Metabolomic Profiling of Asparagine Deprivation in Asparagine Synthetase Deficiency Patient-Derived Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Cell Lines
2.2. Cell Culture and Gas Chromatography-Mass Spectrometry (GC-MS) Analysis
2.3. Identification of Metabolites and Chromatogram Integration
2.4. Statistical Analysis
3. Results
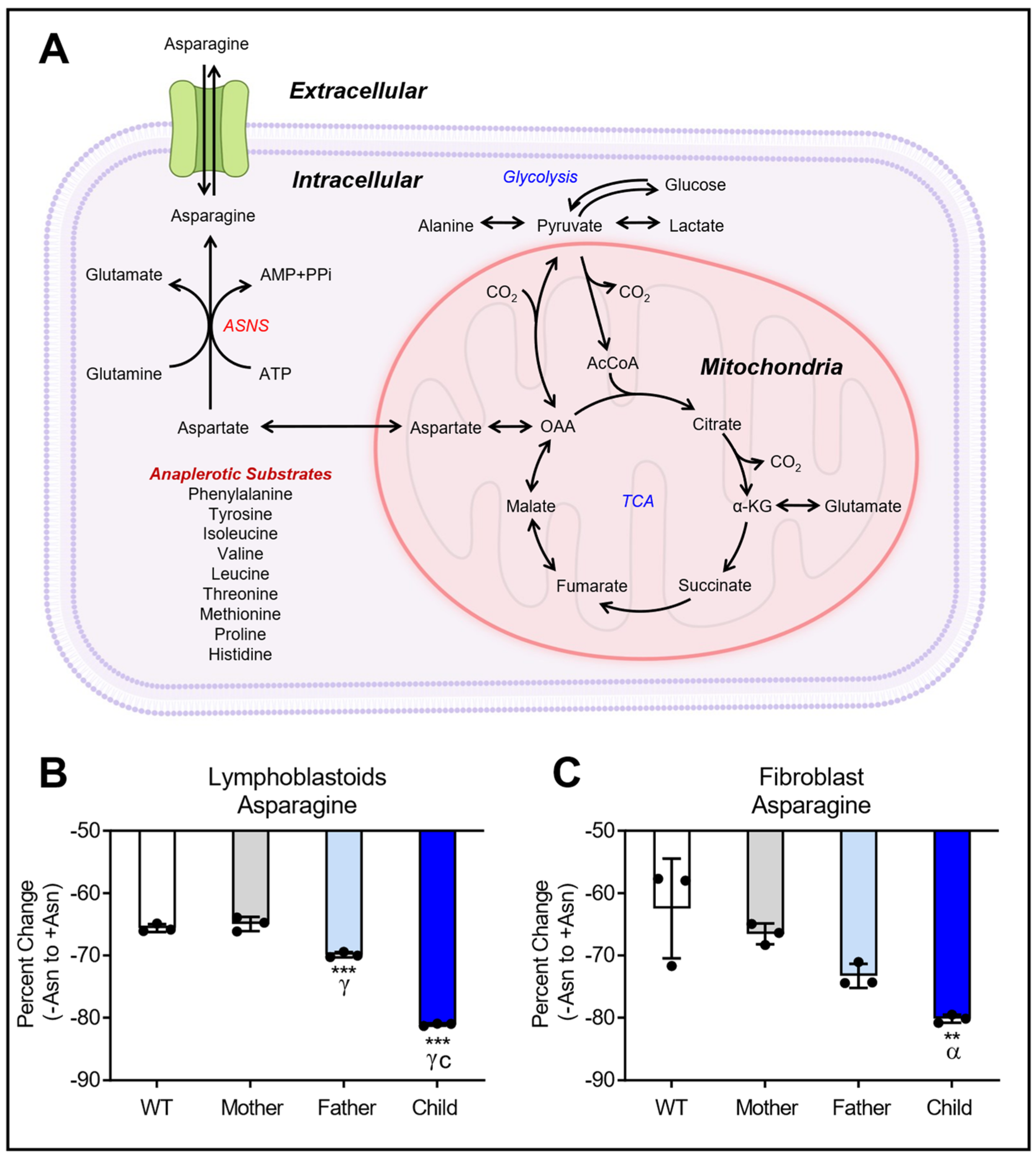
3.1. GC-MS Analysis of Asparagine for Lymphoblastoid and Fibroblast Cell Lines
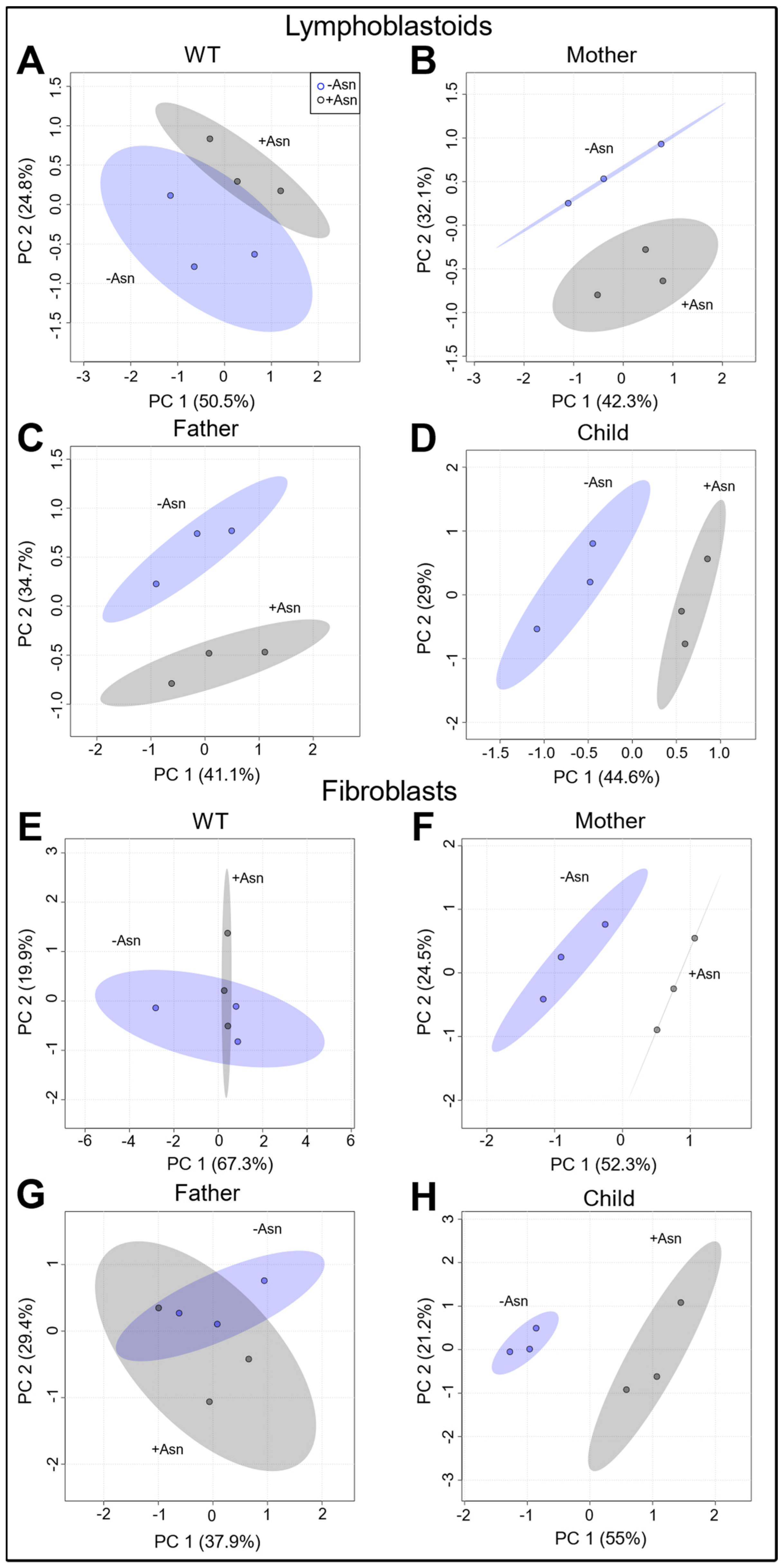
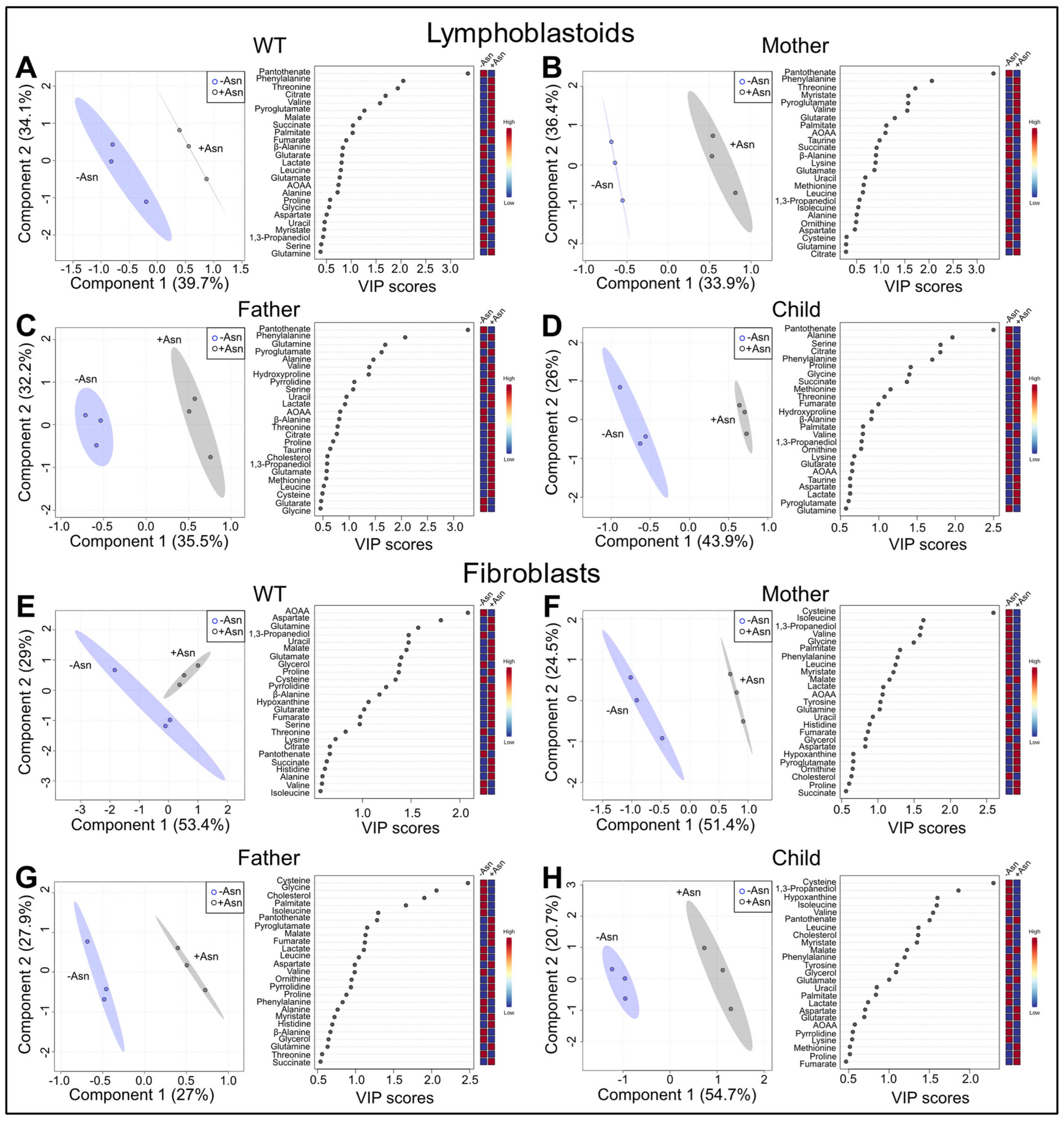
3.2. Analysis of Global Metabolomics
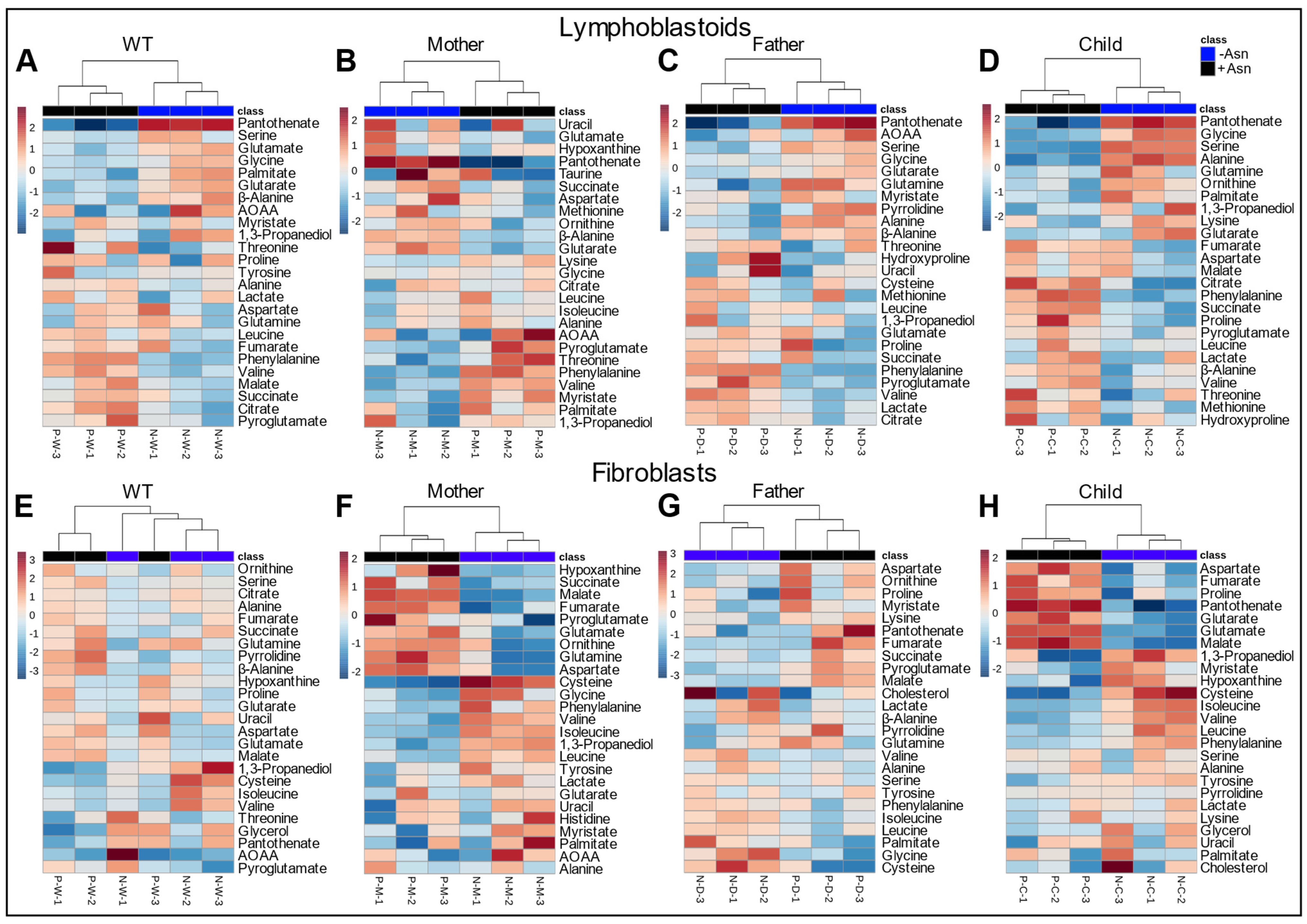
3.3. Hierarchical Clustering Analysis
3.4. Metabolites Identified in Common
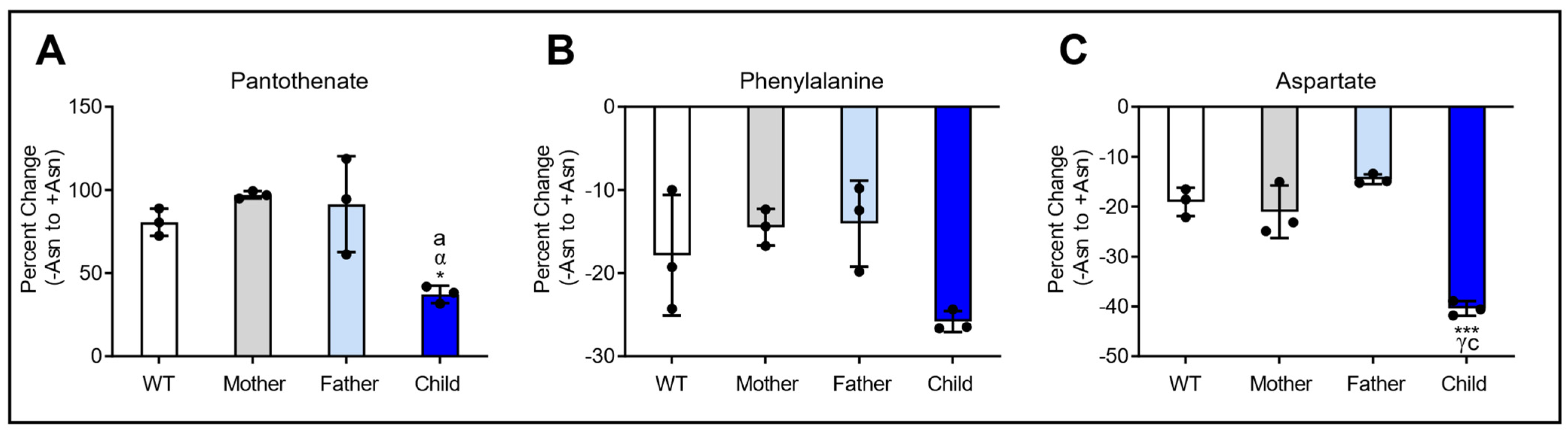
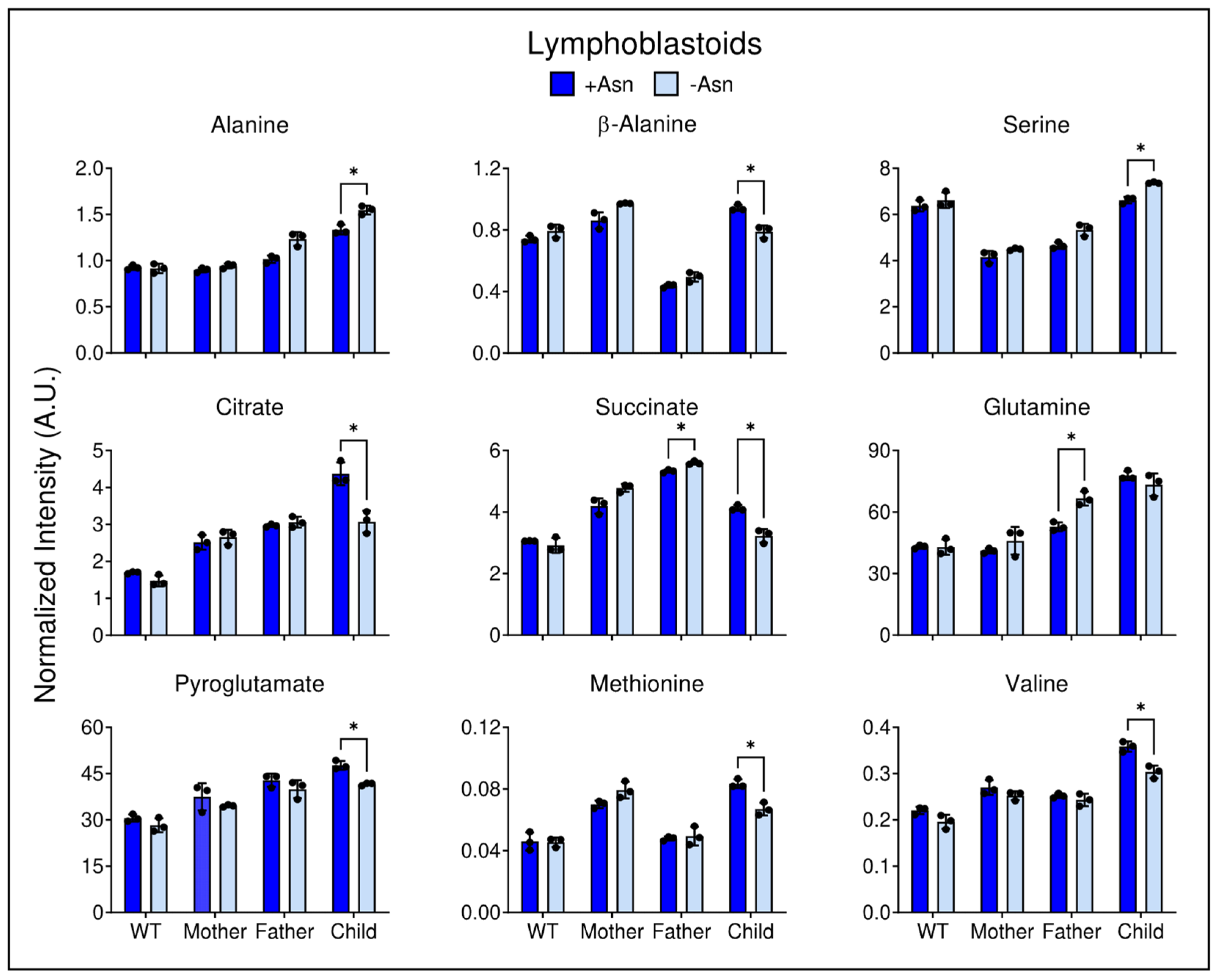
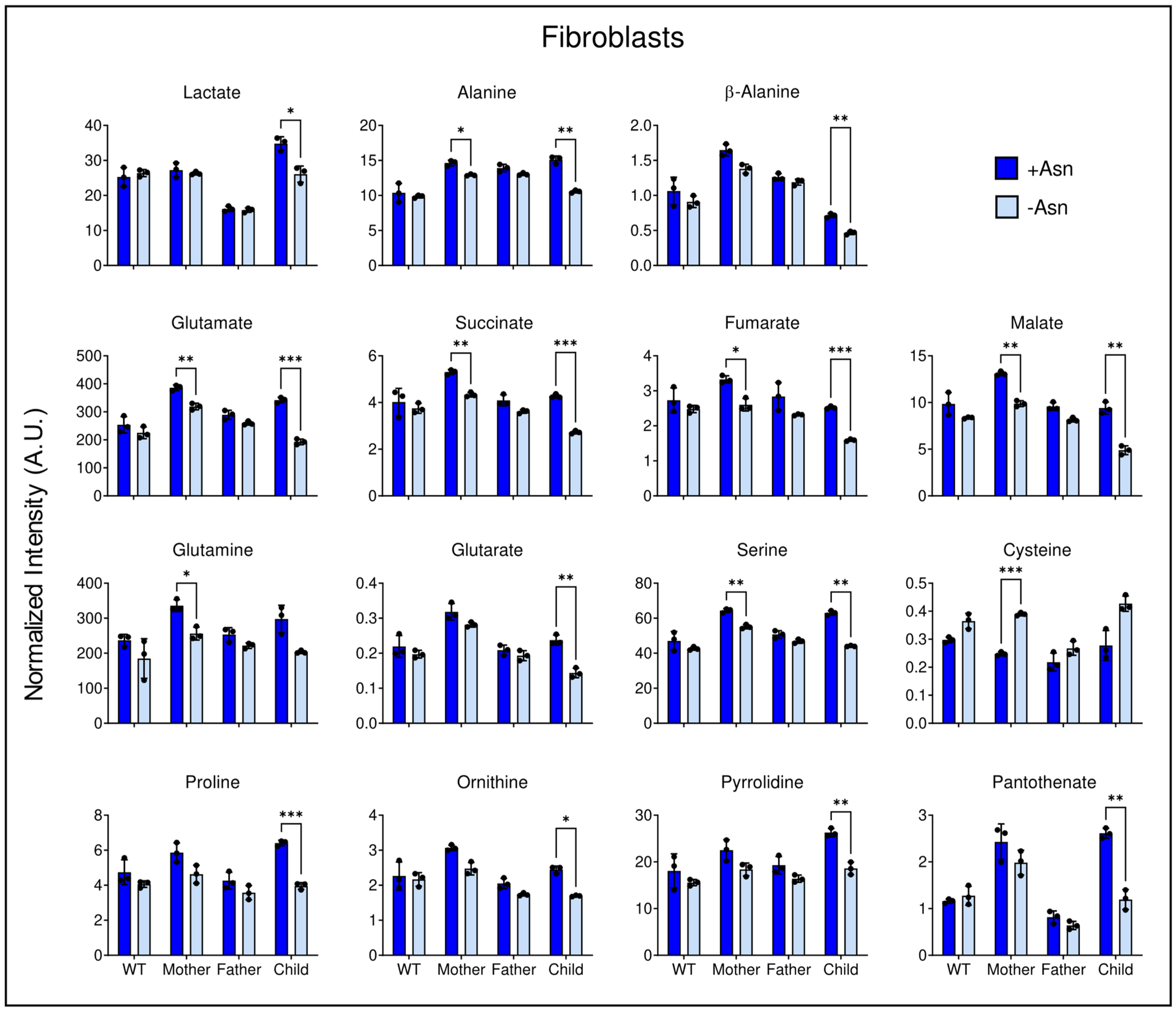
3.5. Asparagine Deprivation Significantly Perturbs Central Carbon Metabolism
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lomelino, C.L.; Andring, J.T.; McKenna, R.; Kilberg, M.S. Asparagine synthetase: Function, structure, and role in disease. J. Biol. Chem. 2017, 292, 19952–19958. [Google Scholar] [CrossRef]
- Richards, N.G.J.; Schuster, S.M. Mechanistic Issues in Asparagine Synthetase Catalysis. In Advances in Enzymology and Related Areas of Molecular Biology; Purich, D.L., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006; pp. 145–198. ISBN 978-0-470-12318-8. [Google Scholar]
