Adverse Maternal Environments Perturb Hepatic DNA Methylome and Transcriptome Prior to the Adult-Onset Non-Alcoholic Fatty Liver Disease in Mouse Offspring
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.1.1. Sex Determination
2.1.2. Hepatic TG Quantification
2.2. Reduced Representation Bisulfite Sequencing (RRBS)
2.3. RNA-Seq
2.4. Real-Time Reverse Transcriptase (RT) -PCR
2.5. Bioinformatics Analysis and Data Integration of RRBS with RNA-Seq
2.6. GO Term Analysis
2.7. Statistical Analysis
3. Results
3.1. AME Male Offspring Did Not Develop Hepatic Steatosis but Were Significantly Heavier Than the Age-Matched Controls at d21
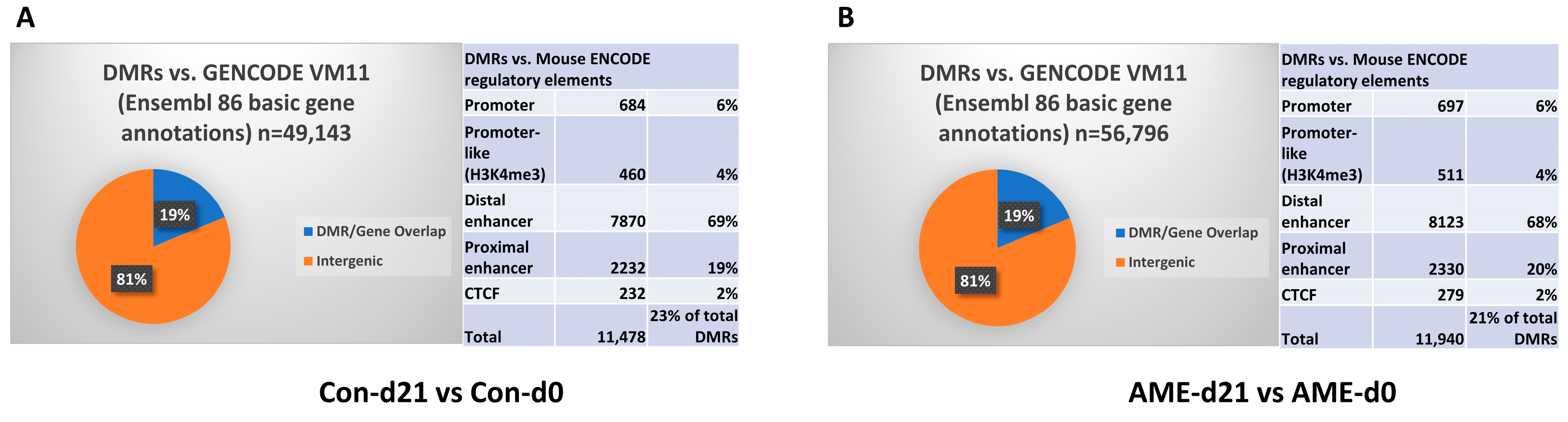
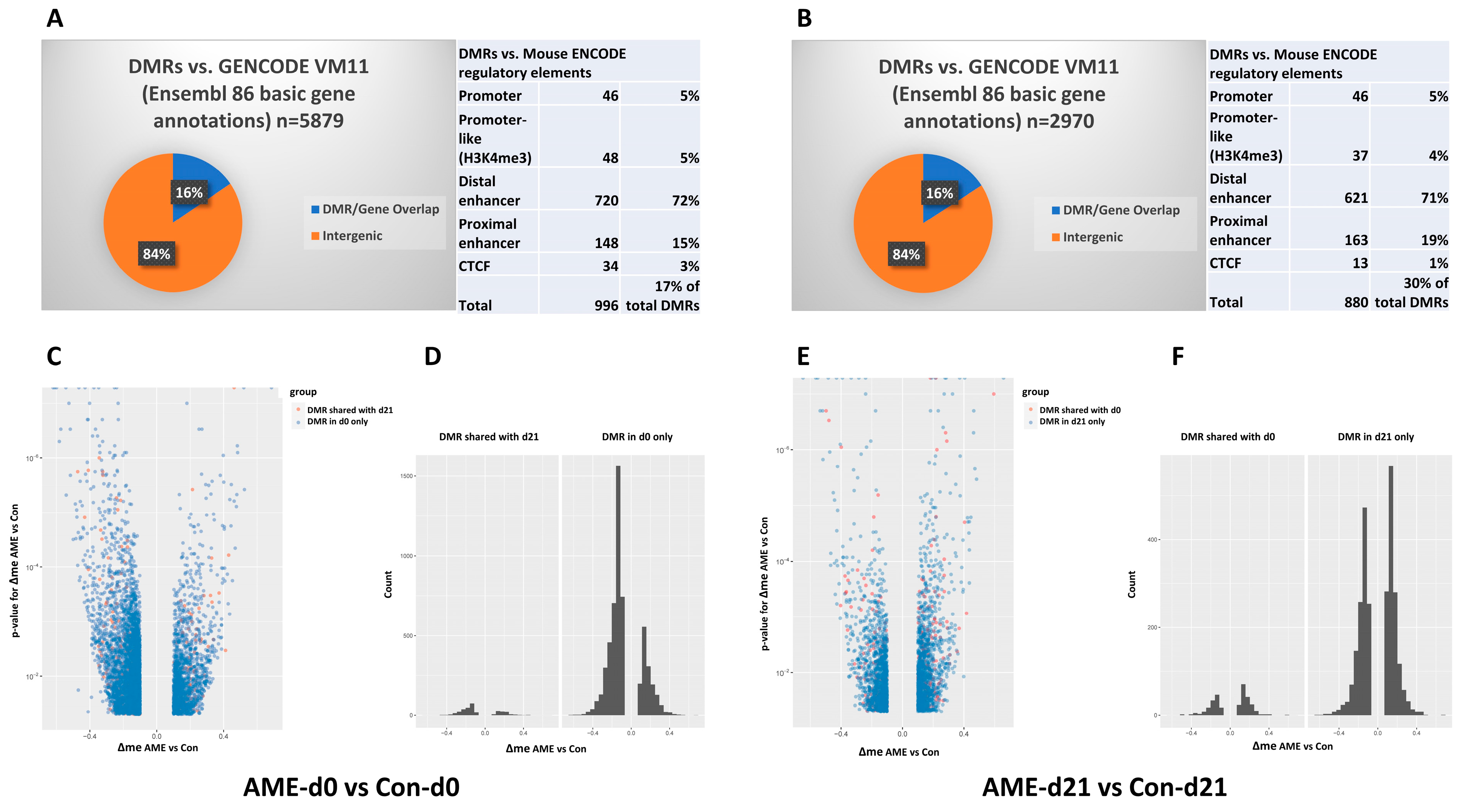
3.2. AME Perturbed the DNA Methylome at Both d0 and d21
3.3. AME Did Not Impact Hepatic Transcriptome at d0 despite the Large Number of DMRs
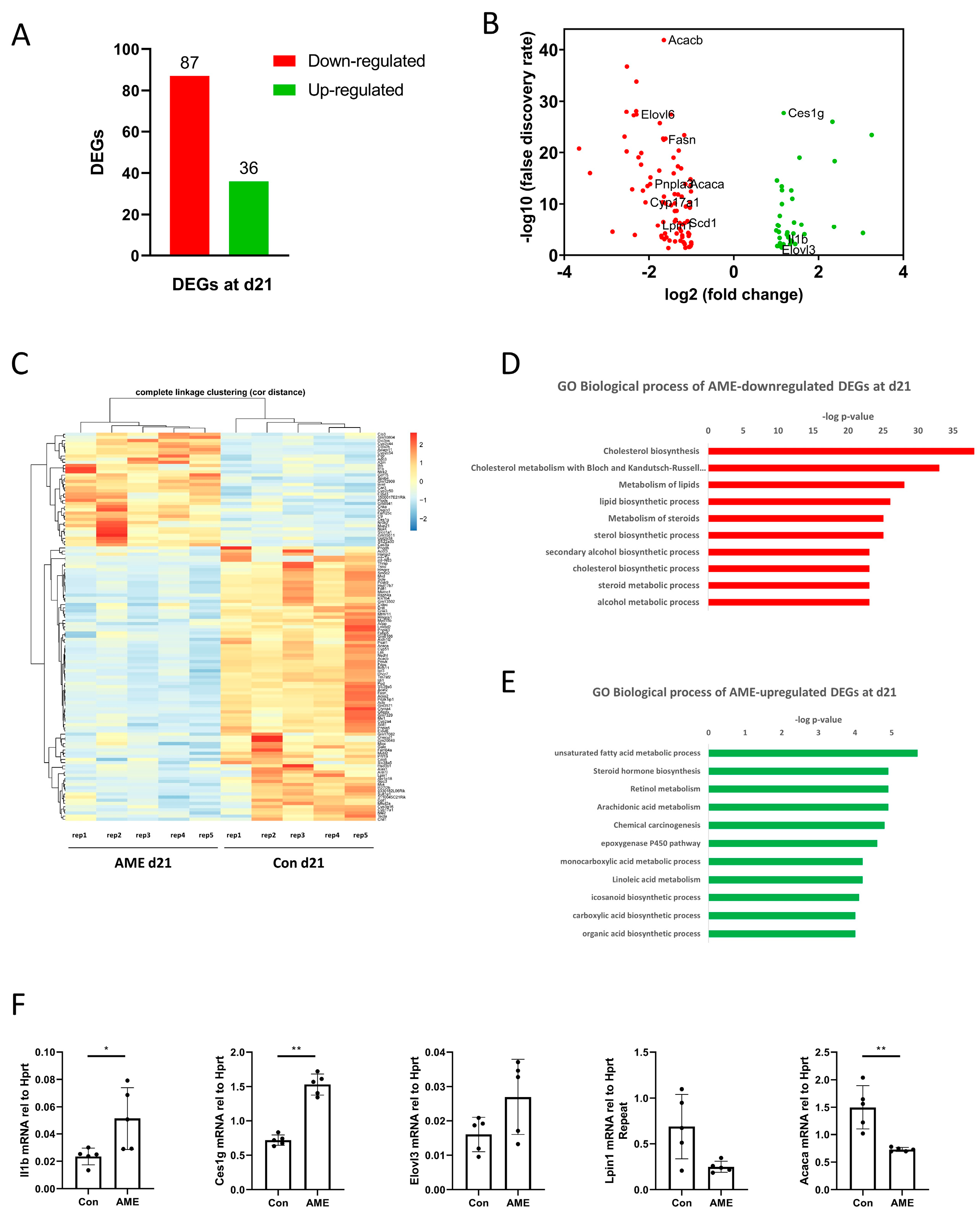
3.4. At d21, AME Perturbed Hepatic Transcriptome, Which Did Not Correlate with DMRs
3.5. AME Significantly Perturbed Genes on Metabolic Pathways at d21
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cohen, J.C.; Horton, J.D.; Hobbs, H.H. Human fatty liver disease: Old questions and new insights. Science 2011, 332, 1519–1523. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; North, P.E.; Ke, X.; Huang, Y.W.; Fritz, K.A.; Majnik, A.V.; Lane, R.H. Adverse Maternal Environment and Postweaning Western Diet Alter Hepatic CD36 Expression and Methylation Concurrently with Nonalcoholic Fatty Liver Disease in Mouse Offspring. J. Nutr. 2021, 151, 3102–3112. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Mao, C.; Li, S.; Zhang, Y.; Lv, J.; Jiang, S.; Xu, Z. Hepatic insulin signaling changes: Possible mechanism in prenatal hypoxia-increased susceptibility of fatty liver in adulthood. Endocrinology 2012, 153, 4955–4965. [Google Scholar] [CrossRef] [PubMed]
- Charlton, M.; Krishnan, A.; Viker, K.; Sanderson, S.; Cazanave, S.; McConico, A.; Masuoko, H.; Gores, G. Fast food diet mouse: Novel small animal model of NASH with ballooning, progressive fibrosis, and high physiological fidelity to the human condition. Am. J. Physiol.-Gastrointest. Liver Physiol. 2011, 301, G825–G834. [Google Scholar] [CrossRef]
