Gut Immunobiosis and Biomodulators
Abstract
:1. Introduction
1.1. One, None, a Hundred Thousand…Self
1.2. In Gut We Trust
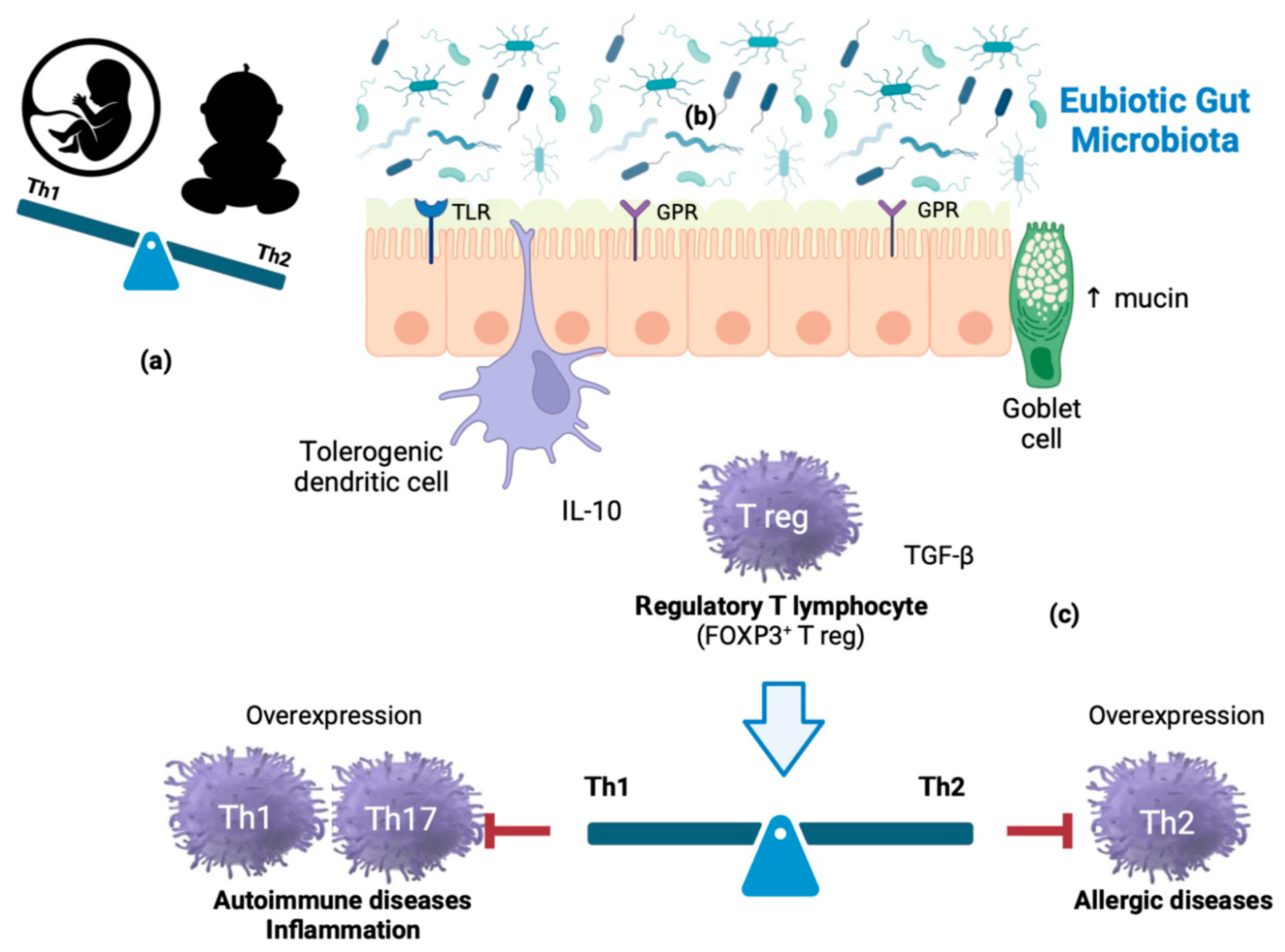
2. Immunobiosis
3. Biomodulators and Immuno-Nutrition
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ma’ayan, A. Complex systems biology. J. R. Soc. Interface 2017, 14, 20170391. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.; Wilson, M. Homo bacteriens and a network of surprises. J. Med. Microbiol. 1996, 45, 393–394. [Google Scholar] [CrossRef]
- Cristofori, F.; Dargenio, V.N.; Dargenio, C.; Miniello, V.L.; Barone, M.; Francavilla, R. Anti-inflammatory and immunomodulatory effects of probiotics in gut inflammation: A door to the body. Front. Immunol. 2021, 12, 578386. [Google Scholar] [CrossRef] [PubMed]
- Fassarella, M.; Blaak, E.E.; Penders, J.; Nauta, A.; Smidt, H.; Zoetendal, E.G. Gut microbiome stability and resilience: Elucidating the response to perturbations in order to modulate gut health. Gut 2021, 70, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Sharon, I.; Quijada, N.M.; Pasolli, E.; Fabbrini, M.; Vitali, F.; Agamennone, V.; Dötsch, A.; Selberherr, E.; Grau, J.H.; Meixner, M.; et al. The core human microbiome: Does it exist and how can we find it? A critical review of the concept. Nutrients 2022, 14, 2872. [Google Scholar] [CrossRef]
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Galazzo, G.; van Best, N.; Bervoets, L.; Dapaah, I.O.; Savelkoul, P.H.; Hornef, M.W.; Hutton, E.K.; Morrison, K.; Holloway, A.C.; McDonald, H.; et al. Development of the microbiota and associations with birth mode, diet, and atopic disorders in a longitudinal analysis of stool samples, collected lrom infancy through early childhood. Gastroenterology 2020, 158, 1584–1596. [Google Scholar] [CrossRef]
- Ionescu, R.F.; Enache, R.M.; Cretoiu, S.M.; Cretoiu, D. The interplay between gut microbiota and miRNAs in cardiovascular diseases. Front. Cardiovasc. Med. 2022, 9, 856901. [Google Scholar] [CrossRef]
- Casado-Bedmar, M.; Viennois, E. MicroRNA and gut microbiota: Tiny but mighty-novel insights into their cross-talk in inflammatory bowel disease pathogenesis and therapeutics. J. Crohn’s Colitis 2022, 16, 992–1005. [Google Scholar] [CrossRef]
- Teng, Y.; Ren, Y.; Sayed, M.; Hu, X.; Lei, C.; Kumar, A.; Hutchins, E.; Mu, J.; Deng, Z.; Luo, C.; et al. Plant-Derived Exosomal MicroRNAs Shape the Gut Microbiota. Cell Host Microbe 2018, 24, 637–652.e8. [Google Scholar] [CrossRef] [PubMed]
