 (9.0) | 2619 (8.84) | 1962 (8.57) | 0.920 (0.825–1.026) |
| Osteoporosis (N, Yes%) | 344 (5.64) | 1568 (5.29) | 1162 (5.08) | 0.920 (0.800–1.057) |
| Low-PRS (n = 14,420) | Middle-PRS (n = 21,641) | High-PRS (n = 4201) | Gene-Nutrient Interaction p Value | |
|---|---|---|---|---|
| Low energy 1 High energy | 1 | 1.032 (0.914–1.165) 1.253 (1.075–1.461) | 1.130 (0.999–1.278) 1.414 (1.210–1.652) | 0.0078 |
| Low KBD 2 High KBD | 1 | 1.092 (0.971–1.228) 1.158 (0.984–1.362) | 1.183 (1.050–1.333) 1.334 (1.132–1.574) | 0.0923 |
| Low PBD 2 High PBD | 1 | 1.056 (0.938–1.188) 1.220 (1.039–1.432) | 1.157 (1.026–1.305) 1.377 (1.170–1.620) | 0.0567 |
| Low WSD 2 High WSD | 1 | 1.070 (0.937–1.222) 1.163 (1.015–1.332) | 1.188 (1.038–1.359) 1.282 (1.117–1.472) | 0.1898 |
| Low RMD 2 High RMD | 1 | 1.157 (1.027–1.303) 1.041 (0.888–1.220) | 1.318 (1.168–1.486) 1.092 (0.929–1.284) | 0.0095 |
| Low alcohol 3 High alcohol | 1 | 1.143 (1.003–1.303) 1.081 (0.941–1.241) | 1.286 (1.125–1.469) 1.175 (1.021–1.353) | 0.6960 |
| Low exercise 4 High exercise | 1 | 1.037 (0.901–1.194) 1.173 (1.031–1.335) | 1.202 (1.042–1.386) 1.253 (1.099–1.429) | 0.2233 |
| Non-smoking Former smoking +smoking | 1 | 1.124 (1.003–1.260) 1.095 (0.920–1.302) | 1.225 (1.091–1.375) 1.260 (1.057–1.502) | 0.0547 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S. Height-Related Polygenic Variants Are Associated with Metabolic Syndrome Risk and Interact with Energy Intake and a Rice-Main Diet to Influence Height in KoGES. Nutrients 2023, 15, 1764. https://doi.org/10.3390/nu15071764
Park S. Height-Related Polygenic Variants Are Associated with Metabolic Syndrome Risk and Interact with Energy Intake and a Rice-Main Diet to Influence Height in KoGES. Nutrients. 2023; 15(7):1764. https://doi.org/10.3390/nu15071764
Chicago/Turabian StylePark, Sunmin. 2023. "Height-Related Polygenic Variants Are Associated with Metabolic Syndrome Risk and Interact with Energy Intake and a Rice-Main Diet to Influence Height in KoGES" Nutrients 15, no. 7: 1764. https://doi.org/10.3390/nu15071764
APA StylePark, S. (2023). Height-Related Polygenic Variants Are Associated with Metabolic Syndrome Risk and Interact with Energy Intake and a Rice-Main Diet to Influence Height in KoGES. Nutrients, 15(7), 1764. https://doi.org/10.3390/nu15071764