- Gutierrez Sanchez, L.H.; Tomita, K.; Guo, Q.; Furuta, K.; Alhuwaish, H.; Hirsova, P.; Baheti, S.; Alver, B.; Hlady, R.; Robertson, K.D.; et al. Perinatal Nutritional Reprogramming of the Epigenome Promotes Subsequent Development of Nonalcoholic Steatohepatitis. Hepatol. Commun. 2018, 2, 1493–1512. [Google Scholar] [CrossRef]
- Gabory, A.; Attig, L.; Junien, C. Developmental programming and epigenetics. Am. J. Clin. Nutr. 2011, 94, 1943S–1952S. [Google Scholar] [CrossRef]
- Lee, H.S. Impact of Maternal Diet on the Epigenome during In Utero Life and the Developmental Programming of Diseases in Childhood and Adulthood. Nutrients 2015, 7, 9492–9507. [Google Scholar] [CrossRef]
- Waterland, R.A.; Jirtle, R.L. Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 2003, 23, 5293–5300. [Google Scholar] [CrossRef]
- Wadhwa, P.D.; Buss, C.; Entringer, S.; Swanson, J.M. Developmental origins of health and disease: Brief history of the approach and current focus on epigenetic mechanisms. Semin. Reprod. Med. 2009, 27, 358–368. [Google Scholar] [CrossRef]
- Tobi, E.W.; Goeman, J.J.; Monajemi, R.; Gu, H.; Putter, H.; Zhang, Y.; Slieker, R.C.; Stok, A.P.; Thijssen, P.E.; Muller, F.; et al. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat. Commun. 2014, 5, 5592. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Stricker, S.H.; Koferle, A.; Beck, S. From profiles to function in epigenomics. Nat. Rev. Genet. 2017, 18, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Stadler, M.B.; Murr, R.; Burger, L.; Ivanek, R.; Lienert, F.; Scholer, A.; van Nimwegen, E.; Wirbelauer, C.; Oakeley, E.J.; Gaidatzis, D.; et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 2011, 480, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, J.; Duan, J.; Gao, X.; Zhu, W.; Lu, X.; Yang, L.; Zhang, J.; Li, G.; Ci, W.; et al. Programming and inheritance of parental DNA methylomes in mammals. Cell 2014, 157, 979–991. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Hariharan, M.; Gorkin, D.U.; Dickel, D.E.; Luo, C.; Castanon, R.G.; Nery, J.R.; Lee, A.Y.; Zhao, Y.; Huang, H.; et al. Spatiotemporal DNA methylome dynamics of the developing mouse fetus. Nature 2020, 583, 752–759. [Google Scholar] [CrossRef] [PubMed]
