Investigating Safety and Technological Traits of a Leading Probiotic Species: Lacticaseibacillus paracasei
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. DNA Extraction from Pure Colonies
2.3. Rep-PCR, RAPD, and Sau-PCR Analyses
2.4. Growth Capabilities
2.5. Antibiotic Resistance
2.6. Antimicrobial Capabilities (Bacteriocin Production)
2.7. Biogenic Amine (BA) Production
2.8. arcABC Gene Presence
2.9. Diaminobenzidine (DAB) Assay and Multicopper Oxidase (MCO) Detection
3. Results and Discussion
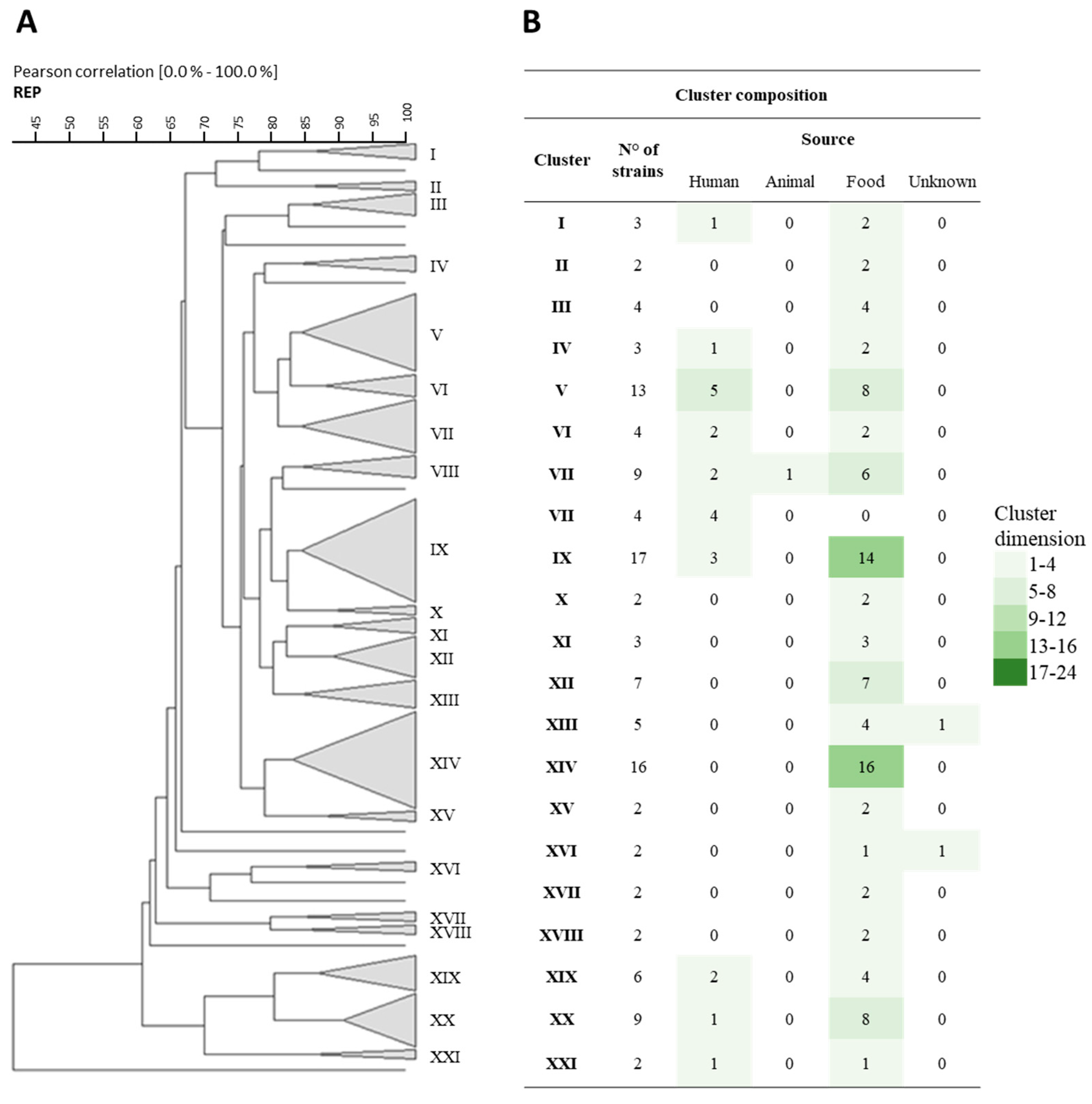
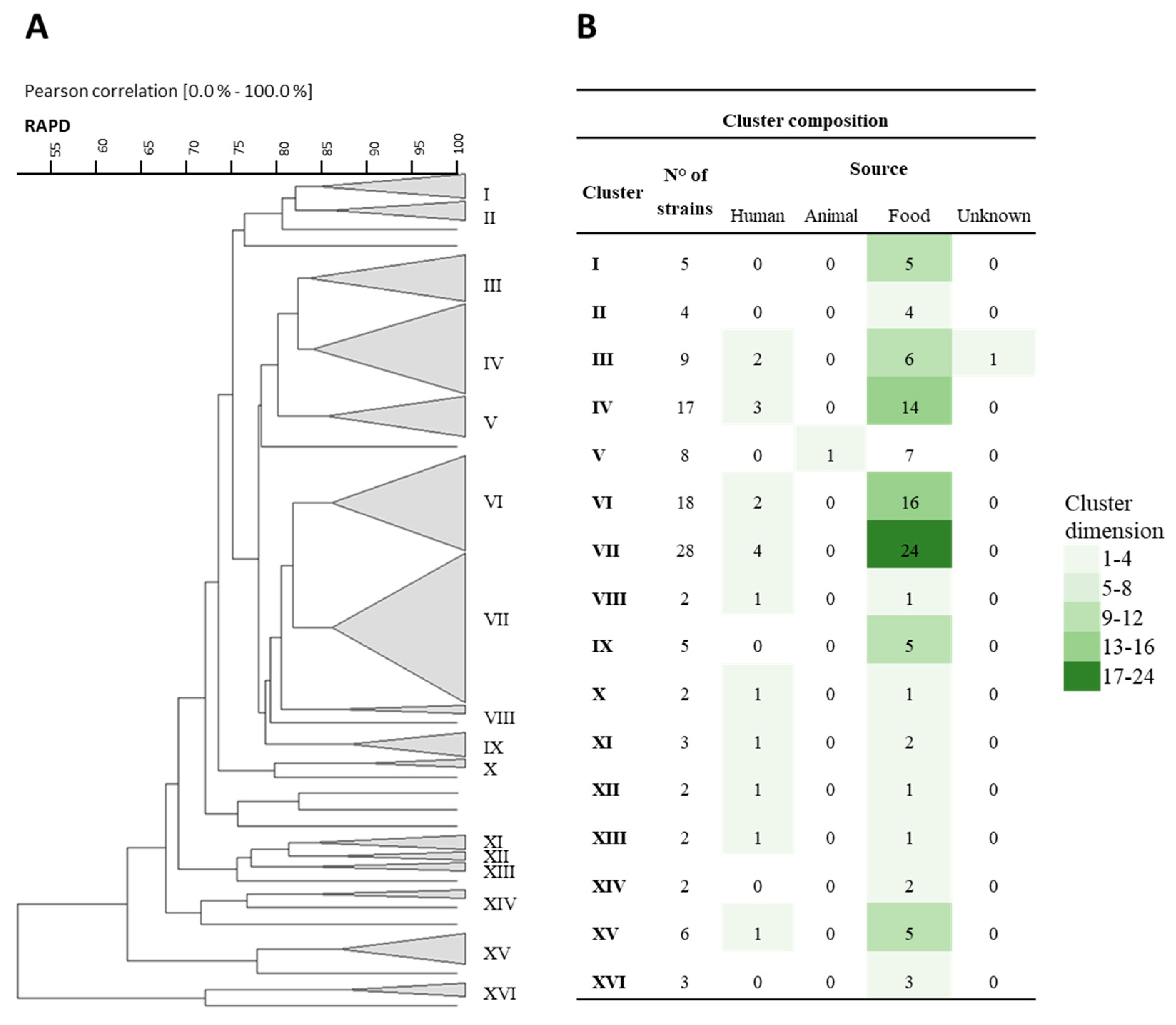
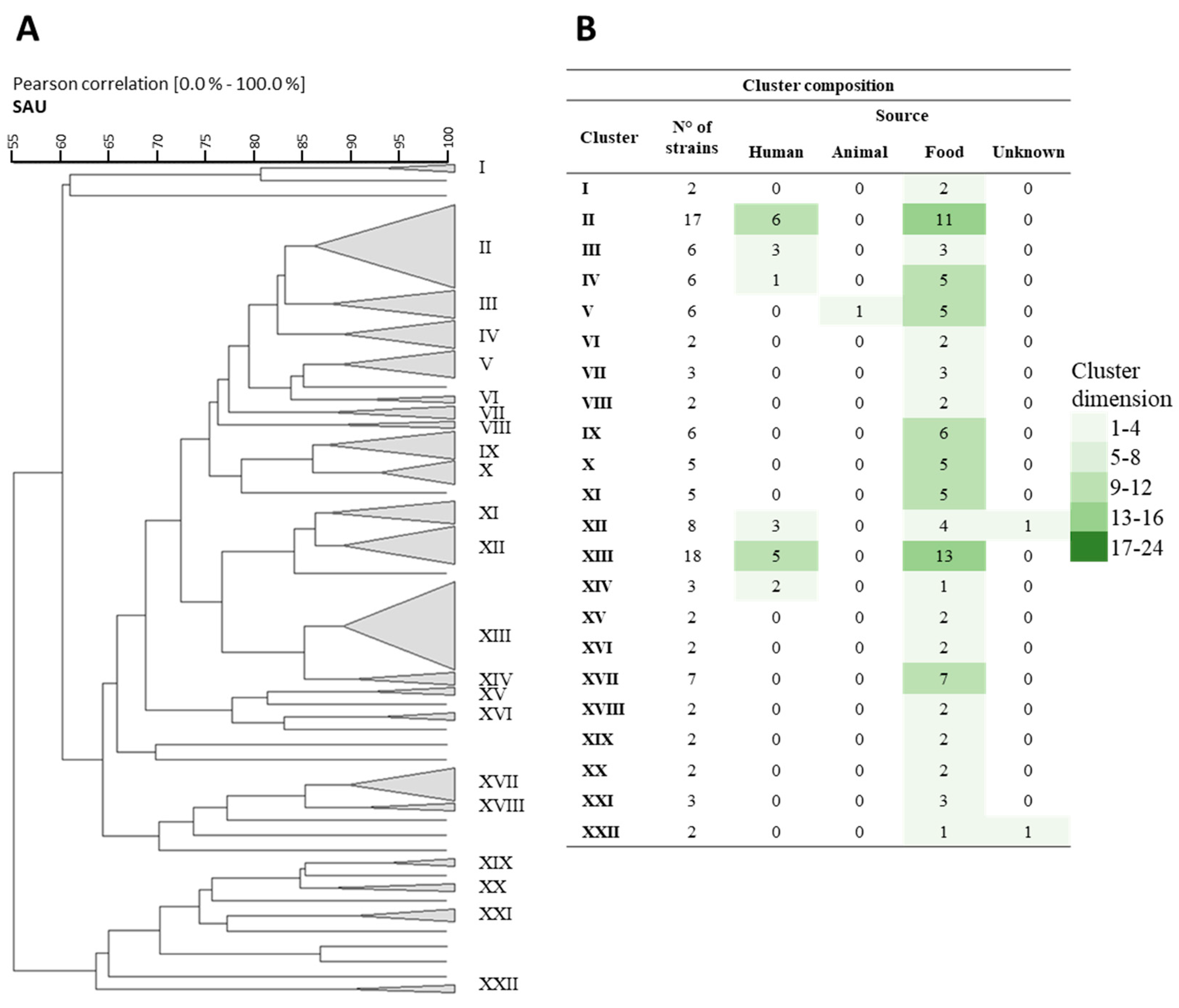
3.1. Rep-PCR, RAPD, and Sau-PCR Molecular Characterization
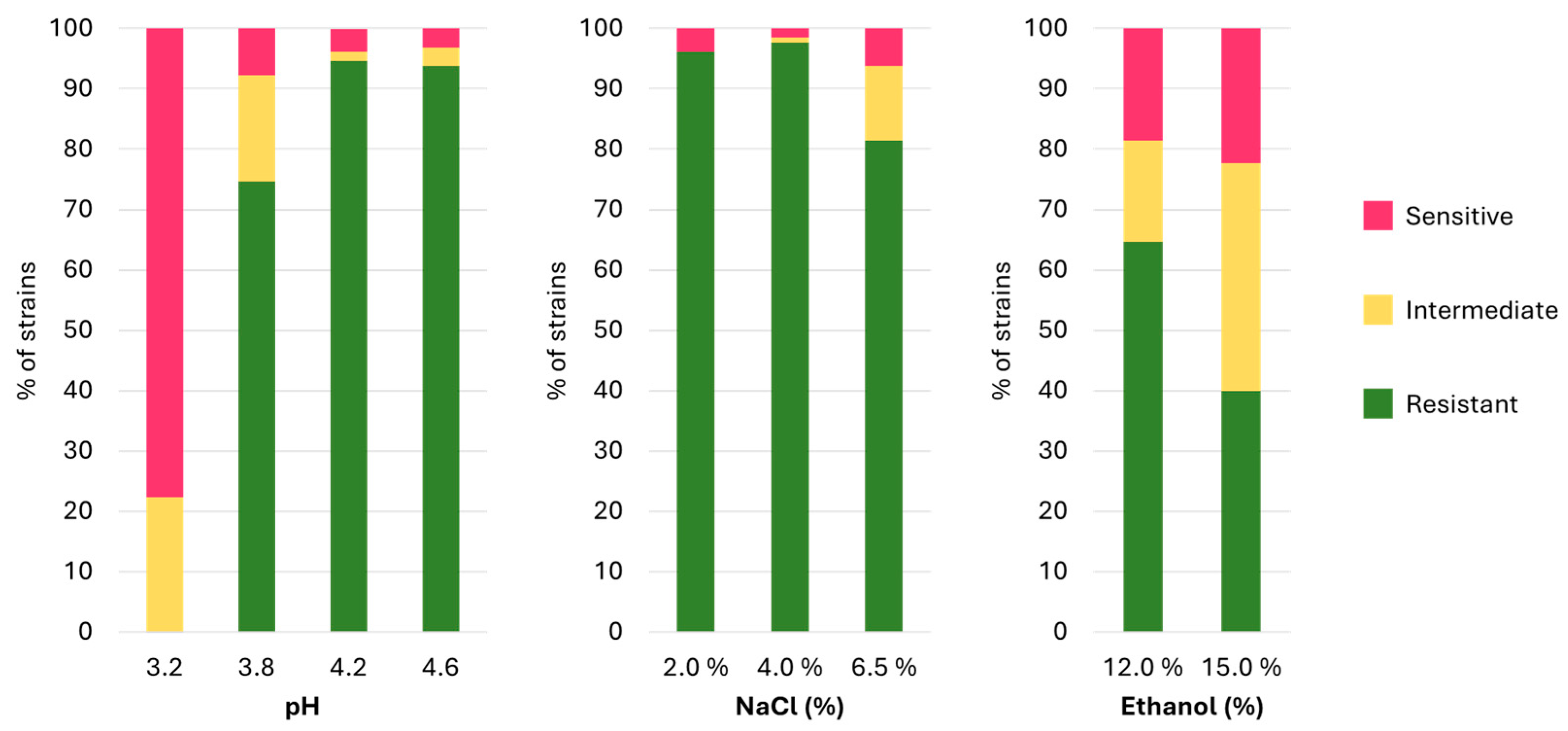
3.2. Stress Resistance
3.3. Antibiotic Resistance
3.4. Antimicrobial Activity and Bacteriocin Production
3.5. Biogenic Amine (BA) Production
3.6. arcABC Gene Presence
3.7. Biogenic Amine (BA)-Degrading Activity: Diaminobenzidine (DAB) Assay and Multicopper Oxidase (MCO) Detection
3.8. Phenotypic Clustering
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A Taxonomic Note on the Genus Lactobacillus: Description of 23 Novel Genera, Emended Description of the Genus Lactobacillus Beijerinck 1901, and Union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Qiao, N.; Wittouck, S.; Mattarelli, P.; Zheng, J.; Lebeer, S.; Felis, G.E.; Gänzle, M.G. After the Storm—Perspectives on the Taxonomy of Lactobacillaceae. JDS Commun. 2022, 3, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.D.; Gu, C.T. Proposal to Reclassify Lactobacillus zhaodongensis, Lactobacillus zeae, Lactobacillus argentoratensis and Lactobacillus buchneri Subsp. silagei as Lacticaseibacillus zhaodongensis Comb. Nov., Lacticaseibacillus zeae Comb. Nov., Lactiplantibacillus argentoratensis comb. nov. and Lentilactobacillus buchneri subsp. silagei comb. nov., respectively and Apilactobacillus kosoi as a later heterotypic synonym of Apilactobacillus micheneri. Int. J. Syst. Evol. Microbiol. 2020, 70, 6414–6417. [Google Scholar] [CrossRef] [PubMed]
- Iacumin, L.; Ginaldi, F.; Manzano, M.; Anastasi, V.; Reale, A.; Zotta, T.; Rossi, F.; Coppola, R.; Comi, G. High Resolution Melting Analysis (HRM) as a New Tool for the Identification of Species Belonging to the Lactobacillus Casei Group Andcomparison with Species-Specific PCRs and Multiplex PCR. Food Microbiol. 2015, 46, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Coudeyras, S.; Marchandin, H.; Fajon, C.; Forestier, C. Taxonomic and Strain-Specific Identification of the Probiotic Strain Lactobacillus rhamnosus 35 within the Lactobacillus Casei Group. Appl. Environ. Microbiol. 2008, 74, 2679–2689. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhu, Z.; Zhang, X.; Chen, L.; Gu, Q.; Li, P. Lactobacillus paracasei ZFM54 Alters the Metabolomic Profiles of Yogurt and the Co-Fermented Yogurt Improves the Gut Microecology of Human Adults. J. Dairy Sci. 2024. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Wang, C.; Chen, A.; Bai, Z.; Tian, Y.; Lv, W. Lactobacillus paracasei Influences the Gut-Microbiota-Targeted Metabolic Modulation of the Immune Status of Diarrheal Mice. Food Funct. 2023, 14, 4368–4379. [Google Scholar] [CrossRef] [PubMed]
- Giraffa, G.; Chanishvili, N.; Widyastuti, Y. Importance of Lactobacilli in Food and Feed Biotechnology. Res. Microbiol. 2010, 161, 480–487. [Google Scholar] [CrossRef] [PubMed]
- EFSA. Introduction of a Qualified Presumption of Safety (QPS) Approach for Assessment of Selected Microorganisms Referred to EFSA—Opinion of the Scientific Committee. EFSA J. 2007, 5, 587. [Google Scholar] [CrossRef]