- Spinler, J.K.; Oezguen, N.; Runge, J.K.; Luna, R.A.; Karri, V.; Yang, J.; Hirschi, K.D. Dietary impact of a plant-derived microRNA on the gut microbiome. ExRNA 2020, 2, 11. [Google Scholar] [CrossRef]
- Netea, M.G.; Domínguez-Andrés, J.; Barreiro, L.B.; Chavakis, T.; Divangahi, M.; Fuchs, E.; Joosten, L.A.B.; van der Meer, J.W.M.; Mhlanga, M.M.; Mulder, W.J.M.; et al. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 2020, 20, 375–388. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhang, Y.; Ge, Y.; Zhu, X.; Pan, J. Regulatory Mechanisms and Promising Applications of Quorum Sensing-Inhibiting Agents in Control of Bacterial Biofilm Formation. Front. Microbiol. 2020, 11, 589640. [Google Scholar] [CrossRef] [PubMed]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Shin, J.H.; Seeley, R.J. REG3 proteins as gut hormones? Endocrinology 2019, 160, 1506–1514. [Google Scholar] [CrossRef] [PubMed]
- Miniello, V.L.; Verga, M.C.; Miniello, A.; Di Mauro, C.; Diaferio, L.; Francavilla, R. Complementary feeding and iron status: “the unbearable lightness of being” infants. Nutrients 2021, 13, 4201. [Google Scholar] [CrossRef]
- Francavilla, R.; Cristofori, F.; Tripaldi, M.E.; Indrio, F. Intervention for dysbiosis in children born by C-section. Ann. Nutr. Metab. 2018, 73, 33–39. [Google Scholar] [CrossRef]
- O’Callaghan, A.; van Sinderen, D. Bifidobacteria and their role as members of the human gut microbiota. Front. Microbiol. 2016, 7, 925. [Google Scholar] [CrossRef]
- Dempsey, E.; Corr, S.C. Lactobacillus spp. for Gastrointestinal Health: Current and Future Perspectives. Front. Immunol. 2022, 13, 840245. [Google Scholar] [CrossRef]
- Simonyté Sjödin, K.; Domellöf, M.; Lagerqvist, C.; Hernell, O.; Lönnerdal, B.; Szymlek-Gay, E.A.; Sjödin, A.; West, C.E.; Lind, T. Administration of ferrous sulfate drops has significant effects on the gut microbiota of iron-sufficient infants: A randomised controlled study. Gut 2019, 68, 2095–2097. [Google Scholar] [CrossRef] [PubMed]
- Georgieff, M.K.; Krebs, N.F.; Cusick, S.E. The Benefits and Risks of Iron Supplementation in Pregnancy and Childhood. Annu. Rev. Nutr. 2019, 39, 121–146. [Google Scholar] [CrossRef] [PubMed]
- McMillen, S.; Thomas, S.; Liang, E.; Nonnecke, E.B.; Slupsky, C.; Lönnerdal, B. Gut microbiome alterations following postnatal iron supplementation depend on iron form and persist into adulthood. Nutrients 2022, 14, 412. [Google Scholar] [CrossRef] [PubMed]
- Björmsjö, M.; Hernell, O.; Lönnerdal, B.; Berglund, S.K. Reducing iron content in infant formula from 8 to 2 mg/l does not increase the risk of iron deficiency at 4 or 6 months of age: A randomized controlled trial. Nutrients 2021, 13, 3. [Google Scholar] [CrossRef]