- Gordon, L.; Joo, J.E.; Powell, J.E.; Ollikainen, M.; Novakovic, B.; Li, X.; Andronikos, R.; Cruickshank, M.N.; Conneely, K.N.; Smith, A.K.; et al. Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Res. 2012, 22, 1395–1406. [Google Scholar] [CrossRef] [PubMed]
- Gluckman, P.D.; Hanson, M.A.; Beedle, A.S. Early life events and their consequences for later disease: A life history and evolutionary perspective. Am. J. Hum. Biol. 2007, 19, 1–19. [Google Scholar] [CrossRef]
- Fu, Q.; Yu, X.; Callaway, C.W.; Lane, R.H.; McKnight, R.A. Epigenetics: Intrauterine growth retardation (IUGR) modifies the histone code along the rat hepatic IGF-1 gene. FASEB J. 2009, 23, 2438–2449. [Google Scholar] [CrossRef]
- Fu, Q.; McKnight, R.A.; Callaway, C.W.; Yu, X.; Lane, R.H.; Majnik, A.V. Intrauterine growth restriction disrupts developmental epigenetics around distal growth hormone response elements on the rat hepatic IGF-1 gene. FASEB J. 2015, 29, 1176–1184. [Google Scholar] [CrossRef]
- Wankhade, U.D.; Zhong, Y.; Kang, P.; Alfaro, M.; Chintapalli, S.V.; Thakali, K.M.; Shankar, K. Enhanced offspring predisposition to steatohepatitis with maternal high-fat diet is associated with epigenetic and microbiome alterations. PLoS ONE 2017, 12, e0175675. [Google Scholar] [CrossRef]
- Abumrad, N.; Harmon, C.; Ibrahimi, A. Membrane transport of long-chain fatty acids: Evidence for a facilitated process. J. Lipid Res. 1998, 39, 2309–2318. [Google Scholar] [CrossRef] [PubMed]
- Ke, X.; Fu, Q.; Sterrett, J.; Hillard, C.J.; Lane, R.H.; Majnik, A. Adverse maternal environment and western diet impairs cognitive function and alters hippocampal glucocorticoid receptor promoter methylation in male mice. Physiol. Rep. 2020, 8, e14407. [Google Scholar] [CrossRef] [PubMed]
- McClive, P.J.; Sinclair, A.H. Rapid DNA extraction and PCR-sexing of mouse embryos. Mol. Reprod. Dev. 2001, 60, 225–226. [Google Scholar] [CrossRef] [PubMed]