- EFSA. EFSA Statement on the Requirements for Whole Genome Sequence Analysis of Microorganisms Intentionally Used in the Food Chain. EFSA J. 2021, 19, e06506. [Google Scholar] [CrossRef]
- EFSA. Guidance on the Characterisation of Microorganisms Used as Feed Additives or as Production Organisms. EFSA J. 2018, 16, e05206. [Google Scholar] [CrossRef]
- Colautti, A.; Arnoldi, M.; Comi, G.; Iacumin, L. Antibiotic Resistance and Virulence Factors in Lactobacilli: Something to Carefully Consider. Food Microbiol. 2021, 103, 103934. [Google Scholar] [CrossRef]
- Colautti, A.; Rossi, N.; Piazza, C.; Comi, G.; Iacumin, L. Draft Genome Sequences of 14 Lacticaseibacillus Spp. Strains, Representatives of a Collection of 200 Strains. Microbiol. Resour. Announc. 2023, 12, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Silla Santos, M.H. Biogenic Amines: Their Importance in Foods. Int. J. Food Microbiol. 1996, 29, 213–231. [Google Scholar] [CrossRef]
- Omer, A.K.; Mohammed, R.R.; Mohammed Ameen, P.S.; Abas, Z.A.; Ekici, K. Presence of Biogenic Amines in Food and Their Public Health Implications: A Review. J. Food Prot. 2021, 84, 1539–1548. [Google Scholar] [CrossRef] [PubMed]
- Pištěková, H.; Jančová, P.; Berčíková, L.; Buňka, F.; Sokolová, I.; Šopík, T.; Maršálková, K.; de Amaral, O.M.R.P.; Buňková, L. Application of QPCR for Multicopper Oxidase Gene (MCO) in Biogenic Amines Degradation by Lactobacillus casei. Food Microbiol. 2020, 91, 103550. [Google Scholar] [CrossRef]
- Iacumin, L.; Osualdini, M.; Bovolenta, S.; Boscolo, D.; Chiesa, L.; Panseri, S.; Comi, G. Microbial, Chemico-Physical and Volatile Aromatic Compounds Characterization of Pitina PGI, a Peculiar Sausage-like Product of North East Italy. Meat Sci. 2020, 163, 108081. [Google Scholar] [CrossRef] [PubMed]
- Gevers, D.; Huys, G.; Swings, J. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 2001, 205, 31–36. [Google Scholar] [CrossRef]
- Huey, B.; Hall, J. Hypervariable DNA Fingerprinting in Escherichia coli: Minisatellite Probe from Bacteriophage M13. J. Bacteriol. 1989, 171, 2528–2532. [Google Scholar] [CrossRef]
- Corich, V.; Mattiazzi, A.; Soldati, E.; Carraro, A.; Giacomini, A. Sau-PCR, a Novel Amplification Technique for Genetic Fingerprinting of Microorganisms. Appl. Environ. Microbiol. 2005, 71, 6401–6406. [Google Scholar] [CrossRef]
- Vauterin, L.; Vauterin, P. Computer-Aided Objective Comparison of Electrophoretic Patterns for Grouping and Identification of Microorganisms. Eur. Microbiol. 1992, 1, 37–41. [Google Scholar]
- Salminen, M.K.; Rautelin, H.; Tynkkynen, S.; Poussa, T.; Saxelin, M.; Valtonen, V.; Järvinen, A. Lactobacillus Bacteremia, Species Identification, and Antimicrobial Susceptibility of 85 Blood Isolates. Clin. Infect. Dis. 2006, 42, e35–e44. [Google Scholar] [CrossRef] [PubMed]
- Charteris, W.P.; Kelly, P.M.; Morelli, L.; Collins, J.K. Antibiotic Susceptibility of Potentially Probiotic Lactobacillus Species. J. Food Prot. 1998, 61, 1636–1643. [Google Scholar] [CrossRef] [PubMed]
- Schillinger, U.; Lucke, F.K. Antimicrobial Activity of Lactobacillus Sake Isolated from Meat. Appl. Environ. Microbiol. 1989, 55, 1901–1906. [Google Scholar] [CrossRef]
- Bover-Cid, S.; Holzapfel, W.H. Improved Screening Procedure for Biogenic Amine Production by Lactic Acid Bacteria. Int. J. Food Microbiol. 1999, 53, 33–41. [Google Scholar] [CrossRef]
- Coton, M.; Romano, A.; Spano, G.; Ziegler, K.; Vetrana, C.; Desmarais, C.; Lonvaud-Funel, A.; Lucas, P.; Coton, E. Occurrence of Biogenic Amine-Forming Lactic Acid Bacteria in Wine and Cider. Food Microbiol. 2010, 27, 1078–1085. [Google Scholar] [CrossRef]