- Richards, N.G.J.; Kilberg, M.S. Asparagine Synthetase Chemotherapy. Annu. Rev. Biochem. 2006, 75, 629–654. [Google Scholar] [CrossRef] [PubMed]
- Ben-Salem, S.; Gleeson, J.G.; Al-Shamsi, A.M.; Islam, B.; Hertecant, J.; Ali, B.R.; Al-Gazali, L. Asparagine synthetase deficiency detected by whole exome sequencing causes congenital microcephaly, epileptic encephalopathy and psychomotor delay. Metab. Brain Dis. 2014, 30, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.H.Q.; Shi, X.-M.; Scherer, S.W.; Andrulis, I.L.; Tsui, L.-C. Refined localization of the asparagine synthetase gene (ASNS) to chromosome 7, region q21.3, and characterization of the somatic cell hybrid line 4AF/106/K015. Cytogenet. Cell Genet. 1994, 66, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.P.; Lambert, M.A.; Cairney, A.E.L.; Wills, D.; Ray, P.N.; Andrulis, I.L. Molecular structure of the human asparagine synthetase gene. Genomics 1989, 4, 259–265. [Google Scholar] [CrossRef]
- Zhu, W.; Radadiya, A.; Bisson, C.; Wenzel, S.; Nordin, B.E.; Martínez-Márquez, F.; Imasaki, T.; Sedelnikova, S.E.; Coricello, A.; Baumann, P.; et al. High-resolution crystal structure of human asparagine synthetase enables analysis of inhibitor binding and selectivity. Commun. Biol. 2019, 2, 345. [Google Scholar] [CrossRef]