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S.; et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P.; Moore, J.E.; Purcaro, M.J.; Pratt, H.E.; Epstein, C.B.; Shoresh, N.; Adrian, J.; Kawli, T.; Davis, C.A.; Dobin, A.; et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 2020, 583, 699–710. [Google Scholar] [CrossRef]
- McLean, C.Y.; Bristor, D.; Hiller, M.; Clarke, S.L.; Schaar, B.T.; Lowe, C.B.; Wenger, A.M.; Bejerano, G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010, 28, 495–501. [Google Scholar] [CrossRef]
- Tanigawa, Y.; Dyer, E.S.; Bejerano, G. WhichTF is functionally important in your open chromatin data? PLoS Comput. Biol. 2022, 18, e1010378. [Google Scholar] [CrossRef]
- Yue, F.; Cheng, Y.; Breschi, A.; Vierstra, J.; Wu, W.; Ryba, T.; Sandstrom, R.; Ma, Z.; Davis, C.; Pope, B.D.; et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature 2014, 515, 355–364. [Google Scholar] [CrossRef]
- Tahan, V.; Eren, F.; Avsar, E.; Yavuz, D.; Yuksel, M.; Emekli, E.; Imeryuz, N.; Celikel, C.; Uzun, H.; Haklar, G.; et al. Rosiglitazone attenuates liver inflammation in a rat model of nonalcoholic steatohepatitis. Dig. Dis. Sci. 2007, 52, 3465–3472. [Google Scholar] [CrossRef]
- Quiroga, A.D.; Li, L.; Trotzmuller, M.; Nelson, R.; Proctor, S.D.; Kofeler, H.; Lehner, R. Deficiency of carboxylesterase 1/esterase-x results in obesity, hepatic steatosis, and hyperlipidemia. Hepatology 2012, 56, 2188–2198. [Google Scholar] [CrossRef]
- Xu, C.; Wang, G.; Hao, Y.; Zhi, J.; Zhang, L.; Chang, C. Correlation analysis between gene expression profile of rat liver tissues and high-fat emulsion-induced nonalcoholic fatty liver. Dig. Dis. Sci. 2011, 56, 2299–2308. [Google Scholar] [CrossRef] [PubMed]
- Maitre, L.; Bustamante, M.; Hernandez-Ferrer, C.; Thiel, D.; Lau, C.E.; Siskos, A.P.; Vives-Usano, M.; Ruiz-Arenas, C.; Pelegri-Siso, D.; Robinson, O.; et al. Multi-omics signatures of the human early life exposome. Nat. Commun. 2022, 13, 7024. [Google Scholar] [CrossRef] [PubMed]