- Araque, I.; Gil, J.; Ramon, C.; Bordons, A.; Reguant, C. Detection of Arc Genes Related with the Ethyl Carbamate Precursors in Wine Lactic Acid Bacteria. J. Agric. Food Chem. 2009, 57, 1841–1847. [Google Scholar] [CrossRef]
- Callejón, S.; Sendra, R.; Ferrer, S.; Pardo, I. Identification of a Novel Enzymatic Activity from Lactic Acid Bacteria Able to Degrade Biogenic Amines in Wine. Appl. Microbiol. Biotechnol. 2014, 98, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Comi, G.; Urso, R.; Iacumin, L.; Rantsiou, K.; Cattaneo, P.; Cantoni, C.; Cocolin, L. Characterisation of Naturally Fermented Sausages Produced in the North East of Italy. Meat Sci. 2005, 69, 381–392. [Google Scholar] [CrossRef]
- Iacumin, L.; Comi, G.; Cantoni, C.; Cocolin, L. Molecular and Technological Characterization of Staphylococcus Xylosus Isolated from Naturally Fermented Italian Sausages by RAPD, Rep-PCR and Sau-PCR Analysis. Meat Sci. 2006, 74, 281–288. [Google Scholar] [CrossRef]
- Amadoro, C.; Rossi, F.; Piccirilli, M.; Colavita, G. Features of Lactobacillus sakei Isolated from Italian Sausages: Focus on Strains from Ventricina Del Vastese. Ital. J. Food Saf. 2015, 4, 5449. [Google Scholar] [CrossRef] [PubMed]
- Torriani, S.; Zapparoli, G.; Dellaglio, F. Use of PCR-Based Methods for Rapid Differentiation of Lactobacillus Delbrueckii Subsp. Bulgaricus and L. Delbrueckii Subsp. Lactis. Appl. Environ. Microbiol. 1999, 65, 4351–4356. [Google Scholar] [CrossRef] [PubMed]
- Turková, K.; Rittich, B.; Španová, A. Identification and Determination of Relatedness of Lactobacilli Using Different DNA Amplification Methods. Chem. Pap. 2012, 66, 842–851. [Google Scholar] [CrossRef]
- Rossetti, L.; Giraffa, G. Rapid Identification of Dairy Lactic Acid Bacteria by M13-Generated, RAPD-PCR Fingerprint Databases. J. Microbiol. Methods 2005, 63, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Jarocki, P.; Komoń-Janczara, E.; Glibowska, A.; Dworniczak, M.; Pytka, M.; Korzeniowska-Kowal, A.; Wzorek, A.; Kordowska-Wiater, M. Molecular Routes to Specific Identification of the Lactobacillus Casei Group at the Species, Subspecies and Strain Level. Int. J. Mol. Sci. 2020, 21, 2694. [Google Scholar] [CrossRef] [PubMed]
- Reale, A.; Di Renzo, T.; Rossi, F.; Zotta, T.; Iacumin, L.; Preziuso, M.; Parente, E.; Sorrentino, E.; Coppola, R. Tolerance of Lactobacillus casei, Lactobacillus paracasei and Lactobacillus rhamnosus Strains to Stress Factors Encountered in Food Processing and in the Gastro-Intestinal Tract. LWT 2015, 60, 721–728. [Google Scholar] [CrossRef]
- Wu, C.; He, G.; Zhang, J. Physiological and Proteomic Analysis of Lactobacillus Casei in Response to Acid Adaptation. J. Ind. Microbiol. Biotechnol. 2014, 41, 1533–1540. [Google Scholar] [CrossRef] [PubMed]
- Rubio, R.; Jofré, A.; Martín, B.; Aymerich, T.; Garriga, M. Characterization of Lactic Acid Bacteria Isolated from Infant Faeces as Potential Probiotic Starter Cultures for Fermented Sausages. Food Microbiol. 2014, 38, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, C.N.; Nielsen, V.R.; Hayford, A.E.; Møller, P.L.; Michaelsen, K.F.; Pærregaard, A.; Sandström, B.; Tvede, M.; Jakobsen, M. Screening of Probiotic Activities of Forty-Seven Strains of Lactobacillus Spp. by in Vitro Techniques and Evaluation of the Colonization Ability of Five Selected Strains in Humans. Appl. Environ. Microbiol. 1999, 65, 4949–4956. [Google Scholar] [CrossRef]