- Larsen, T.M.; Boehlein, S.K.; Schuster, S.M.; Richards, N.G.J.; Thoden, J.B.; Holden, H.M.; Rayment, I. Three-Dimensional Structure of Escherichia coli Asparagine Synthetase B: A Short Journey from Substrate to Product. Biochemistry 1999, 38, 16146–16157. [Google Scholar] [CrossRef]
- Tesson, A.R.; Soper, T.S.; Ciustea, M.; Richards, N.G.J. Revisiting the steady state kinetic mechanism of glutamine-dependent asparagine synthetase from Escherichia coli. Arch. Biochem. Biophys. 2003, 413, 23–31. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Hui, S.; Ghergurovich, J.M.; Fan, J.; Intlekofer, A.M.; White, R.M.; Rabinowitz, J.D.; Thompson, C.B.; Zhang, J. As Extracellular Glutamine Levels Decline, Asparagine Becomes an Essential Amino Acid. Cell Metab. 2018, 27, 428–438.e5. [Google Scholar] [CrossRef]
- Krall, A.S.; Xu, S.; Graeber, T.G.; Braas, D.; Christofk, H.R. Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nat. Commun. 2016, 7, 11457. [Google Scholar] [CrossRef]
- Krall, A.S.; Mullen, P.J.; Surjono, F.; Momcilovic, M.; Schmid, E.W.; Halbrook, C.J.; Thambundit, A.; Mittelman, S.D.; Lyssiotis, C.A.; Shackelford, D.B.; et al. Asparagine couples mitochondrial respiration to ATF4 activity and tumor growth. Cell Metab. 2021, 33, 1013–1026.e6. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, B.; Meister, A. Glutamine-dependent Asparagine Synthetase from Leukemia Cells. Chloride Dependence, Mechanism of Action, and Inhibition. J. Biol. Chem. 1972, 247, 6708–6719. [Google Scholar] [CrossRef]
- Horowitz, B.; Madras, B.K.; Meister, A.; Old, L.J.; Boyes, E.A.; Stockert, E. Asparagine Synthetase Activity of Mouse Leukemias. Science 1968, 160, 533–535. [Google Scholar] [CrossRef]
- Broome, J.D. Evidence that the L-asparaginase of guinea pig serum is responsible for its antilymphoma effects. J. Exp. Med. 1963, 118, 99–120. [Google Scholar] [CrossRef]
- Broome, J.D. Studies on the mechanism of tumor inhibition by l-asparaginase. J. Exp. Med. 1968, 127, 1055–1072. [Google Scholar] [CrossRef]
- Ruzzo, E.K.; Capo-Chichi, J.-M.; Ben-Zeev, B.; Chitayat, D.; Mao, H.; Pappas, A.L.; Hitomi, Y.; Lu, Y.-F.; Yao, X.; Hamdan, F.F.; et al. Deficiency of Asparagine Synthetase Causes Congenital Microcephaly and a Progressive Form of Encephalopathy. Neuron 2013, 80, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Radha Rama Devi, A.; Naushad, S.M. Molecular diagnosis of asparagine synthetase (ASNS) deficiency