- Everson, T.M.; Vives-Usano, M.; Seyve, E.; Cardenas, A.; Lacasana, M.; Craig, J.M.; Lesseur, C.; Baker, E.R.; Fernandez-Jimenez, N.; Heude, B.; et al. Placental DNA methylation signatures of maternal smoking during pregnancy and potential impacts on fetal growth. Nat. Commun. 2021, 12, 5095. [Google Scholar] [CrossRef] [PubMed]
- Joubert, B.R.; Felix, J.F.; Yousefi, P.; Bakulski, K.M.; Just, A.C.; Breton, C.; Reese, S.E.; Markunas, C.A.; Richmond, R.C.; Xu, C.J.; et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. Am. J. Hum. Genet. 2016, 98, 680–696. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Su, J.; Chen, K.; Rodriguez, B.; Li, W. Sparse conserved under-methylated CpGs are associated with high-order chromatin structure. Genome Biol. 2017, 18, 163. [Google Scholar] [CrossRef] [PubMed]
- Roadmap Epigenomics, C.; Kundaje, A.; Meuleman, W.; Ernst, J.; Bilenky, M.; Yen, A.; Heravi-Moussavi, A.; Kheradpour, P.; Zhang, Z.; Wang, J.; et al. Integrative analysis of 111 reference human epigenomes. Nature 2015, 518, 317–330. [Google Scholar] [CrossRef]
- Baylin, S.B.; Jones, P.A. A decade of exploring the cancer epigenome—Biological and translational implications. Nat. Rev. Cancer 2011, 11, 726–734. [Google Scholar] [CrossRef]
- Yang, L.; Rodriguez, B.; Mayle, A.; Park, H.J.; Lin, X.; Luo, M.; Jeong, M.; Curry, C.V.; Kim, S.B.; Ruau, D.; et al. DNMT3A Loss Drives Enhancer Hypomethylation in FLT3-ITD-Associated Leukemias. Cancer Cell 2016, 30, 363–365. [Google Scholar] [CrossRef]
- Park, Y.J.; Lee, S.; Lim, S.; Nahmgoong, H.; Ji, Y.; Huh, J.Y.; Alfadda, A.A.; Kim, S.; Kim, J.B. DNMT1 maintains metabolic fitness of adipocytes through acting as an epigenetic safeguard of mitochondrial dynamics. Proc. Natl. Acad. Sci. USA 2021, 118, e2021073118. [Google Scholar] [CrossRef]
- Ankill, J.; Aure, M.R.; Bjorklund, S.; Langberg, S.; Oslo Breast Cancer, C.; Kristensen, V.N.; Vitelli, V.; Tekpli, X.; Fleischer, T. Epigenetic alterations at distal enhancers are linked to proliferation in human breast cancer. NAR Cancer 2022, 4, zcac008. [Google Scholar] [CrossRef]