- Agrawal, S.; Berggren, K.L.; Marks, E.; Fox, J.H. Impact of high iron intake on cognition and neurodegeneration in humans and in animal models: A systematic review. Nutr. Rev. 2017, 75, 456–470. [Google Scholar] [CrossRef]
- Parmanand, B.A.; Kellingray, L.; Le Gall, G.; Basit, A.W.; Fairweather-Tait, S.; Narbad, A. A decrease in iron availability to human gut microbiome reduces the growth of potentially pathogenic gut bacteria; an in vitro colonic fermentation study. J. Nutr. Biochem. 2019, 67, 20–27. [Google Scholar] [CrossRef]
- Li, W.; Liu, B.; Lin, Y.; Xue, P.; Lu, Y.; Song, S.; Li, Y.; Szeto, I.M.; Ren, F.; Guo, H. The application of lactoferrin in infant formula: The past, present and future. Crit. Rev. Food Sci. Nutr. 2022, 19, 1–20. [Google Scholar] [CrossRef]
- Kell, D.B.; Heyden, E.L.; Pretorius, E. The Biology of Lactoferrin, an Iron-Binding Protein That Can Help Defend Against Viruses and Bacteria. Front. Immunol. 2020, 11, 1221. [Google Scholar] [CrossRef]
- Björmsjö, M.; Hernell, O.; Lönnerdal, B.; Berglund, S.K. Immunological Effects of Adding Bovine Lactoferrin and Reducing Iron in Infant Formula: A Randomized Controlled Trial. J. Pediatr. Gastroenterol. Nutr. 2022, 74, e65–e72. [Google Scholar] [CrossRef]
- Miniello, V.L.; Colasanto, A.; Cristofori, F.; Diaferio, L.; Ficele, L.; Lieggi, M.S.; Santoiemma, V.; Francavilla, R. Gut microbiota biomodulators, when the stork comes by the scalpel. Clin. Chim. Acta 2015, 451, 88–96. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Swanson, K.S.; Gibson, G.R.; Hutkins, R.; Reimer, R.A.; Reid, G.; Verbeke, K.; Scott, K.P.; Holscher, H.D.; Azad, M.B.; Delzenne, N.M.; et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 687–701. [Google Scholar] [CrossRef]
- Salminen, S.; Collado, M.C.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.M.M.; Sanders, M.E.; Shamir, R.; Swann, J.R.; Szajewska, H.; et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 649–667. [Google Scholar] [CrossRef] [PubMed]
- Fabiano, V.; Indrio, F.; Verduci, E.; Calcaterra, V.; Pop, T.L.; Mari, A.; Zuccotti, G.V.; Cokugras, F.C.; Pettoello-Mantovani, M.; Goulet, O. Term infant formulas influencing gut microbiota: An overview. Nutrients 2021, 13, 4200. [Google Scholar] [CrossRef]