in two Indian families and literature review of 29 ASNS deficient cases. Gene 2019, 704, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Sprute, R.; Ardicli, D.; Oguz, K.K.; Malenica-Mandel, A.; Daimagüler, H.-S.; Koy, A.; Coskun, T.; Wang, H.; Topcu, M.; Cirak, S. Clinical outcomes of two patients with a novel pathogenic variant in ASNS: Response to asparagine supplementation and review of the literature. Hum. Genome Var. 2019, 6, 24. [Google Scholar] [CrossRef]
- Matsumoto, H.; Kawashima, N.; Yamamoto, T.; Nakama, M.; Otsuka, H.; Ago, Y.; Sasai, H.; Kubota, K.; Ozeki, M.; Kawamoto, N.; et al. In vitro functional analysis of four variants of human asparagine synthetase. J. Inherit. Metab. Dis. 2021, 44, 1226–1234. [Google Scholar] [CrossRef]
- Liu, L.; Wang, J.; Li, H.; Dong, Y.; Li, Y.; Xia, L.; Yang, B.; Wang, H.; Xu, Y.; Cheng, G.; et al. An intractable epilepsy phenotype of ASNS novel mutation in two patients with asparagine synthetase deficiency. Clin. Chim. Acta 2022, 531, 331–336. [Google Scholar] [CrossRef]
- Staklinski, S.J.; Chang, M.C.; Yu, F.; Ruff, K.C.; Franz, D.N.; Qian, Z.; Bloom, L.B.; Merritt, M.E.; McKenna, R.; Kilberg, M.S. Cellular and molecular characterization of two novel asparagine synthetase gene mutations linked to Asparagine Synthetase Deficiency. J. Biol. Chem. 2022, 298, 102385. [Google Scholar] [CrossRef] [PubMed]