- Federici, S.; Ciarrocchi, F.; Campana, R.; Ciandrini, E.; Blasi, G.; Baffone, W. Identification and Functional Traits of Lactic Acid Bacteria Isolated from Ciauscolo Salami Produced in Central Italy. Meat Sci. 2014, 98, 575–584. [Google Scholar] [CrossRef]
- Kou, X.; Chen, Q.; Ju, X.; Xue, Z.; Chen, W.; Xue, Z. A Tolerant Lactic Acid Bacteria, Lactobacillus paracasei, and Its Immunoregulatory Function. Can. J. Microbiol. 2014, 60, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, N.; Gu, Q.; Li, P. Whole Genome Sequence Analysis and in Vitro Probiotic Characteristics of a Lactobacillus Strain Lactobacillus paracasei ZFM54. J. Appl. Microbiol. 2020, 129, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, J.; Xia, X.; Zhao, Y.; Shao, W. Probiotic Potential of Lactobacillus paracasei FM-LP-4 Isolated from Xinjiang Camel Milk Yoghurt. Int. Dairy J. 2016, 62, 28–34. [Google Scholar] [CrossRef]
- Radulović, Z.; Petrović, T.; Nedović, V.; Dimitrijević, S.; Mirković, N.; Petrušić, M.; Paunović, D. Characterization of Autochthonous Lactobacillus paracasei Strains on Potential Probiotic Ability. Mljekarstvo 2010, 60, 86–93. [Google Scholar]
- Muna, M.A.; Adel, M.M. Isolation of Lactobacillus Strains with Probiotic Potential from Camels Milk. Afr. J. Microbiol. Res. 2014, 8, 1645–1655. [Google Scholar] [CrossRef]
- De Albuquerque, T.M.R.; Garcia, E.F.; De Oliveira Araújo, A.; Magnani, M.; Saarela, M.; De Souza, E.L. In Vitro Characterization of Lactobacillus Strains Isolated from Fruit Processing By-Products as Potential Probiotics. Probiotics Antimicrob. Proteins 2018, 10, 704–716. [Google Scholar] [CrossRef]
- Dishan, A.; Gönülalan, Z. Lacticaseibacillus paracasei AD22 Stress Response in Brined White Cheese Matrix: In Vitro Probiotic Profiles and Molecular Characterization. Probiotics Antimicrob. Proteins 2024. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Z.; Vohra, M.S.; Khan, M.N.; Ahmed, A.; Khan, T.A. Antimicrobial Role of Lactobacillus Species as Potential Probiotics against Enteropathogenic Bacteria in Chickens. J. Infect. Dev. Ctries. 2019, 13, 130–136. [Google Scholar] [CrossRef]
- Piuri, M.; Sanchez-Rivas, C.; Ruzal, S.M. Cell Wall Modifications during Osmotic Stress in Lactobacillus Casei. J. Appl. Microbiol. 2005, 98, 84–95. [Google Scholar] [CrossRef]
- Pittet, V.; Morrow, K.; Ziola, B. Ethanol Tolerance of Lactic Acid Bacteria, Including Relevance of the Exopolysaccharide Gene Gtf. J. Am. Soc. Brew. Chem. 2011, 69, 57–61. [Google Scholar] [CrossRef]
- Lee, H.I.; Kim, M.H.; Kim, K.Y.; So, J.S. Screening and Selection of Stress Resistant Lactobacillus Spp. Isolated from the Marine Oyster (Crassostrea gigas). Anaerobe 2010, 16, 522–526. [Google Scholar] [CrossRef]
- Sharma, P.; Tomar, S.K.; Goswami, P.; Sangwan, V.; Singh, R. Antibiotic Resistance among Commercially Available Probiotics. Food Res. Int. 2014, 57, 176–195. [Google Scholar] [CrossRef]
- Belletti, N.; Gatti, M.; Bottari, B.; Neviani, E.; Tabanelli, G.; Gardini, F. Antibiotic Resistance of Lactobacilli Isolated from Two Italian Hard Cheeses. J. Food Prot. 2009, 72, 2162–2169. [Google Scholar] [CrossRef]
- European Commission—HEALTH & CONSUMER PROTECTION DIRECTORATE-GENERAL—Directorate C—Scientific Opinions C2—Management of Scientific Committees; Scientific Co-Operation and Networks. Position Paper of the Scientific Committee on Animal Nutrition on Safety Assessment and Regulatory Aspects of Micro-organisms in Feed and Food Applications. Available online: https://food.ec.europa.eu/document/download/b2beaff9-5fb3-45eb-9e63-d5396ec85154_en?filename=sci-com_scan-old_report_out85.pdf (accessed on 7 July 2024).