- Mennella, J.A.; Li, Y.; Bittinger, K.; Friedman, E.S.; Zhao, C.; Li, H.; Wu, G.D.; Trabulsi, J.C. The Macronutrient Composition of Infant Formula Produces Differences in Gut Microbiota Maturation That Associates with Weight Gain Velocity and Weight Status. Nutrients 2022, 14, 1241. [Google Scholar] [CrossRef]
- Skórka, A.; Pieścik-Lech, M.; Kołodziej, M.; Szajewska, H. Infant formulae supplemented with prebiotics: Are they better than unsupplemented formulae? An updated systematic review. Br. J. Nutr. 2018, 119, 810–825. [Google Scholar] [CrossRef]
- Ivakhnenko, O.S.; Nyankovskyy, S.L. Effect of the specific infant formula mixture of oligosaccharides on local immunity and development of allergic and infectious disease in young children: Randomized study. Pediatr. Pol. 2013, 88, 398–404. [Google Scholar] [CrossRef]
- Borewicz, K.; Suarez-Diez, M.; Hechler, C.; Beijers, R.; de Weerth, C.; Arts, I.; Penders, J.; Thijs, C.; Nauta, A.; Lindner, C.; et al. The effect of prebiotic fortified infant formulas on microbiota composition and dynamics in early life. Sci. Rep. 2019, 9, 2434. [Google Scholar] [CrossRef]
- Mitselou, N.; Hallberg, J.; Stephansson, O.; Almqvist, C.; Melén, E.; Ludvigsson, J.F. Cesarean delivery, preterm birth, and risk of food allergy: Nationwide Swedish cohort study of more than 1 million children. J. Allergy Clin. Immunol. 2018, 142, 1510–1514.e2. [Google Scholar] [CrossRef]
- Martín-Peláez, S.; Cano-Ibáñez, N.; Pinto-Gallardo, M.; Amezcua-Prieto, C. The Impact of Probiotics, Prebiotics, and Synbiotics during Pregnancy or Lactation on the Intestinal Microbiota of Children Born by Cesarean Section: A Systematic Review. Nutrients 2022, 14, 341. [Google Scholar] [CrossRef] [PubMed]
- Sprenger, N.; Tytgat, H.L.P.; Binia, A.; Austin, S.; Singhal, A. Biology of human milk oligosaccharides: From basic science to clinical evidence. J. Hum. Nutr. Diet. 2022, 35, 280–299. [Google Scholar] [CrossRef] [PubMed]
- Wiciński, M.; Sawicka, E.; Gębalski, J.; Kubiak, K.; Malinowski, B. Human milk oligosaccharides: Health benefits, potential applications in infant formulas, and pharmacology. Nutrients 2020, 12, 266. [Google Scholar] [CrossRef] [PubMed]
- Pero-Gascon, R.; Hemeryck, L.Y.; Poma, G.; Falony, G.; Nawrot, T.S.; Raes, J.; Vanhaecke, L.; De Boevre, M.; Covaci, A.; De Saeger, S. FLEXiGUT: Rationale for exposomics associations with chronic low-grade gut inflammation. Environ. Int. 2022, 158, 106906. [Google Scholar] [CrossRef] [PubMed]
- Canani, R.B.; Paparo, L.; Nocerino, R.; Di Scala, C.; Della Gatta, G.; Maddalena, Y.; Buono, A.; Bruno, C.; Voto, L.; Ercolini, D. Gut microbiome as target for innovative strategies against food allergy. Front. Immunol. 2019, 10, 191. [Google Scholar] [CrossRef] [PubMed]