- Qin, Y.; Grimm, S.A.; Roberts, J.D.; Chrysovergis, K.; Wade, P.A. Alterations in promoter interaction landscape and transcriptional network underlying metabolic adaptation to diet. Nat. Commun. 2020, 11, 962. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.D.; Gu, H.; Bock, C.; Gnirke, A.; Meissner, A. High-throughput bisulfite sequencing in mammalian genomes. Methods 2009, 48, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Meissner, A.; Mikkelsen, T.S.; Gu, H.; Wernig, M.; Hanna, J.; Sivachenko, A.; Zhang, X.; Bernstein, B.E.; Nusbaum, C.; Jaffe, D.B.; et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008, 454, 766–770. [Google Scholar] [CrossRef]
- Zhao, H.; Sun, Z.; Wang, J.; Huang, H.; Kocher, J.P.; Wang, L. CrossMap: A versatile tool for coordinate conversion between genome assemblies. Bioinformatics 2014, 30, 1006–1007. [Google Scholar] [CrossRef]
- Pinero, J.; Ramirez-Anguita, J.M.; Sauch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Q.; Cheung, W.A.; Majnik, A.V.; Ke, X.; Pastinen, T.; Lane, R.H. Adverse Maternal Environments Perturb Hepatic DNA Methylome and Transcriptome Prior to the Adult-Onset Non-Alcoholic Fatty Liver Disease in Mouse Offspring. Nutrients 2023, 15, 2167. https://doi.org/10.3390/nu15092167
Fu Q, Cheung WA, Majnik AV, Ke X, Pastinen T, Lane RH. Adverse Maternal Environments Perturb Hepatic DNA Methylome and Transcriptome Prior to the Adult-Onset Non-Alcoholic Fatty Liver Disease in Mouse Offspring. Nutrients. 2023; 15(9):2167. https://doi.org/10.3390/nu15092167
Chicago/Turabian StyleFu, Qi, Warren A. Cheung, Amber V. Majnik, Xingrao Ke, Tomi Pastinen, and Robert H. Lane. 2023. "Adverse Maternal Environments Perturb Hepatic DNA Methylome and Transcriptome Prior to the Adult-Onset Non-Alcoholic Fatty Liver Disease in Mouse Offspring" Nutrients 15, no. 9: 2167. https://doi.org/10.3390/nu15092167
APA StyleFu, Q., Cheung, W. A., Majnik, A. V., Ke, X., Pastinen, T., & Lane, R. H. (2023). Adverse Maternal Environments Perturb Hepatic DNA Methylome and Transcriptome Prior to the Adult-Onset Non-Alcoholic Fatty Liver Disease in Mouse Offspring. Nutrients, 15(9), 2167. https://doi.org/10.3390/nu15092167