- Coton, M.; Lebreton, M.; Leyva Salas, M.; Garnier, L.; Navarri, M.; Pawtowski, A.; Le Blay, G.; Valence, F.; Coton, E.; Mounier, J. Biogenic Amine and Antibiotic Resistance Profiles Determined for Lactic Acid Bacteria and a Propionibacterium Prior to Use as Antifungal Bioprotective Cultures. Int. Dairy J. 2018, 85, 21–26. [Google Scholar] [CrossRef]
- Huys, G.; D’Haene, K.; Danielsen, M.; Mättö, J.; Egervärn, M.; Vandamme, P. Phenotypic and Molecular Assessment of Antimicrobial Resistance in Lactobacillus paracasei Strains of Food Origin. J. Food Prot. 2008, 71, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Gómez, N.C.; Ramiro, J.M.P.; Quecan, B.X.V.; de Melo Franco, B.D.G. Use of Potential Probiotic Lactic Acid Bacteria (LAB) Biofilms for the Control of Listeria monocytogenes, Salmonella typhimurium, and Escherichia coli O157: H7 Biofilms Formation. Front. Microbiol. 2016, 7, 863. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zhang, Y.; Zhou, H.; Tian, F.; Ni, Y. Antimicrobial Activities and in Vitro Properties of Cold-Adapted Lactobacillus Strains Isolated from the Intestinal Tract of Cold Water Fishes of High Latitude Water Areas in Xinjiang, China. BMC Microbiol. 2019, 19, 247. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Tomar, S.K.; Sangwan, V.; Goswami, P.; Singh, R. Antibiotic Resistance of Lactobacillus Sp. Isolated from Commercial Probiotic Preparations. J. Food Saf. 2016, 36, 38–51. [Google Scholar] [CrossRef]
- Todorov, S.D.; Perin, L.M.; Carneiro, B.M.; Rahal, P.; Holzapfel, W.; Nero, L.A. Safety of Lactobacillus plantarum ST8Sh and Its Bacteriocin. Probiotics Antimicrob. Proteins 2017, 9, 334–344. [Google Scholar] [CrossRef]
- Ammor, M.S.; Belén Flórez, A.; Mayo, B. Antibiotic Resistance in Non-Enterococcal Lactic Acid Bacteria and Bifidobacteria. Food Microbiol. 2007, 24, 559–570. [Google Scholar] [CrossRef]
- Goldstein, E.J.C.; Tyrrell, K.L.; Citron, D.M. Lactobacillus Species: Taxonomic Complexity and Controversial Susceptibilities. Clin. Infect. Dis. 2015, 60, S98–S107. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Tian, Y.; Cao, Y.; Li, J.; Guo, H.; Su, Y.; Tian, Y.; Wang, C.; Wang, T.; Zhang, L. Probiotic Properties of Lactobacillus paracasei Subsp. Paracasei L1 and Its Growth Performance-Promotion in Chicken by Improving the Intestinal Microflora. Front. Physiol. 2019, 10, 937. [Google Scholar] [CrossRef] [PubMed]
- Lozo, J.; Jovcic, B.; Kojic, M.; Dalgalarrondo, M.; Chobert, J.M.; Haertlé, T.; Topisirovic, L. Molecular Characterization of a Novel Bacteriocin and an Unusually Large Aggregation Factor of Lactobacillus paracasei Subsp. Paracasei BGSJ2-8, a Natural Isolate from Homemade Cheese. Curr. Microbiol. 2007, 55, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Verdenelli, M.C.; Ghelfi, F.; Silvi, S.; Orpianesi, C.; Cecchini, C.; Cresci, A. Probiotic Properties of Lactobacillus rhamnosus and Lactobacillus paracasei Isolated from Human Faeces. Eur. J. Nutr. 2009, 48, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, R.; Yocheva, L.; Tserovska, L.; Zhelezova, G.; Stefanova, N.; Atanasova, A.; Danguleva, A.; Ivanova, G.; Karapetkov, N.; Rumyan, N.; et al. Antimicrobial Activity and Antibiotic Susceptibility of Lactobacillus and Bifidobacterium Spp. Intended for Use as Starter and Probiotic Cultures. Biotechnol. Biotechnol. Equip. 2015, 29, 84–91. [Google Scholar] [CrossRef]
- Peng, M.; Tabashsum, Z.; Patel, P.; Bernhardt, C.; Biswas, D. Linoleic Acids Overproducing Lactobacillus Casei Limits Growth, Survival, and Virulence of Salmonella Typhimurium and Enterohaemorrhagic Escherichia coli. Front. Microbiol. 2018, 9, 2663. [Google Scholar] [CrossRef] [PubMed]