- Berni Canani, R.; Di Costanzo, M.; Bedogni, G.; Amoroso, A.; Cosenza, L.; Di Scala, C.; Granata, V.; Nocerino, R. Extensively hydrolyzed casein formula containing Lactobacillus rhamnosus GG reduces the occurrence of other allergic manifestations in children with cow’s milk allergy: 3-year randomized controlled trial. J. Allergy Clin. Immunol. 2017, 139, 1906–1913.e4. [Google Scholar] [CrossRef]
- Akobeng, A.K.; Singh, P.; Kumar, M.; Al Khodor, S. Role of the gut microbiota in the pathogenesis of coeliac disease and potential therapeutic implications. Eur. J. Nutr. 2020, 59, 3369–3390. [Google Scholar] [CrossRef]

| Function | |
|---|---|
| Protective |
|
| Trophic Metabolic Structural |
|
| Human TLR | Ligands |
|---|---|
| TLR1 |
|
| TLR2 |
|
| TLR3 |
|
| TLR4 |
|
| TLR5 |
|
| TLR6 |
|
| TLR7 |
|
| TLR8 |
|
| TLR9 |
|
| TLR10 |
|
| Biomodulator | Definition |
|---|---|
| Probiotic | Live microorganisms which confer a health benefit on the host when administered in adequate amounts |
| Prebiotic | A substrate that is selectively utilized by host microorganisms conferring health benefits on the host |
| Synbiotic | A mixture comprising live microorganisms and substrate(s) selectively utilized by host microorganisms that confers health benefits on the host |
| Postbiotic | Preparation of inanimate microorganisms and/or their components that confers a health benefit on the host |
| Mechanism of Action | |
|---|---|
| Microbiological |
|
| Structural |
|
| Immunologic |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miniello, V.L.; Miniello, A.; Ficele, L.; Skublewska-D’Elia, A.; Dargenio, V.N.; Cristofori, F.; Francavilla, R. Gut Immunobiosis and Biomodulators. Nutrients 2023, 15, 2114. https://doi.org/10.3390/nu15092114
Miniello VL, Miniello A, Ficele L, Skublewska-D’Elia A, Dargenio VN, Cristofori F, Francavilla R. Gut Immunobiosis and Biomodulators. Nutrients. 2023; 15(9):2114. https://doi.org/10.3390/nu15092114
Chicago/Turabian StyleMiniello, Vito Leonardo, Andrea Miniello, Laura Ficele, Aleksandra Skublewska-D’Elia, Vanessa Nadia Dargenio, Fernanda Cristofori, and Ruggiero Francavilla. 2023. "Gut Immunobiosis and Biomodulators" Nutrients 15, no. 9: 2114. https://doi.org/10.3390/nu15092114
APA StyleMiniello, V. L., Miniello, A., Ficele, L., Skublewska-D’Elia, A., Dargenio, V. N., Cristofori, F., & Francavilla, R. (2023). Gut Immunobiosis and Biomodulators. Nutrients, 15(9), 2114. https://doi.org/10.3390/nu15092114