- Sacharow, S.J.; Dudenhausen, E.E.; Lomelino, C.L.; Rodan, L.; El Achkar, C.M.; Olson, H.E.; Genetti, C.A.; Agrawal, P.B.; McKenna, R.; Kilberg, M.S. Characterization of a novel variant in siblings with Asparagine Synthetase Deficiency. Mol. Genet. Metab. 2018, 123, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Palmer, E.E.; Hayner, J.; Sachdev, R.; Cardamone, M.; Kandula, T.; Morris, P.; Dias, K.-R.; Tao, J.; Miller, D.; Zhu, Y.; et al. Asparagine Synthetase Deficiency causes reduced proliferation of cells under conditions of limited asparagine. Mol. Genet. Metab. 2015, 116, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Staklinski, S.J.; Chang, M.C.; Ahrens-Nicklas, R.C.; Kaur, S.; Stefanatos, A.K.; Dudenhausen, E.E.; Merritt, M.E.; Kilberg, M.S. Characterizing asparagine synthetase deficiency variants in lymphoblastoid cell lines. JIMD Rep. 2023, 64, 167–179. [Google Scholar] [CrossRef]
- Staklinski, S.J.; Snanoudj, S.; Guerrot, A.-M.; Vanhulle, C.; Lecoquierre, F.; Bekri, S.; Kilberg, M.S. Analysis of Enzyme Activity and Cellular Function for the N80S and S480F Asparagine Synthetase Variants Expressed in a Child with Asparagine Synthetase Deficiency. Int. J. Mol. Sci. 2022, 24, 559. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics by Gas Chromatography–Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr. Protoc. Mol. Biol. 2016, 114, 30.4.1–30.4.32. [Google Scholar] [CrossRef]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. FiehnLib: Mass Spectral and Retention Index Libraries for Metabolomics Based on Quadrupole and Time-of-Flight Gas Chromatography/Mass Spectrometry. Anal. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef]
- Mahar, R.; Donabedian, P.L.; Merritt, M.E. HDO production from [2H7]glucose Quantitatively Identifies Warburg Metabolism. Sci. Rep. 2020, 10, 8885. [Google Scholar] [CrossRef]
- Chang, M.C.; Mahar, R.; McLeod, M.A.; Giacalone, A.G.; Huang, X.; Boothman, D.A.; Merritt, M.E. Synergistic Effect of β-Lapachone and Aminooxyacetic Acid on Central Metabolism in Breast Cancer. Nutrients 2022, 14, 3020. [Google Scholar] [CrossRef]
- Mahar, R.; Ragavan, M.; Chang, M.C.; Hardiman, S.; Moussatche, N.; Behar, A.; Renne, R.; Merritt, M.E. Metabolic signatures associated with oncolytic myxoma viral infections. Sci. Rep. 2022, 12, 12599. [Google Scholar] [CrossRef]
- Siderius, D. NIST Standard Reference Simulation Website—SRD 173; National Institute of Standards and Technology: Gaithersburg, MD, USA, 2017.