- Iacumin, L.; Cappellari, G.; Colautti, A.; Comi, G. Listeria Monocytogenes Survey in Cubed Cooked Ham Packaged in Modified Atmosphere and Bioprotective Effect of Selected Lactic Acid Bacteria. Microorganisms 2020, 8, 898. [Google Scholar] [CrossRef]
- Reis, J.A.; Paula, A.T.; Casarotti, S.N.; Penna, A.L.B. Lactic Acid Bacteria Antimicrobial Compounds: Characteristics and Applications. Food Eng. Rev. 2012, 4, 124–140. [Google Scholar] [CrossRef]
- Capra, M.L.; Tibaldo, M.M.; Vinderola, G.; Reinheimer, J.A.; Quiberoni, A. Technological and Probiotic Characterisation of Lactobacillus Casei/Paracasei Strains and Their Phage-Resistant Mutants. Int. Dairy J. 2014, 37, 39–47. [Google Scholar] [CrossRef]
- Campana, R.; Van Hemert, S.; Baffone, W. Strain-Specific Probiotic Properties of Lactic Acid Bacteria and Their Interference with Human Intestinal Pathogens Invasion. Gut Pathog. 2017, 9, 12. [Google Scholar] [CrossRef]
- Muyyarikkandy, M.S.; Amalaradjou, M.A. Lactobacillus bulgaricus, Lactobacillus rhamnosus and Lactobacillus paracasei Attenuate Salmonella Enteritidis, Salmonella Heidelberg and Salmonella Typhimurium Colonization and Virulence Gene Expression in Vitro. Int. J. Mol. Sci. 2017, 18, 2381. [Google Scholar] [CrossRef]
- Bartkiene, E.; Lele, V.; Ruzauskas, M.; Domig, K.J.; Starkute, V.; Zavistanaviciute, P.; Bartkevics, V.; Pugajeva, I.; Klupsaite, D.; Juodeikiene, G.; et al. Lactic Acid Bacteria Isolation from Spontaneous Sourdough and Their Characterization Including Antimicrobial and Antifungal Properties Evaluation. Microorganisms 2020, 8, 64. [Google Scholar] [CrossRef]
- Coman, M.M.; Verdenelli, M.C.; Cecchini, C.; Silvi, S.; Orpianesi, C.; Boyko, N.; Cresci, A. In Vitro Evaluation of Antimicrobial Activity of Lactobacillus rhamnosus IMC 501®, Lactobacillus paracasei IMC 502® and SYNBIO® against Pathogens. J. Appl. Microbiol. 2014, 117, 518–527. [Google Scholar] [CrossRef]
- Colautti, A.; Orecchia, E.; Comi, G.; Iacumin, L. Lactobacilli, a Weapon to Counteract Pathogens through the Inhibition of Their Virulence Factors. J. Bacteriol. 2022, 204, e00272-22. [Google Scholar] [CrossRef] [PubMed]
- Suzzi, G.; Gardini, F. Biogenic Amines in Dry Fermented Sausages: A Review. Int. J. Food Microbiol. 2003, 88, 41–54. [Google Scholar] [CrossRef]
- Buňková, L.; Buňka, F.; Hlobilová, M.; Vaňátková, Z.; Nováková, D.; Dráb, V. Tyramine Production of Technological Important Strains of Lactobacillus, Lactococcus and Streptococcus. Eur. Food Res. Technol. 2009, 229, 533–538. [Google Scholar] [CrossRef]
- Coton, E.; Coton, M. Multiplex PCR for Colony Direct Detection of Gram-Positive Histamine- and Tyramine-Producing Bacteria. J. Microbiol. Methods 2005, 63, 296–304. [Google Scholar] [CrossRef]
- Landete, J.M.; de las Rivas, B.; Marcobal, A.; Muñoz, R. Molecular Methods for the Detection of Biogenic Amine-Producing Bacteria on Foods. Int. J. Food Microbiol. 2007, 117, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Landete, J.M.; Ferrer, S.; Pardo, I. Biogenic Amine Production by Lactic Acid Bacteria, Acetic Bacteria and Yeast Isolated from Wine. Food Control 2007, 18, 1569–1574. [Google Scholar] [CrossRef]
- Ladero, V.; Coton, M.; Fernández, M.; Buron, N.; Martín, M.C.; Guichard, H.; Coton, E.; Alvarez, M.A. Biogenic Amines Content in Spanish and French Natural Ciders: Application of QPCR for Quantitative Detection of Biogenic Amine-Producers. Food Microbiol. 2011, 28, 554–561. [Google Scholar] [CrossRef]
- Costantini, A.; Pietroniro, R.; Doria, F.; Pessione, E.; Garcia-Moruno, E. Putrescine Production from Different Amino Acid Precursors by Lactic Acid Bacteria from Wine and Cider. Int. J. Food Microbiol. 2013, 165, 11–17. [Google Scholar] [CrossRef] [PubMed]
- García-Ruiz, A.; González-Rompinelli, E.M.; Bartolomé, B.; Moreno-Arribas, M.V. Potential of Wine-Associated Lactic Acid Bacteria to Degrade Biogenic Amines. Int. J. Food Microbiol. 2011, 148, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Landete, J.M.; Ferrer, S.; Pardo, I. Which Lactic Acid Bacteria Are Responsible for Histamine Production in Wine? J. Appl. Microbiol. 2005, 99, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Postupolski, J.; Stasiak, M.; Maćkiw, E.; Kowalska, J.; Kucharek, K. Silent Genes: Antimicrobial Resistance and Antibiotic Production. Pol. J. Microbiol. 2021, 70, 421–429. [Google Scholar] [CrossRef]
- Tonon, T.; Lonvaud-Funel, A. Arginine Metabolism by Wine Lactobacilli Isolated from Wine. Food Microbiol. 2002, 19, 451–461. [Google Scholar] [CrossRef]
- Fang, R.S.; Dong, Y.C.; Xu, T.Y.; He, G.Q.; Chen, Q.H. Ethyl Carbamate Formation Regulated by Ornithine Transcarbamylase and Urea Metabolism in the Processing of Chinese Yellow Rice Wine. Int. J. Food Sci. Technol. 2013, 48, 2551–2556. [Google Scholar] [CrossRef]
- Arena, M.E.; Saguir, F.M.; Manca De Nadra, M.C. Arginine, Citrulline and Ornithine Metabolism by Lactic Acid Bacteria from Wine. Int. J. Food Microbiol. 1999, 52, 155–161. [Google Scholar] [CrossRef] [PubMed]
- du Toit, M.; Engelbrecht, L.; Lerm, E.; Krieger-Weber, S. Lactobacillus: The Next Generation of Malolactic Fermentation Starter Cultures-an Overview. Food Bioprocess Technol. 2011, 4, 876–906. [Google Scholar] [CrossRef]
- Mira De Orduña, R.; Liu, S.Q.; Patchett, M.L.; Pilone, G.J. Ethyl Carbamate Precursor Citrulline Formation from Arginine Degradation by Malolactic Wine Lactic Acid Bacteria. FEMS Microbiol. Lett. 2000, 183, 31–35. [Google Scholar] [CrossRef]
- Mtshali, P.S.; Divol, B.; Du Toit, M. Identification and Characterization of Lactobacillus Florum Strains Isolated from South African Grape and Wine Samples. Int. J. Food Microbiol. 2012, 153, 106–113. [Google Scholar] [CrossRef]
- Li, B.; Lu, S. The Importance of Amine-Degrading Enzymes on the Biogenic Amine Degradation in Fermented Foods: A Review. Process Biochem. 2020, 99, 331–339. [Google Scholar] [CrossRef]
- Naila, A.; Flint, S.; Fletcher, G.; Bremer, P.; Meerdink, G. Control of Biogenic Amines in Food—Existing and Emerging Approaches. J. Food Sci. 2010, 75, R139–R150. [Google Scholar] [CrossRef] [PubMed]
- Tabanelli, G. Biogenic Amines and Food Quality: Emerging Challenges and Public Health Concerns. Foods 2020, 9, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Kongkiattikajorn, J. Potential of Starter Culture to Reduce Biogenic Amines Accumulation in Som-Fug, a Thai Traditional Fermented Fish Sausage. J. Ethn. Foods 2015, 2, 186–194. [Google Scholar] [CrossRef]
- Fadda, S.; Vignolo, G.; Oliver, G. Tyramine Degradation and Tyramine/Histamine Production by Lactic Acid Bacteria and Kocuria Strains. Biotechnol. Lett. 2001, 23, 2015–2019. [Google Scholar] [CrossRef]
- Guarcello, R.; de Angelis, M.; Settanni, L.; Formiglio, S.; Gaglio, R.; Minervini, F.; Moschetti, G.; Gobbetti, M. Selection of Amine-Oxidizing Dairy Lactic Acid Bacteria and Identification of the Enzyme and Gene Involved in the Decrease of Biogenic Amines. Appl. Environ. Microbiol. 2016, 82, 6870–6880. [Google Scholar] [CrossRef]
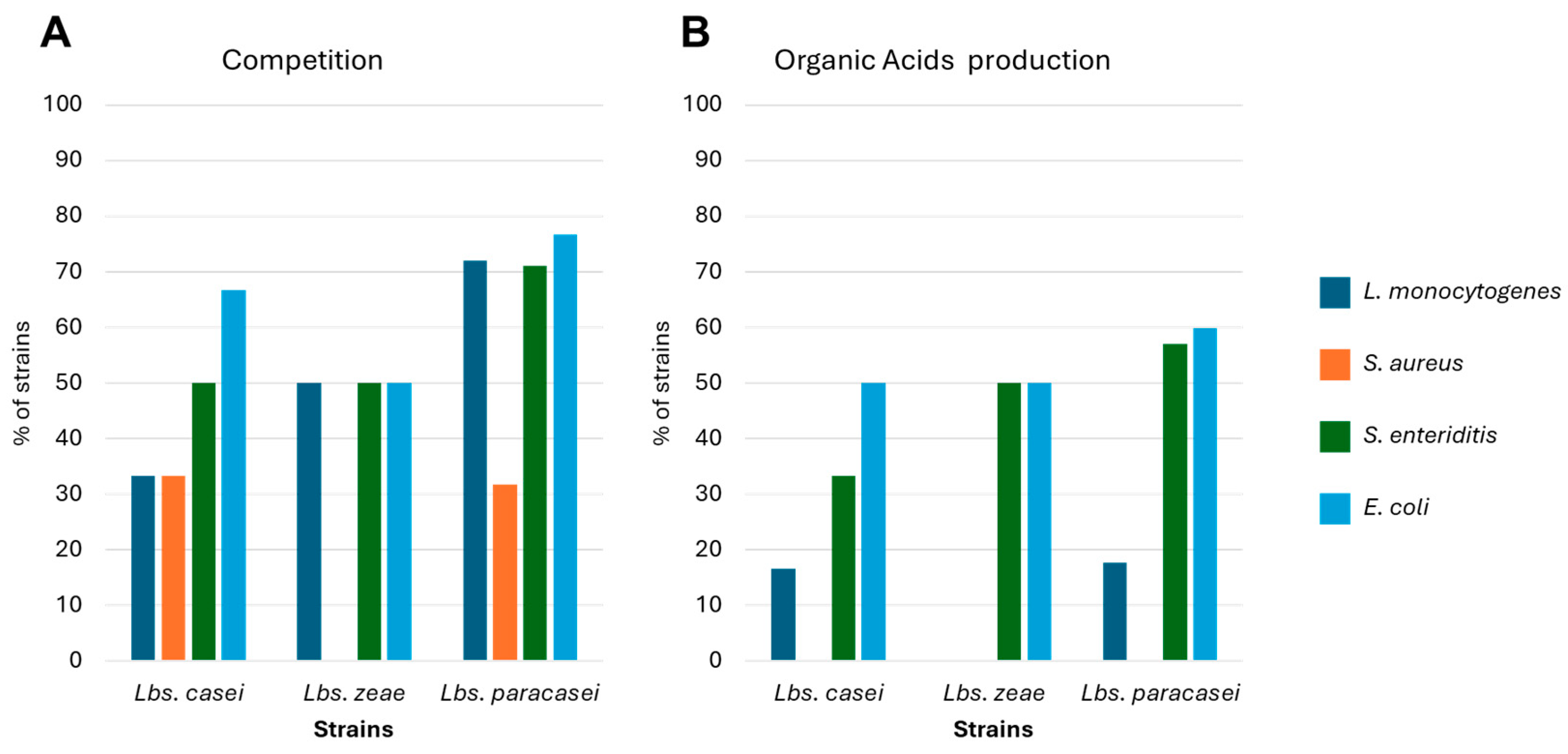
 ), B (
), B ( ), C (
), C ( ), and D (
), and D ( ).
).
 ), B (
), B ( ), C (
), C ( ), and D (
), and D ( ).