- Zhang, J.; Fan, J.; Venneti, S.; Cross, J.R.; Takagi, T.; Bhinder, B.; Djaballah, H.; Kanai, M.; Cheng, E.H.; Judkins, A.R.; et al. Asparagine Plays a Critical Role in Regulating Cellular Adaptation to Glutamine Depletion. Mol. Cell 2014, 56, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Batra, S.; Zhang, J. Asparagine: A Metabolite to Be Targeted in Cancers. Metabolites 2021, 11, 402. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.; Taurino, G.; Bianchi, M.G.; Kilberg, M.S.; Bussolati, O. Asparagine Synthetase in Cancer: Beyond Acute Lymphoblastic Leukemia. Front. Oncol. 2020, 9, 1480. [Google Scholar] [CrossRef]
- Arnstein, H.R.V.; Barwick, C.W.; Lange, J.D.; Thomas, H.D.J. Control of Protein Synthesis by Amino Acid Supply: The Effect of Asparagine Deprivation on the Translation of Messenger RNA in Reticulocyte Lysates. FEBS Lett. 1986, 194, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Nambu, T.; Ebara, S.; Hasegawa, Y.; Toyoshima, K.; Tsuchiya, Y.; Tomita, D.; Fujimoto, J.; Kurasawa, O.; Takahara, C.; et al. Inhibition of GCN2 sensitizes ASNS-low cancer cells to asparaginase by disrupting the amino acid response. Proc. Natl. Acad. Sci. USA 2018, 115, E7776–E7785. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.; van der Meer, L.T.; van Leeuwen, F.N. Amino Acid Depletion Therapies: Starving Cancer Cells to Death. Trends Endocrinol. Metab. 2021, 32, 367–381. [Google Scholar] [CrossRef]
- Bauer, C.; Quante, M.; Breunis, W.B.; Regina, C.; Schneider, M.; Andrieux, G.; Gorka, O.; Groß, O.; Boerries, M.; Kammerer, B.; et al. Lack of Electron Acceptors Contributes to Redox Stress and Growth Arrest in Asparagine-Starved Sarcoma Cells. Cancers 2021, 13, 412. [Google Scholar] [CrossRef]
- Alfadhel, M.; El-Hattab, A.W. Asparagine Synthetase Deficiency. In GeneReviews®; Adam, M.P., Everman, D.B., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Mahar, R.; Zeng, H.; Giacalone, A.; Ragavan, M.; Mareci, T.H.; Merritt, M.E. Deuterated water imaging of the rat brain following metabolism of [2H7]glucose. Magn. Reson. Med. 2021, 85, 3049–3059. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, M.C.; Staklinski, S.J.; Malut, V.R.; Pierre, G.L.; Kilberg, M.S.; Merritt, M.E. Metabolomic Profiling of Asparagine Deprivation in Asparagine Synthetase Deficiency Patient-Derived Cells. Nutrients 2023, 15, 1938. https://doi.org/10.3390/nu15081938
Chang MC, Staklinski SJ, Malut VR, Pierre GL, Kilberg MS, Merritt ME. Metabolomic Profiling of Asparagine Deprivation in Asparagine Synthetase Deficiency Patient-Derived Cells. Nutrients. 2023; 15(8):1938. https://doi.org/10.3390/nu15081938
Chicago/Turabian StyleChang, Mario C., Stephen J. Staklinski, Vinay R. Malut, Geraldine L. Pierre, Michael S. Kilberg, and Matthew E. Merritt. 2023. "Metabolomic Profiling of Asparagine Deprivation in Asparagine Synthetase Deficiency Patient-Derived Cells" Nutrients 15, no. 8: 1938. https://doi.org/10.3390/nu15081938
APA StyleChang, M. C., Staklinski, S. J., Malut, V. R., Pierre, G. L., Kilberg, M. S., & Merritt, M. E. (2023). Metabolomic Profiling of Asparagine Deprivation in Asparagine Synthetase Deficiency Patient-Derived Cells. Nutrients, 15(8), 1938. https://doi.org/10.3390/nu15081938