).
| Origin | Given Identification |
|---|---|
| Raw and heat-treated milk, yogurt, milking machines | Lbs. paracasei: LMG9192 1, DSM5622 2, P1E6 3, P2P3 3, Mo4 14, DialYac1 14, DialYac2 14, DialYac3 14, DialYac4 14, DialYac5 14, DialYac6 14, DialYac7 14, DialYac8 14, DialDan1 14, DialDan2 14, DialDan3 14, DialDan4 14, DialDan5 14, DialDan6 14, DialDan7 14, DialDan8 14 |
| Lbs. paracasei subsp. tolerans: LMG9191 1, DSM20258 2 | |
| Green, creamy, seasoned cheeses (Italian cheeses: Scamorza, Parmigiano Reggiano, Grana Padano, Spressa, Asiago, Montasio, Canestrato di Moliterno, Morlacco, Bellunese, Pecorino, Caciocavallo, Provolone, Emmenthal, Raclette de Savoie; Chinese and Tunisian cheeses) | Lbs. zeae: CI4368 11 |
| Lbs. paracasei: LMG 6904 1, LMG25880 1, LMG25883 1, LMG12164 1, DBPZ0420 7, DBPZ0421 7, DBPZ0422 7, DBPZ0424 7, DBPZ0434 7, DBPZ0435 7, DBPZ0450 7, DBPZ0451 7, DBPZ0472 7, DBPZ0475 7, DBPZ0476 7, DBPZ0477 7, DBPZ0478 7, DBPZ0635 7, DBPZ0733 7, DBPZ0734 7, M266 7, M268 7, M299 7, M308 7, M348 7, M354 7, M359 7, S1 7, S3 7, V3 7, W11 7, DSG03 7, DSG05 7, DSG07 7, ESG10 7, HSG09 7, PSG06 7, PSG09 7, PSG10 7, P71 8, TH1229 8, SP57 8, L24 8, TH406 8, FSL346 9, FSL451 9, DBPZ0436 7, TMW1.1444 5, TMW1.1259 5, LACcas7 6, Cst7 10, 3LC 10, DBPZ0718 7, CF143 11, R61 12, F17 12, N24 14, H12 12 | |
| Fermented sausages | Lbs. paracasei: CTC1675 13 |
| Sourdoughs | Lbs. casei: DBPZ0571 7 |
| Lbs. paracasei: DBPZ0561 7, DBPZ0572 7, Q2 7, Q4 7, I1 4, I2 15, DBPZ0563 7, DBPZ0564 7, DBPZ0579 7, I3 15 | |
| Wine, must, cellar equipment | Lbs. paracasei: LMG11961 1, LMG11963 1, LMG13717 1, LMG13731 1, B061 16, B161 16, B169 16, B171 16, B195 16, B196 16, B350 16 |
| Beer, malt | Lbs. paracasei: LACcas25 6, LACcas29 6, TMW 1.300 5 |
| Corn steep liquor | Lbs. zeae: DSM20178 2 |
| Humans (saliva, dental caries, blood, urethra, faeces of infants and adults) | Lbs. casei: LMG23516 1, N87 15, N811 15, DSM4905 2, N2014 15 |
| Lbs. paracasei: DSM20020 2, LMG9438 1, LMG11459 1, LMG23511 1, LMG23518 1, LMG23523 1, LMG23538 1, LMG23543 1, LMG24098 1, LMG24101 1, LMG24132 1, DBTA34 17, N161 15, N42 15, N44 15, N76 15, | |
| Unknown | Lbs. paracasei: NRRL B-456 18 |
| Strain | Origin | Antibiotics | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CFP30 | KZ30 | C10 | DA10 | E30 | K30 | OFX5 | QD15 | RD30 | S25 | TE10 | TOB10 | VA30 | ||
| Lbs. casei LMG 23516 | Human faeces | S | S | S | S | S | R | R | S | S | R | S | R | R |
| Lbs. paracasei DBPZ0478 | Caciocavallo | R | R | R | S | R | R | R | S | S | R | R | R | R |
| Lbs. paracasei M308 | Canestrato Moliterno | S | S | S | I | S | R | R | I | S | I | S | R | S |
| Lbs. paracasei DialDan8 | Yogurt | S | S | S | S | I | R | S | S | S | S | S | I | S |
| Lbs. paracasei DialYak4 | Yogurt | S | S | S | S | R | R | I | S | S | R | S | S | R |
| Lbs. paracasei DialYak5 | Yogurt | S | S | S | S | R | R | I | S | S | R | S | S | R |
| Lbs. paracasei DialYak6 | Yogurt | S | S | S | S | S | R | I | S | S | R | S | R | R |
| Lbs. paracasei DialYak7 | Yogurt | S | S | S | S | R | R | I | S | S | R | S | S | R |
| Lbs. paracasei DialYak8 | Yogurt | S | S | S | S | R | R | I | S | S | R | S | S | R |
| Gene | Biogenic Amine | |||||||
|---|---|---|---|---|---|---|---|---|
| Strain | tyrdc | odc | agdi | hdc | Tym | Agm or Putr | Putr | Cad |
| NRRL B-456 | + | − | − | − | − | − | − | − |
| B085 | − | − | + | − | − | − | − | − |
| B161 | − | − | + | − | − | − | − | − |
| B169 | − | − | + | − | + | + | − | − |
| B195 | − | − | + | − | + | − | − | − |
| LacCas7 | − | − | + | − | − | − | − | − |
| B350 | − | − | − | + | − | − | − | − |
| PSG10 | + | − | − | + | − | + | − | − |
| N2014 | − | − | + | + | − | − | − | − |
| Strain | Source | Strain | Source |
|---|---|---|---|
| Lbs. zeae DSM20178 | Corn steep liquor | Lbs. paracasei R61 | Pecorino cheese |
| Lbs. casei N87 | Body excreta | Lbs. paracasei F17 | Pecorino cheese |
| Lbs. casei N2014 | Body excreta | Lbs. paracasei H12 | Pecorino cheese |
| Lbs. paracasei NRRL B-456 | Unknown | Lbs. paracasei N76 | Body excreta |
| Lbs. paracasei DBPZ0564 | Altamura sourdough | Lbs. paracasei B195 | Wine |
| Lbs. paracasei DBPZ0572 | Altamura sourdough | Lbs. paracasei LACcas25 | Elixir |
| Lbs. paracasei V3 | Canestrato Moliterno | Lbs. paracasei TMW 1.1444 | Pecorino cheese |
| Lbs. paracasei ESG10 | Parmigiano Reggiano | Lbs. paracasei TMW 1.1259 | Parmesan cheese |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colautti, A.; Ginaldi, F.; Camprini, L.; Comi, G.; Reale, A.; Iacumin, L. Investigating Safety and Technological Traits of a Leading Probiotic Species: Lacticaseibacillus paracasei. Nutrients 2024, 16, 2212. https://doi.org/10.3390/nu16142212
Colautti A, Ginaldi F, Camprini L, Comi G, Reale A, Iacumin L. Investigating Safety and Technological Traits of a Leading Probiotic Species: Lacticaseibacillus paracasei. Nutrients. 2024; 16(14):2212. https://doi.org/10.3390/nu16142212
Chicago/Turabian StyleColautti, Andrea, Federica Ginaldi, Lucia Camprini, Giuseppe Comi, Anna Reale, and Lucilla Iacumin. 2024. "Investigating Safety and Technological Traits of a Leading Probiotic Species: Lacticaseibacillus paracasei" Nutrients 16, no. 14: 2212. https://doi.org/10.3390/nu16142212
APA StyleColautti, A., Ginaldi, F., Camprini, L., Comi, G., Reale, A., & Iacumin, L. (2024). Investigating Safety and Technological Traits of a Leading Probiotic Species: Lacticaseibacillus paracasei. Nutrients, 16(14), 2212. https://doi.org/10.3390/nu16142212









